Significance
A major challenge in modern biological studies is in the determination of the 3D molecular architecture of cellular organelles. In recent years, much progress in nanoscale imaging has been made because of the advent of superresolution optical microscopy. However, many superresolution techniques are still limited to 2D acquisition. Here, we show a volumetric approach for superresolution imaging based on the simultaneous imaging of multiple sample planes using multifocal microscopy. The depth over which structures can be reconstructed reaches 4 µm, comparable with the thickness of many cellular organelles or even whole cells.
Keywords: superresolution, 3D localization, microscopy, multiplane imaging, single-molecule fluorescence
Abstract
Single molecule-based superresolution imaging has become an essential tool in modern cell biology. Because of the limited depth of field of optical imaging systems, one of the major challenges in superresolution imaging resides in capturing the 3D nanoscale morphology of the whole cell. Despite many previous attempts to extend the application of photo-activated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM) techniques into three dimensions, effective localization depths do not typically exceed 1.2 µm. Thus, 3D imaging of whole cells (or even large organelles) still demands sequential acquisition at different axial positions and, therefore, suffers from the combined effects of out-of-focus molecule activation (increased background) and bleaching (loss of detections). Here, we present the use of multifocus microscopy for volumetric multicolor superresolution imaging. By simultaneously imaging nine different focal planes, the multifocus microscope instantaneously captures the distribution of single molecules (either fluorescent proteins or synthetic dyes) throughout an ∼4-µm-deep volume, with lateral and axial localization precisions of ∼20 and 50 nm, respectively. The capabilities of multifocus microscopy to rapidly image the 3D organization of intracellular structures are illustrated by superresolution imaging of the mammalian mitochondrial network and yeast microtubules during cell division.
Because of its specificity and ability to image live samples, fluorescence microscopy is the most widely used imaging tool for biological studies. In recent years, several methods have been introduced to increase the resolution of fluorescence microscopy beyond the diffraction limit (1, 2). These methods include stimulated emission depletion (3), structured illumination (4), and single-molecule localization microscopy (LM) (5, 6). In the latter approach, precise control over illumination conditions enables sparse activation of individual fluorescent molecules, permitting determination of their positions with an accuracy of a few tens of nanometers. The sequential photoactivation, imaging, and bleaching (or photoswitching) of large numbers of fluorophores then allow the reconstruction of the investigated structure, embodying the principles of photo-activated localization microscopy (PALM) (5), fluorescence photo-activation localization microscopy (FPALM) (7), and stochastic optical reconstruction microscopy (STORM) (6, 8).
LM techniques have garnered significant interest in biological studies (9, 10) but are still predominantly implemented using evanescent-wave (or total internal reflection fluorescence) microscopy (11), which limits its application to 2D or thin structures close to the cell membrane. However, in many biological contexts, it is highly desirable to access the 3D intracellular organization of the cell with subdiffraction resolution.
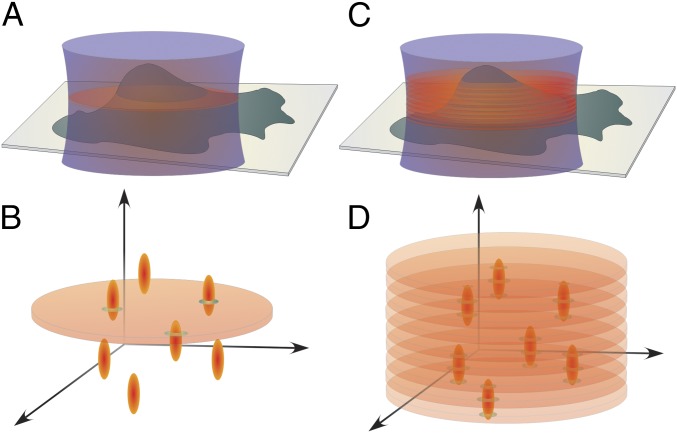
For efficient 3D LM in cultured cells, two issues need to be resolved. First, single molecules must be localized with subdiffraction accuracy both laterally and axially. Second, the axial depth over which localizations are made should be comparable with the thickness of the whole cell. Approaches developed to address the former issue usually rely on encrypting axial information into 2D images by engineering the point-spread function (PSF) of the microscope (8, 12–15), single-photon interferometry (16), or biplane imaging (17–19). However, the typical imaging depth of most of these methods does not exceed 1.2 μm and hence, is insufficient for whole-cell imaging. Furthermore, because of the wide-field excitation configuration predominantly used for imaging in 3D LM, the activation and emission of out-of-focus molecules lead to increased background in the fluorescence image and their unnecessary bleaching (Fig. 1 A and B). In other words, information outside the imaging plane is lost, whereas the signal-to-noise ratio in the image is reduced. Selective plane excitation (20) or activation (21, 22) can circumvent this problem but requires sequential scanning to image entire cell volume.
