Significance
Nutrient uptake is a central property of ocean biogeochemistry, but our understanding of this process is based on laboratory cultures or bulk environmental studies. Thus, mathematical descriptions of nutrient uptake, at the heart of most biogeochemical models, must rely on this limited information. Hence, we have little knowledge of how natural phytoplankton populations vary in their abilities to take up key nutrients. Using advanced analytical techniques, this study provides the first comprehensive in situ quantification of nutrient uptake capabilities among dominant phytoplankton groups. Supported by a model that considers plastic ecological responses in an evolutionary context, this work further provides a fundamentally new framework for the integration of microbial diversity to describe and understand the controls of ocean nutrient assimilation.
Keywords: phosphate kinetics, cyanobacteria, adaptive dynamics, eco-evolutionary dynamics
Abstract
We have a limited understanding of the consequences of variations in microbial biodiversity on ocean ecosystem functioning and global biogeochemical cycles. A core process is macronutrient uptake by microorganisms, as the uptake of nutrients controls ocean CO2 fixation rates in many regions. Here, we ask whether variations in ocean phytoplankton biodiversity lead to novel functional relationships between environmental variability and phosphate (Pi) uptake. We analyzed Pi uptake capabilities and cellular allocations among phytoplankton groups and the whole community throughout the extremely Pi-depleted western North Atlantic Ocean. Pi uptake capabilities of individual populations were well described by a classic uptake function but displayed adaptive differences in uptake capabilities that depend on cell size and nutrient availability. Using an eco-evolutionary model as well as observations of in situ uptake across the region, we confirmed that differences among populations lead to previously uncharacterized relationships between ambient Pi concentrations and uptake. Supported by novel theory, this work provides a robust empirical basis for describing and understanding assimilation of limiting nutrients in the oceans. Thus, it demonstrates that microbial biodiversity, beyond cell size, is important for understanding the global cycling of nutrients.
The composition of microbial communities varies among different ocean regions and along environmental gradients (e.g., refs. 1 and 2). This variation includes phylogenetic, genomic, and functional diversity among and between heterotrophic or autotrophic groups. Presently, we have a limited understanding of the consequences of these different levels of microbial biodiversity on specific processes and more broadly on global ocean biogeochemical cycles (3). An important process is macronutrient uptake by microorganisms, as the uptake of nitrate and/or inorganic phosphate (Pi) controls ocean CO2 fixation rates in many regions (4). Indeed, mathematical descriptions of nutrient uptake are at the heart of most marine ecosystem models (5). The ability of microorganisms to assimilate nutrients as a function of concentration is commonly described by a hyperbolic uptake kinetics curve (6, 7). Analogous to the classical Michaelis–Menten curves for enzyme kinetics (8), the parameters quantifying this relationship are the maximum uptake rate (Vmax), the half-saturation concentration (Ks), and the ratio of the two parameters named the nutrient affinity (α). Despite the importance of accurate descriptions of nutrient uptake capabilities for the understanding of competition and ocean biogeochemistry (7), our knowledge of these properties is mostly limited to laboratory studies of cultured strains (9). However, culture-based kinetics estimates would suggest plankton are proliferating at <25% of the growth rates observed in the oligotrophic subtropical gyres. Thus, we need to quantify this key process in naturally competing populations (10–12) and explain the discrepancies. Furthermore, we have a limited quantitative knowledge of in situ uptake capabilities under conditions where the focal nutrient is extremely depleted. The latter is important as marine microorganisms like Prochlorococcus often have unique genomic adaptions to maximize nutrient assimilation under such conditions (13, 14).
To address this lack of knowledge for a globally relevant ecosystem process, we here aimed at identifying the influence of different levels of microbial biodiversity on in situ Pi uptake in the western subtropical North Atlantic Ocean. Phosphate plays a central role in regulating the functioning of microbial communities in this region as the surface waters likely have the lowest Pi concentration observed anywhere in the ocean (15). We used a combination of shipboard cell sorting and isotopically labeled Pi to quantify nutrient uptake capabilities for the whole field community and four phytoplankton groups of different sizes—Prochlorococcus, Synechococcus, small eukaryotes (<20 μm), and the nitrogen fixer Trichodesmium. We asked the following: (i) do the in situ Pi uptake capabilities differ among abundant phytoplankton groups, (ii) what is the variation in uptake capabilities within each group between environments, and (iii) what is the integrative effect of marine microbial diversity and environmental variability on nutrient uptake across the region? The answers to these questions will provide both a theoretical and empirical basis for describing how microbial diversity affects a core ocean ecosystem process.