Fig. 1.
Comparison between conventional wide-field detection and multifocus detection. (A and B) The whole volume is excited in the wide-field configuration, and fluorescence from out-of-focus molecules constitutes the background of the recorded in-focus signal. (C and D) In the multifocus configuration, the 3D extent of the PSF is used to localize molecules within the volume as information is obtained from multiple focal planes.
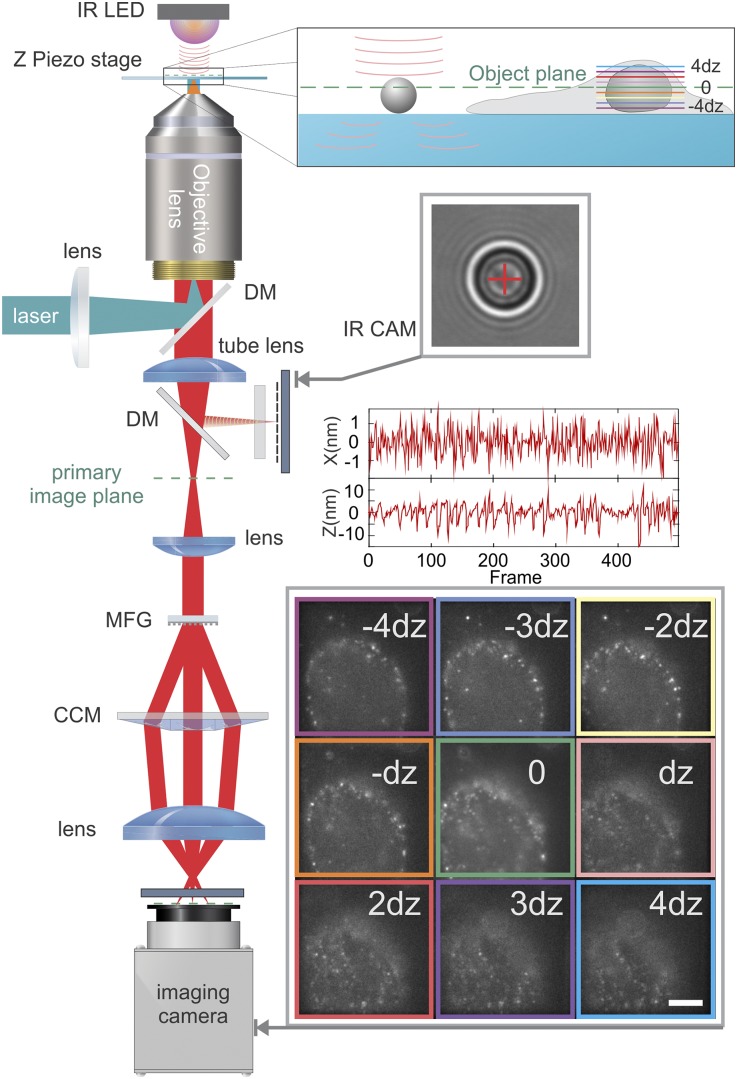
We report the successful implementation of a volumetric PALM/STORM superresolution method that avoids the aforementioned problems. Our method relies on the recently developed multifocus microscope (MFM) (23), which achieves simultaneous acquisition of nine equally spaced focal planes (Fig. 1 C and D) on a single camera through the combination of a specialized diffractive grating and chromatic correction elements placed in the microscope emission path (Fig. 2). The spacing between consecutive focal planes is ∼440 nm, allowing accurate 3D localization of single fluorescent molecules with 3D Gaussian fitting of resulting PSFs. We show that such volumetric acquisition is compatible with two-color superresolution PALM/STORM imaging of mammalian and yeast cells, with lateral and axial localization precisions of ∼20 and 50 nm, respectively. The imaging depth is ∼4 µm, well beyond the capabilities of other 3D superresolution techniques (review in ref. 24), and notably, it permits complete 3D imaging of many cellular organelles or whole cells (Movies S1 and S2).
Fig. 2.
MFM setup. The excitation lasers are combined in a fiber through an acousto-optic tunable filter, collimated, reflected on a dichroic mirror (DM), and focused at the back aperture of a high-N.A. objective to achieve wide-field excitation. The collected emission is transmitted through the DMs and passes through the MFG placed in a plane conjugated to the back pupil plane of the objective. The diffraction orders pass through the chromatic correction module before being separately focused on the detector. To record sample and stage drift, polystyrene beads (4-μm diameter) were immobilized on the cover glass, illuminated by an infrared light-emitting diode (IR LED), and their diffraction pattern was recorded with an IR camera (IR CAM) through an additional beamsplitter inserted into the emission path. The graphs are the recorded positions of the bead along the x and z axes. (Lower Inset) Example of emitting molecules recorded at different z planes corresponding to labeled nucleopores (the raw data are in Movie S1, and a reconstructed image is in Movie S2). (Scale bar: 5 μm.)