Results
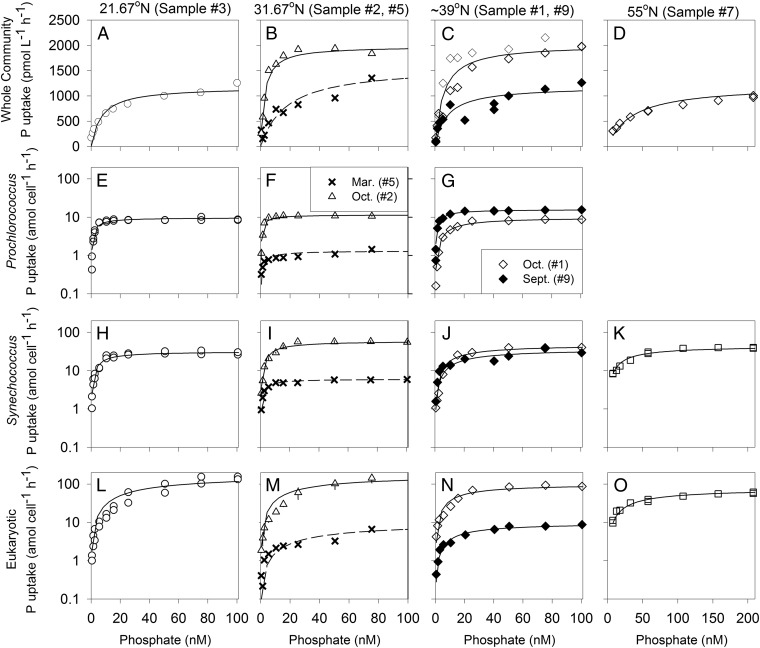
We first examined the uptake capabilities for the whole community and four phytoplankton groups—Prochlorococcus, Synechococcus, small eukaryotes (<20 μm), and the nitrogen fixer Trichodesmium (Fig. 1 and Fig. S1) across a range of environments (Fig. S2). When we experimentally added increasing concentrations of Pi, the nutrient uptake response closely resembled a hyperbolic shape for all discrete populations as well as the whole community (R2 > 0.9, Fig. 1 and Fig. S1). We then estimated the parameters Ks, Vmax, and affinity (α) (Table S1) and found significant (one-way ANOVA, P < 0.05) differences in Ks among phytoplankton groups (Fig. S3 and Table S1). Prochlorococcus had the lowest average Ks followed by Synechococcus, small eukaryotic phytoplankton, and Trichodesmium, respectively. In comparison, the whole microbial community was characterized by Ks values between those of Prochlorococcus and Synechococcus, the most abundant autotrophs. There was also significant variation in Vmax among phytoplankton lineages (one-way ANOVA, P < 0.05), and the order was analogous to Ks.
Fig. 1.
In situ phosphate uptake curves for the whole community (A–D), Prochlorococcus (E–G), Synechococcus (H–K), and eukaryotic phytoplankton (L–O). The lines represent the best fit of a hyperbolic curve. Each row represents the whole community or specific population and each column represents a discrete station as listed in Table S1 and noted at the top of the panels. In B, F, I, and M, data from both October and March are shown as denoted in the legend in F. C, G, J, and N show samples from 39°N taken ∼1 y apart. Triangle symbols and associated error bars represent the mean ± SD of duplicate experiments at this station.