Results
Precision of Localization.
We first investigated the dependence of the localization precision as a function of the spacing between consecutive planes when using the MFM. To this end, 200-nm fluorescent beads immobilized on a coverslip were imaged using four different multifocus gratings (MFGs) corresponding to interplane spacing of 110, 220, 440, and 880 nm (at λ = 670 nm) (Fig. 3A). The microscope stage was shifted in steps of 60 nm (120 nm for the case of 880-nm spacing) along the optical axis, and 20 frames were acquired per step (Fig. 3B). The excitation intensity and the image acquisition parameters were adjusted such that the bead signal was comparable with that of an individual fluorophore in a PALM/STORM acquisition. For each frame, the signal was fitted with a 3D integrated Gaussian function (SI Text and Fig. S1), and the SD of the estimated positions was calculated for each step. For all gratings with a below 500 nm, we observed that the axial localization precision was less than 50 nm over the full imaging depth (Fig. 3C and Fig. S2).
Fig. 3.
Characterization of 3D MFM. (A) Relative in-focus position of consecutive z planes for a sample of diffraction MFM gratings tested at the design wavelength of 515 nm. The spacing was confirmed to be 880, 440, 220, and 110 nm for gratings I, II, III, and IV, respectively. (B) Measurements of the axial position of 200-nm fluorescent beads compared with the displacement of the piezoelectric stage. Twenty frames were acquired at each stage position, and bead localization along the z axis was performed as described in the text. An excellent agreement between the stage position and the experimentally detected bead localization is apparent over an ∼4-µm range when using grating II (440-nm spacing). (C) SD of the axial localization of fluorescent beads as a function of the z position recorded with gratings of various spacing. (D) Relative in-focus positions of consecutive z planes are measured for the grating II at three different wavelengths. (Inset) Wavelength dependence of plane separation of this grating. (E) Distribution of lateral localization precision (SD) of fluorescent beads imaged in a 2D wide-field configuration using 2D Gaussian fitting (average = 2.6 nm). (F) Distribution of lateral localization precision of the same beads imaged in the MFM configuration and localized by 2D Gaussian fitting using only one (in-focus) image plane (average = 13 nm). (G) Distribution of lateral localization precision achieved by 3D Gaussian fitting of the same MFM image as in F (average = 9 nm).
For multicolor superresolution imaging, the MFM grating is used at wavelengths different from the design value (∼515 nm), thus affecting the total diffraction efficiency and uniformity between diffraction orders. In practice, the former effect leads to a modest decrease in signal to noise, whereas the latter effect can be compensated with a postacquisition procedure (Methods, System Calibration and Data Analysis). In addition, the use of diffraction gratings introduces chromatic dispersion, which can lead to chromatic aberration if not accounted for. Although lateral chromatic dispersion is efficiently corrected with a blazed grating and prism placed after the MFG along the emission pathway, the axial chromatic dispersion manifests itself as a change in the effective between consecutive planes.
In the Fourier plane of an imaging system, the wavefront of light at wavelength λ, emanating from emitters located an axial distance of away from the focal plane in a medium of index of refraction nλ, is characterized by a phase-shift profile that scales as (25). The distortion of the MFG acts on the emission wavefront to counterbalance this phase-shift profile (23). The induced phase shift scales as (a is an integer between −4 and +4 specific to the order of diffraction). The axial position and the wavelength are incorporated in the design of the grating. Imaging at a different wavelength results in a slight modification of the corrected phase-shift profile that, in turn, corresponds to a different axial position. As shown experimentally in Fig. 3, for a given grating, the effective scales linearly with the wavelength (SI Text has a complete analytical derivation).
To achieve coregistration of the different spectral channels, multicolor beads were imaged sequentially through different filters during stepwise axial displacements. The separate spatial transformation matrices, , and diffraction efficiency were then computed and served as calibration to realign the image stacks in the different spectral channels. We estimate the overall two-color registration accuracy as better than 10 nm laterally and 30 nm axially (SI Text and Figs. S3 and S4).