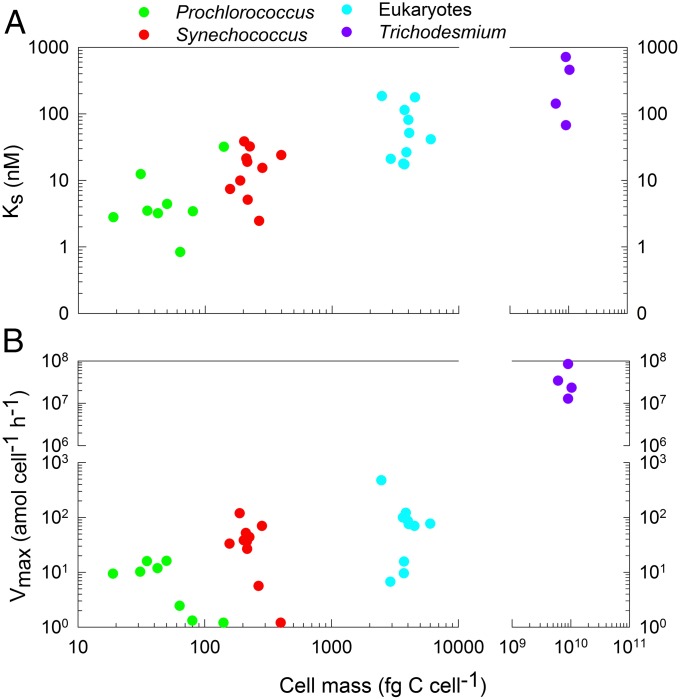
We then examined whether differences in uptake abilities were related to cell size and found a significant positive relationship for both Ks and Vmax (Fig. 2 A and B, PSpearman < 0.05), but not affinity. The latter would suggest that small cells do not have a distinct competitive advantage at very low substrate concentrations. However, we also measured the Pi cell quota (Qp) for all groups (Table S1) and observed that affinity normalized to Qp ranked Prochlorococcus > Synechococcus > eukaryotic phytoplankton > Trichodesmium. An identical pattern was observed for Vmax normalized to Qp. Thus, Prochlorococcus had the highest potential for uptake in relation to demand at low concentrations, despite having a low absolute Vmax.
Fig. 2.
Relationship between Ks, Vmax, and cell mass across phytoplankton groups. Due to difficulties of accurately estimating cell volume, we used cellular carbon biomass as a proxy for cell size (31).
In addition to size-dependent variations across phytoplankton groups, we also observed differences in nutrient uptake capabilities within each group. For example, samples 2 and 10 at the Bermuda Atlantic Time-series during the highly stratified late summer/early fall period consistently had a higher Vmax but not Ks for the whole community and three discrete phytoplankton lineages in comparison with samples from the less stratified springtime (numbers 4 and 5) (Fig. 1 and Table S1). Similarly, we observed a higher Vmax for a surface (number 5) vs. 80 m sample (number 6) (Fig. S4). We hypothesized that these differences were related to Pi availability. To investigate this result further, we compared uptake capabilities to ambient Pi concentration at the time of sampling and found Vmax, and especially affinity, were negatively correlated to Pi (Fig. 3, PANCOVA < 0.05). In further support, Vmax was lower in Prochlorococcus field populations from samples with higher Pi from the North Pacific Ocean (10). Thus, populations growing in low Pi environments showed significantly enhanced uptake capabilities.
Fig. 3.
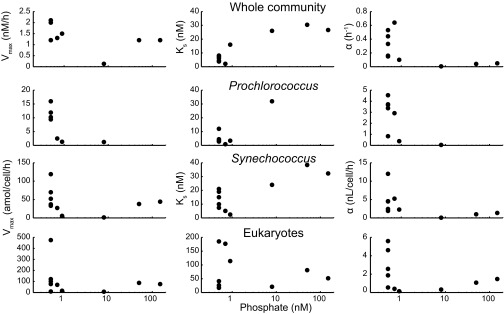
Relationships between the ambient Pi concentration and uptake capabilities (i.e., Ks, Vmax, and α) for the whole community and Prochlorococcus, Synechococcus, and eukaryotic phytoplankton populations.