Compared with imaging a single plane, an unavoidable drawback of the multiplane detection is a lower signal-to-noise ratio, because the total number of photons is split between different image planes. Because of losses caused by the multifocal grating (65% diffraction efficiency) and chromatic correction (85% efficiency at the imaging wavelength) elements, we estimate that ∼5% of the emitted photons are detected in each of nine planes (at 670-nm wavelength). Thus, the SD of the lateral emitter position is expected to increase by . To investigate the consequences of the signal splitting, we imaged fluorescent beads over a few hundred frames using the MFM as well as with a single-plane configuration, where all excitation and acquisition parameters were identical. We compare the lateral localization SD for both scenarios in Fig. 3 E–G. As expected, splitting significantly decreases the lateral localization precision when planes are analyzed individually (Fig. 3E vs. Fig. 3F). However, the MFM permits z sampling of the PSF at different focal planes, which effectively improves the lateral SD by the square root of the number of sampling planes. With 440-nm spacing, the PSF typically shows up on two planes, indicating an improvement by , which was confirmed experimentally in Fig. 3G. Therefore, the 440-nm grating spacing was chosen as a good compromise between accuracy and imaging depth (∼4 µm) (Fig. 3C).
Imaging Mitochondria Network in HeLa Cells.
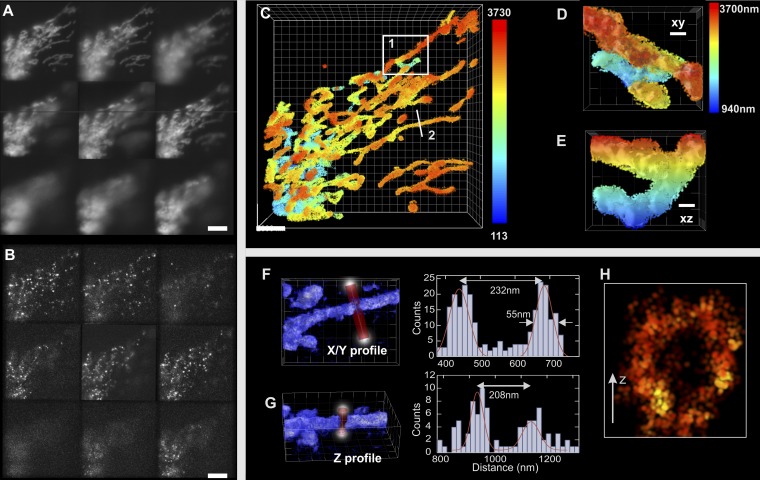
To test the robustness and the performance of 3D MFM superresolution microscopy, we first applied it to STORM imaging of mitochondria in HeLa cells transfected with TOM20 (translocase of outer mitochondrial membrane) fused to GFP. After fixation with paraformaldehyde (PFA), TOM20-GFP was labeled with anti-GFP nanobody–Alexa 647 conjugate (26) in the presence of an oxygen scavenger (SI Text). To achieve sparse detection (∼0.08 molecules/µm2 per frame), a low-power 488-nm (or 405-nm) laser was used together with high-power 640-nm laser illumination (1 kW/cm2). Because image acquisition in STORM requires data collection over a long time (∼15 min), we corrected the thermal drift of the microscope. To this end, we used a custom-made sample tracking system that analyzes the diffraction rings of a 4-µm bead attached to the sample coverslip and finds its center with a precision of 1 nm in x/y and 5 nm in z (27, 28) (Figs. S5–S7). Fig. 4 shows a typical superresolved image of mitochondria over a depth of ∼4 µm reconstructed with the localization of ∼106 molecules detected over 30,000 frames. Note that comparable imaging depths are not attainable with other methods unless sequential z scanning is used combined with selective plane illumination (20, 21). To directly evaluate the localization precision for individual Alexa 647 dyes (bound to TOM20 proteins) in our imaging and sample conditions, we isolated single-molecule events that spread over a few consecutive frames. We found that the localization accuracy was 16 nm laterally and 35 nm axially over the range of the detection depth (SI Text and Figs. S8–S10). The image, obtained with the 3D superresolution reconstruction/visualization software ViSP (29), shows a highly interconnected spatial network of mitochondria that is particularly crowded near the nucleus of the cell (Fig. 4C, lower left corner and Movies S3–S5). Because of the continuous fission and fusion in live mitochondria, such networks constantly change their shape; 3D MFM localization accuracy was sufficient to clearly distinguish the outer membrane of the mitochondria in the lateral and axial directions separated by 230 and 208 nm, respectively (Fig. 4 F–H).
Fig. 4.
Multifocal superresolved (MFM STORM) image of mitochondria. TOM20–GFP-expressing HeLa cells were labeled with anti-GFP nanobody Alexa 647 conjugate. (A) Image recorded by wide-field MFM. (Scale bar: 5 μm.) (B) Image recorded by MFM STORM. Single Alexa 647 fluorophores are detectable at different focal planes. (Scale bar: 5 μm.) (C) Reconstructed 3D superresolution image (the depth is color-coded) (Movies S3–S5). The localizations are represented as isotropic 3D Gaussian profiles of 20-nm SD using the 3D superresolution reconstruction software ViSP (29). (Scale bar: 2 μm.) (D and E) Enlarged view of area 1 in C showing the lateral and axial views, respectively. (Scale bar: 500 nm.) (F and G) Detection density profile of structure 2 in C plotted laterally and axially, respectively. (H) Cross-section of the structure shown in F and G showing mitochondrial lumen (arrow = 100 nm).