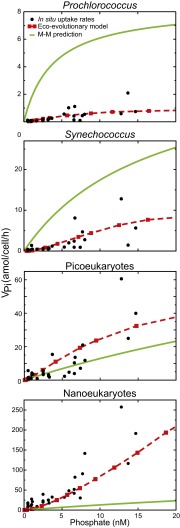
We finally asked whether the presence of the observed physiologically (and possibly genetically) diverse populations would influence the link between nutrient availability and in situ uptake (VPi) across environments. To address this, we developed an eco-evolutionary model in which, according to our observations, each lineage was influenced by a size-dependent scaling of Ks and Vmax (resulting from adaptation) as well as a regulation of the concentration of transport proteins (and associated Vmax) in response to ambient nutrient availability (i.e., acclimation) (Fig. S5). This theoretical model predicted a relationship between ambient Pi and VPi that was very different from a traditional Michaelis-Menten–type curve. Moreover, in contrast to a classic hyperbolic model, the emergent uptake curves accurately replicated our measurements of VPi of four phytoplankton groups in samples collected across the whole Western North Atlantic region (Fig. 4 and Fig. S2). However, our model required specific allometries for each phytoplankton group, which suggested that size alone could not describe differences in Pi uptake between the lineages. Overall, these biodiversity effects also manifested themselves on the whole-community VPi, where a linear fit replicated our observations better than a hyperbolic one (Fig. S6). These results highlight how the interaction of size and lineage diversity with physiological plasticity of phytoplankton had a direct impact on in situ nutrient uptake patterns in this region.
Fig. 4.
Relationship between in situ phosphate uptake rates (VPi, black dots) and the ambient Pi concentration. The dashed lines are predictions from our eco-evolutionary model, and the solid lines are traditional Michaelis–Menten functions applied to each phytoplankton group. The Michaelis–Menten curves are based on the mean values for Ks and Vmax (Table S1).
Discussion
Theoretical studies and culture data have both suggested that differences in microbial biodiversity can have an impact on nutrient uptake capabilities (5, 9, 16). Our results support culture studies showing an allometric scaling of Ks and Vmax (9) including the lowest values in the small Prochlorococcus and Synechococcus. A recent compilation of available marine culture data does not report data for organisms as small as Prochlorococcus and Synechococcus (9), but based on their size, the values for Prochlorococcus and Synechococcus cells fall well below the predicted allometric line. Indeed, the best possible match between our eco-evolutionary model output and observations could only be achieved by using lineage-specific allometries for the traits involved. As a result, uptake capabilities of a given lineage cannot solely be described by specific cell size-dependent Ks and Vmax values.
Biodiversity may also influence nutrient uptake by a taxonomic group via differences in genomic content (14, 17, 18) and associated physiological capabilities of the cells (19, 20). We see strong support for a variation in uptake capabilities within populations that is likely linked to acclimation through the regulation of nutrient transporters in response to changes in the nutrient environment. To illustrate this further, we examined the ratio of Vmax to Qp, which can be interpreted as a proxy for the maximum growth rate (if we assume no leakage). However, we find values up to 27 d−1 for Prochlorococcus and 7.7 d−1 for Synechococcus, which are much higher than previously described maximum growth rates for these groups (21, 22). This suggests that at least Prochlorococcus and Synechococcus have highly induced active Pi transporters at very low substrate levels. A maintenance of high Vmax under strongly nutrient-limited conditions has been observed in marine diatom cultures (20), but this is the first demonstration (to our knowledge) of such Vmax response mechanism in natural phytoplankton populations from the open ocean.
Identifying the linkages between marine biodiversity, environmental variation, and nutrient uptake rates has significant biogeochemical implications. A Prochlorococcus Ks of 0.8 nM reported here is the lowest value detected for any group yet, and we generally see high uptake rates for the whole community at low Pi. Thus, our data suggest that abundant phytoplankton groups can readily satisfy their P requirements, whether directly from Pi or from hydrolysed dissolved organic phosphorus, at less than 10 nM, and thus lower the threshold for when Pi becomes limiting for growth. Our nutrient kinetics values are consistent with past studies of Trichodesmium (11) as well as the whole community (23) but add important quantitative information for specific unicellular lineages. Another biogeochemical consequence of our work concerns the parameterization of nutrient uptake in ocean models and associated skills in predicting future ocean chemical conditions, competition for limiting nutrients, and estimates of primary production. Several ocean biochemical models use Ks for Pi above 0.5 μM (24, 25), which results in gross model overpredictions of dissolved Pi concentrations in many oligotrophic regions. As a corollary, this results in underestimation of primary production, which is important given the interest in predicting future rates of biological productivity in ocean gyres. Furthermore, given the hypothesis that open ocean gyres will continue to expand into the future due to increasing stratification (26), these data suggest that a priori assumptions about reductions in ocean productivity need to be reevaluated.