Yeast Imaging.
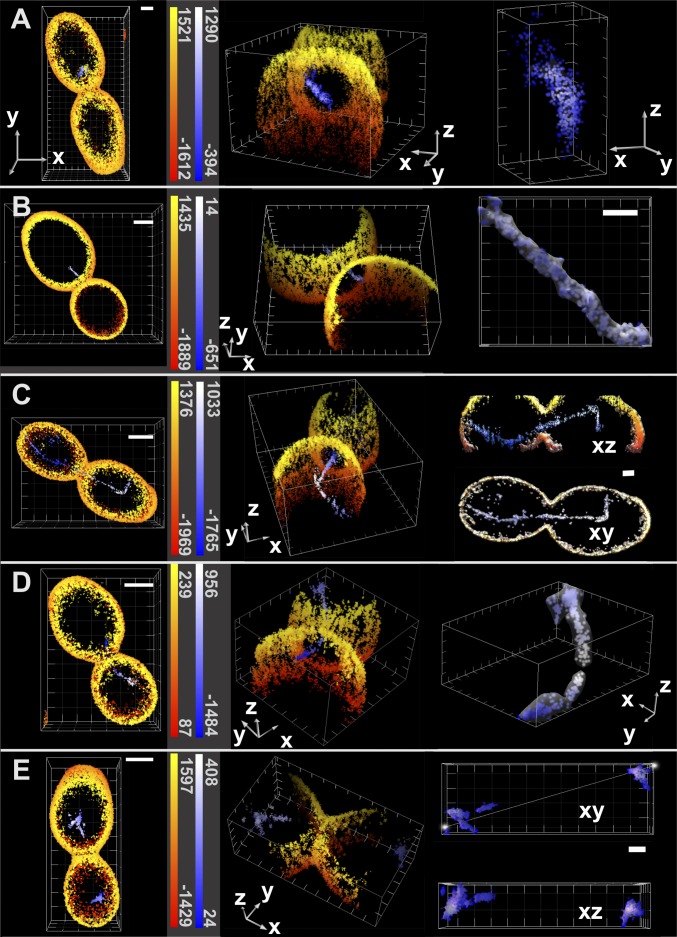
In a second example, we applied the 3D MFM PALM/STORM approach to budding yeast (Saccharomyces cerevisiae) cells. Despite their frequent use as model system in cell biology, yeast cells have rarely been imaged using superresolution microscopy (26). In particular, their rounded shape and thick cell wall constitute a significant obstacle to PALM applications, making imaging in total internal reflection mode unsuitable. The imaging depth of the MFM almost matches the ∼5-µm size of yeast cells and thus, is ideal for whole-cell superresolution imaging. We genetically introduced a photoconvertible tdEos (the tandem dimer form of Eos fluorescent protein) tag at the C terminus of one of the α-tubulin genes in the diploid strain. PFA-fixed cells were then coated with concanavaline A–Alexa 647 conjugate to label the cell walls. Two-color superresolution data were acquired sequentially. First, Alexa 647 was switched [in the presence of 100 mM cysteamine hydrochloride (MEA)] into dark state with intense 640-nm illumination. Second, individual molecules of Alexa 647 were observed during their transient return into the fluorescent state (STORM mode). After acquiring ∼20,000 frames (when Alexa 647 fluorescent events became rare), we switched to PALM imaging of tdEos-tubulin. Single molecules were activated into orange-emitting fluorophores by low-intensity photoconversion with 405-nm laser light, detected, and bleached with intense 561-nm illumination (2 kW/cm2). For analysis, we applied neighborhood-density filtering to tdEos data to exclude the free α-tubulin that is abundantly present in the cytoplasm and immobilized by fixation (SI Text). This approach allowed better visualization of mitotic spindles and microtubules (Movie S6). Cells in different stages of the cell cycle were analyzed, and typical examples are presented in Fig. 5. In the case of cells undergoing telophase, we could readily localize α-tubulin–tdEos-decorated spindles, because they thread through a narrow neck connecting the mother cell and the bud (Fig. 5 C and D). At that stage of mitosis, the spindle consists of about two to four microtubules. Additional examples of yeast cells imaged in different stages of the cell cycle are shown in Fig. 5. In all observed cases, it was possible to identify a spindle or microtubule inside the cell enclosed by the cell wall. Observed patterns correspond to known spindle–microtubule orientations (30) (conventional wide-field images shown for comparison in Supporting Information and Fig. S11). In addition to the mitotic spindle, we could detect cytoplasmic microtubules extending from both ends of the spindle throughout the axial range (Fig. 5C). In this particular example, the measured tubulin fiber profiles in the unfiltered localization data showed a full-width half-max (FWHM) of ∼65 nm laterally and ∼108 nm axially (Fig. S12). These values are larger than the reported values of 25 nm in ref. 26, but the difference might be because of the reduced localization precision in our measurements and contribution of tdEos tag size.