We find strong support for a hyperbolic link between Pi and uptake for individual populations, but the summed outcomes for Pi uptake by specific microbial lineages across environmental gradients in Pi have a unique functional form. These results likely apply to a large fraction (∼30%) of the global ocean surface area where Pi is similarly low. Thus, static Ks and Vmax parameters for individual populations do not adequately describe the uptake rates across the region. Therefore, we recommend including these quantitative responses (e.g., much lower Ks values, feedback from plastic or adaptive responses, etc.) in ocean models if the aim is to accurately identify ecosystem processes in oligotrophic regions. This may be particularly pertinent if the goal is to predict future ocean biogeochemistry where increased warming may lead to decreases in Pi concentration (26) but not necessarily in phytoplankton abundances (27).
Methods
Sample Collection.
The data presented in this study were collected on seven cruises throughout the western North Atlantic Ocean (cruise X0606, X0705, X0804, BVAL 39, BVAL 46, AE1206, and AE1319). All samples for Pi uptake rates and kinetics experiments were collected in acid-cleaned Niskin bottles and kept in subdued lighting until experiments were initiated (<1 h). Samples for whole-community ambient uptake rates were collected from approximately four depths in the upper 60 m, whereas samples for taxon-specific ambient uptake rates were collected from 5 m, 40 m, and the deep chlorophyll maximum (ranging from 80 to 120 m) (28). Trichodesmium colonies were collected from the near surface (roughly within the top 20 m) by vertically hauling a handheld 100-µm net. Single colonies were transferred a second time into fresh 0.2-µm–filtered water to reduce contamination of closely associated organisms, and subsequently separated by morphotype (either “puff” with radial trichomes or “raft” with parallel trichomes); only data for rafts are presented here.
33Phosphate Incubations.
The approach for ambient whole community and population-specific uptake rate measurements were previously published (28). Briefly, duplicate aliquots of 10 mL of seawater were amended with 0.15 μCi (∼80 pmol⋅L−1) additions of H333PO4 (3,000 Ci⋅mol−1; PerkinElmer), and incubated for 30–60 min in subdued lighting (∼100 µmol photons⋅m−2⋅s−1) at ∼23 °C. This temperature was within ∼3 °C of the coolest/warmest in situ temperature from which the samples were collected. The duration of each incubation varied depending on turnover time of the added isotope, such that efforts were made to keep uptake to <25% of the tracer added. Duplicate killed control incubations were conducted for each station. Killed controls were amended with paraformaldehyde (0.5% final concentration) for 30 min before the addition of isotopic tracer and incubation. Whole-community incubations were terminated by filtration onto 0.2-µm polycarbonate filters that were subsequently placed in glass scintillation vials. Population-specific ambient uptake incubations were terminated by the addition of paraformaldehyde (0.5% final concentration), and stored at 4 °C until sorting (<12 h) as described in the next section.
Whole-community and population-specific kinetics experiments were conducted by adding 0.15 μCi (∼80 pM) of H333PO4 to ∼10 replicate 10-mL seawater samples that were further amended by increasing additions of “cold” KH2PO4 up to 100 nM. Samples were incubated as above, but the incubations were terminated by the addition of KH2PO4 to a final concentration of 100 µM (29). Whole-community samples were filtered onto 0.2-µm polycarbonate filters and rinsed with an oxalate wash (30). Surface-bound phosphate in population-specific samples was accounted for by subtracting 33P counts for sorted populations to which 100 µM phosphate had been added before addition of the isotopic tracer. It is assumed that addition of such a high level of phosphate would result in negligible uptake of radioactive phosphate, and thus any signal was attributed to surface absorption; this correction was always <2–3%. Population-specific kinetics experiments for samples collected in the deep chlorophyll maximum were first gravity concentrated and resuspended in phosphate-free Sargasso Sea surface water before incubation as described. Population-specific samples were stored at 4 °C in the dark until sorting (<3 h) as described in the next section. Kinetics experiments for Trichodesmium spp. were conducted in the same manner as above for whole community samples but with picked and rinsed colonies and increasing additions of cold KH2PO4 up to 1,000 nM.