Fig. 5.
Two color superresolution MFM PALM/STORM images of yeast cells containing α-tubulin–tdEos- and Con A–Alexa 647-labeled cell wall. The red–yellow scale codes the z positions of Alexa 647 detections, whereas the blue–white scale corresponds to tdEos. (Left) Cells are shown in the xy view with color-coded depth. (Scale bars: 1 μm.) (Center) A tilted view reveals the mitotic spindle going through the neck of the bud during division. (Axes lengths: 1 μm.) (Right) Surface rendering of tdEos localizations in blue–white. Here, the color code represents molecule density. Snap shots present the budding yeast at different stages of cell division. (Scale bars and axes lengths: 1 μm.) (A) Metaphase cell with a short intranuclear spindle not oriented toward the neck. (B) Cell entering anaphase with a spindle beginning to penetrate through the neck opening. (C) Late anaphase/telophase cell with the mitotic spindle going through the neck of the bud (Movie S6). (D) Cell at the end of telophase with a disintegrating spindle breaking up between mother and daughter cells. (E) Postmitotic G1 cells without spindles.
Conclusion
In the investigation of cell organelles, 3D superresolution microscopy is becoming an essential tool. Our results show that multifocus microscopy combined with PALM/STORM imaging enables volumetric superresolution imaging over a depth of ∼4 µm, much larger than other methods, and achieves localization precisions of ∼20 and ∼50 nm along the lateral and axial directions, respectively. Because the precision depends directly on the number of detected photons, it will likely see improvement in the future through the use of brighter fluorophores and more efficient grating designs. In particular, the use of a multiphase grating instead of a binary one may increase the diffraction efficiency into nine central orders by upward of ∼90% (31). Modified designs with diffracted light concentrated into 25 central orders (instead of the reported 9 orders) may also increase imaging depth without compromising axial sampling precision. In contrast to sequential z scanning for 3D imaging, our approach is able to image relevant out-of-focus molecules before they photobleach. Capturing a maximum number of single-molecule events over the cellular extent is an important advantage, especially when low copy numbers of fluorescent proteins are involved. A challenge for 3D PALM/STORM microscopy is the large number of molecules that need to be localized to properly reconstruct an image, which results in longer acquisition times and potentially overlapping PSFs. These difficulties may be alleviated by means of computational tools, such as multiemitter fitting (32) or compressed sensing (33, 34), that enable the localization of single molecules at higher densities. In summary, we believe that multifocus microscopy opens new and exciting possibilities for whole-cell superresolution imaging in biological sciences.
Methods
Setup.
To permit multifocal imaging, custom optical elements are introduced in the emission pathway of a conventional wide-field microscope (Nikon Ti-Eclipse) (Fig. 2) (23). First, a diffraction grating conjugated with the back pupil plane of the objective (100×, 1.4 N.A.; Nikon) acts to direct the incoming light into nine central orders with ∼65% efficiency. The grating pattern is distorted to introduce a focal distance shift between diffraction orders. Second, a combination of a blazed grating and prism is used to correct for residual chromatic dispersion. Third, an imaging lens forms the image of nine orders (corresponding to nine different imaging planes) on a single EM–CCD detector (DU-897; Andor). Illumination for excitation and activation is provided with a set of lasers (405, 488, 514, 560, and 640 nm) controlled by an acousto-optic tunable filter (AA Opto-Electronic) and coupled into a single-mode fiber. The output light of the fiber is recollimated and directed to the objective of the microscope by a multiband dichroic mirror (LF405/488/561/635–4 × 4M-A-000; Semrock). The region of excitation and activation is delimited laterally by a slit placed in the excitation path, whereas a second slit in the emission path is used to limit the imaged area and avoid overlapping of nine planes on the detector. We additionally placed a band-pass filter before the camera (FF01-607/36 for PALM imaging of tdEos and FF01-670/30 for STORM imaging of Alexa 647; Semrock).
Because of the long recording time (typically ≥20 min) to acquire raw data for 3D superresolution image reconstruction, the sample drift needs to be corrected. To this end, polystyrene beads (4 μm in diameter; Dynabeads) were placed on the coverslip next to the target cells and observed with an IR camera to function as fiduciary markers (Fig. 2, SI Text, and Figs. S5–S7). Analysis of the position of beads and their diffraction patterns permitted precise detection of lateral and axial shifts of the sample with an SD of less than 1 nm laterally and 5 nm axially. Those coordinates were subsequently used to correct the 3D sample drift.
System Calibration and Data Analysis.
Multicolor fluorescent beads (TetraSpeck Fluorescent Microspheres Kit; T14792; Invitrogen), immobilized on a coverslip were used to calibrate the 3D imaging setup and ensure precise reconstruction of raw images into a z stack with minimal loss of resolution (23). When translating the microscope stage with a piezo stage in fixed steps along the optical axis, the beads appear consecutively in focus on the different panels (z planes) of the image. A maximum intensity projection of the acquired stack shows beads appearing in focus throughout nine panels. After the center of the beads are determined by Gaussian fitting, a transformation matrix is computed to realign the different panels with a precision on the order of 2 nm (SD).
Furthermore, the binary phase grating used in our MFM diffracts about 65% of the incoming light in nine (3 × 3) central orders at the design wavelength. The relative fluctuations between the intensities are on the order of ∼5% at the grating design wavelength (515 nm) and increase up to ∼50% at 670 nm, mostly because of a brighter (0, 0) order. A postacquisition intensity correction of the different panels is, therefore, applied to ensure proper reconstruction of the image volume and achieve an accurate estimate of the axial position of individual molecules.
In summary, the raw images (an example is shown in Movie S1) are processed as follows. First, the camera offset is subtracted. Second, the image panels are split and aligned using the transformation matrix generated in the bead calibration step. Third, the relative intensities of the different panels are corrected; 3D Gaussian fitting of individual 3D PSFs was performed with the freely available FishQUANT software (35) to retrieve the emitter position, and 3D superresolution images were visualized with ViSP software (29).
Two-Color Colocalization and z-Position Correction.
Calibration is done by moving multicolor fluorescent beads mounted on a coverslip along the optical axis while sequentially recording images in the two spectral channels at each position. The maximum intensity projection image of each channel was reconstructed, and the beads were localized in the different panels of the image in the different channels.
The position of the beads on each panel of the images was then compared with the central (undistorted) panel of the red channel. The relative positions served as basis of two calibration matrices that allowed a sensitive reconstruction of the 3D volume as well as an accurate alignment between the planes of the two channels, taking into consideration any possible magnification, rotation, or translation transformation.
Any shift in the nominal plane between the two channels was also determined and accounted for in a postprocessing step as well as the plane spacing as a function of the imaging wavelength (SI Text and Fig. S3).
The overall accuracy of the spectral coregistration is estimated to be 10 nm in the lateral dimension and 30 nm in the axial dimension. The accuracy was determined experimentally where multicolor beads were imaged in two channels sequentially (Fig. S4).
Supplementary Material
Acknowledgments
We thank the late Mats Gustaffson for the original design of the multifocus microscope; Sara Abrahamsson for her assistance while designing and building the microscope; Florian Mueller for his assistance with the localization software; Gleb Schtengel, Davide Normanno, and Ignacio Izeddin for useful discussions; the members of the Transcription Imaging Consortium (TIC) at the Howard Hughes Medical Institute Janelia Farm for their help and discussion; Dr. Karin Busch for the TOM20-GFP plasmid; Luke Lavis and Jonathan B. Grimm for sharing dyes and useful discussions; the information technology (IT) Department of Janelia Farm Research Campus for setting up the cluster analysis pipeline and helping with the software; and Vasili Gurchenkov for his input on figures and graphical representation. B.H. acknowledges the financial support of the TIC, the Pierre Gilles de Gennes Foundation, and Agence pour la Recherche sur le Cancer (ARC Foundation). We acknowledge financial support from French National Research Agency (ANR) Paris-Science-Lettres Program (ANR-10- IDEX-0001-02 PSL), Labex CelTisPhyBio (N° ANR-10-LBX-0038), France-BioImaging infrastructure supported by ANR Grant ANR-10-INSB-04 (Investments for the Future), and ANR Grant TRIDIMIC.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412396111/-/DCSupplemental.
References
- 1.Galbraith CG, Galbraith JA. Super-resolution microscopy at a glance. J Cell Sci. 2011;124(Pt 10):1607–1611. doi: 10.1242/jcs.080085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: Super-resolution imaging of cells. Cell. 2010;143(7):1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19(11):780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 4.Gustafsson MG. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA. 2005;102(37):13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 6.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3(10):793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hess ST, Girirajan TP, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91(11):4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319(5864):810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szymborska A, et al. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science. 2013;341(6146):655–658. doi: 10.1126/science.1240672. [DOI] [PubMed] [Google Scholar]
- 10.Shim SH, et al. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc Natl Acad Sci USA. 2012;109(35):13978–13983. doi: 10.1073/pnas.1201882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Löschberger A, et al. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J Cell Sci. 2012;125(Pt 3):570–575. doi: 10.1242/jcs.098822. [DOI] [PubMed] [Google Scholar]
- 12.Izeddin I, et al. PSF shaping using adaptive optics for three-dimensional single-molecule super-resolution imaging and tracking. Opt Express. 2012;20(5):4957–4967. doi: 10.1364/OE.20.004957. [DOI] [PubMed] [Google Scholar]
- 13.Grover G, DeLuca K, Quirin S, DeLuca J, Piestun R. Super-resolution photon-efficient imaging by nanometric double-helix point spread function localization of emitters (SPINDLE) Opt Express. 2012;20(24):26681–26695. doi: 10.1364/OE.20.026681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavani SR, et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci USA. 2009;106(9):2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia S, Vaughan JC, Zhuang X. Isotropic three-dimensional super-resolution imaging with a self-bending point spread function. Nat Photonics. 2014;8(4):302–306. doi: 10.1038/nphoton.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shtengel G, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci USA. 2009;106(9):3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prabhat P, et al. Simultaneous imaging of different focal planes in fluorescence microscopy for the study of cellular dynamics in three dimensions. IEEE Trans Nanobiosci. 2004;3(4):237–242. doi: 10.1109/tnb.2004.837899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ram S, Kim D, Ober RJ, Ward ES. 3D single molecule tracking with multifocal plane microscopy reveals rapid intercellular transferrin transport at epithelial cell barriers. Biophys J. 2012;103(7):1594–1603. doi: 10.1016/j.bpj.2012.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juette MF, et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5(6):527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 20.Cella Zanacchi F, et al. Live-cell 3D super-resolution imaging in thick biological samples. Nat Methods. 2011;8(12):1047–1049. doi: 10.1038/nmeth.1744. [DOI] [PubMed] [Google Scholar]
- 21.Vaziri A, Tang J, Shroff H, Shank CV. Multilayer three-dimensional super resolution imaging of thick biological samples. Proc Natl Acad Sci USA. 2008;105(51):20221–20226. doi: 10.1073/pnas.0810636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.York AG, Ghitani A, Vaziri A, Davidson MW, Shroff H. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nat Methods. 2011;8(4):327–333. doi: 10.1038/nmeth.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abrahamsson S, et al. Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat Methods. 2013;10(1):60–63. doi: 10.1038/nmeth.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajj B, El Beheiry M, Izeddin I, Darzacq X, Dahan M. Accessing the third dimension in localization-based super-resolution microscopy. Phys Chem Phys. 2014;16(31):16340–16348. doi: 10.1039/c4cp01380h. [DOI] [PubMed] [Google Scholar]
- 25.Botcherby EJ, Juskaitis R, Booth MJ, Wilson T. Aberration-free optical refocusing in high numerical aperture microscopy. Opt Lett. 2007;32(14):2007–2009. doi: 10.1364/ol.32.002007. [DOI] [PubMed] [Google Scholar]
- 26.Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat Methods. 2012;9(6):582–584. doi: 10.1038/nmeth.1991. [DOI] [PubMed] [Google Scholar]
- 27.Gosse C, Croquette V. Magnetic tweezers: Micromanipulation and force measurement at the molecular level. Biophys J. 2002;82(6):3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revyakin A, et al. Transcription initiation by human RNA polymerase II visualized at single-molecule resolution. Genes Dev. 2012;26(15):1691–1702. doi: 10.1101/gad.194936.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Beheiry M, Dahan M. ViSP: Representing single-particle localizations in three dimensions. Nat Methods. 2013;10(8):689–690. doi: 10.1038/nmeth.2566. [DOI] [PubMed] [Google Scholar]
- 30.Winey M, Bloom K. Mitotic spindle form and function. Genetics. 2012;190(4):1197–1224. doi: 10.1534/genetics.111.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mait JN. Understanding diffractive optic design in the scalar domain. J Opt Soc Am A. 1995;12(10):2145–2158. [Google Scholar]
- 32.Holden SJ, Uphoff S, Kapanidis AN. DAOSTORM: An algorithm for high- density super-resolution microscopy. Nat Methods. 2011;8(4):279–280. doi: 10.1038/nmeth0411-279. [DOI] [PubMed] [Google Scholar]
- 33.Gu L, et al. High-density 3D single molecular analysis based on compressed sensing. Biophys J. 2014;106(11):2443–2449. doi: 10.1016/j.bpj.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu L, Zhang W, Elnatan D, Huang B. Faster STORM using compressed sensing. Nat Methods. 2012;9(7):721–723. doi: 10.1038/nmeth.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller F, et al. FISH-quant: Automatic counting of transcripts in 3D FISH images. Nat Methods. 2013;10(4):277–278. doi: 10.1038/nmeth.2406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.