Flow Cytometry Analysis and Cell Sorting.
Samples were sorted on an InFlux cell sorter (BD) at an average flow rate of ∼40 µL⋅min−1. Samples were sorted for Prochlorococcus, Synechococcus, and an operationally defined eukaryotic algae size fraction (eukaryotes >2-µm). A 100-mW blue (488-nm) excitation laser was used. After exclusion of laser noise gated on pulse width and forward scatter, autotrophic cells were discriminated by chlorophyll fluorescence (>650 nm), phycoerythrin (585/30 nm), and granularity (side scatter). Sheath fluid was made fresh daily from distilled deionized water (Millipore) and molecular-grade NaCl (Mallinckrodt Baker), prefiltered through a 0.2-µm capsule filter (Pall), and a STERIVEX sterile 0.22-µm inline filter (Millipore). Mean coincident abort rates were <1% and mean recovery from secondary sorts (n = 25) was 97.5 ± 1.1% (data not shown). Spigot (BD) and FCS Express V3 (DeNovo Software) were used for data acquisition and postacquisition analysis, respectively. Sorted cells from each sample were gently filtered onto 0.2-µm Nucleopore polycarbonate filters, rinsed with copious amounts of 0.2-µm–filtered seawater, an oxalate wash (30), and placed in a 7-mL scintillation vial for liquid scintillation counting.
Data Analysis.
Parameters for the hyperbolic nutrient uptake curves from all samples were estimated in SigmaPlot (Systat Software; version 10), and the ANCOVA analysis was done with R. All other statistical analyses were done in Matlab (MathWorks).
Biodiversity Uptake Model with Adaptation and Acclimation.
To develop a theoretical model capable of predicting phosphate uptake and kinetic parameters Vmax and Ks observed in the field across diverse populations, we used standard expressions for growth (Droop) and uptake (Michaelis–Menten). To these expressions, we added the possibility for phytoplankton to regulate kinetic parameters in reaction to environmental changes. We explicitly did not include the option of shifting expression between high- and low-affinity transporters as at least Prochlorococcus and Synechococcus only contain one type of Pi transporter system (14, 18). We then considered this ecological description within an evolutionary framework, which allowed us to calculate the most competitive within-taxon strain for each environmental setup. For each taxon, the compilation of winning strains in different locations provided the data we then contrasted with our observations (see SI Text for further details and Table S2 and Fig. S7 for model results). We did not include Trichodesmium in this comparison, as we did not measure ambient uptake rates for this lineage.
Supplementary Material
Acknowledgments
We thank Stacey Goldberg and Céline Mouginot for assistance with cell sorting and field sampling, and Steven Allison and Jennifer Martiny for many helpful comments. Financial support for this work was provided by the National Science Foundation Dimensions of Biodiversity and Biological Oceanography programs.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420760111/-/DCSupplemental.
References
- 1.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5(3):e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zinger L, et al. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS One. 2011;6(9):e24570. doi: 10.1371/journal.pone.0024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arrigo KR. Marine microorganisms and global nutrient cycles. Nature. 2005;437(7057):349–355. doi: 10.1038/nature04159. [DOI] [PubMed] [Google Scholar]
- 4.Moore JK, Doney SC, Kleypas JA, Glover DM, Fung IY. An intermediate complexity marine ecosystem model for the global domain. Deep Sea Res Part II Top Stud Oceanogr. 2002;49(1):403–462. [Google Scholar]
- 5.Franks PJS. Planktonic ecosystem models: Perplexing parameterizations and a failure to fail. J Plankton Res. 2009;31(11):1299–1306. [Google Scholar]
- 6.Dugdale RC. Nutrient limitation in the sea: Dynamics, identification and significance. Limnol Oceanogr. 1967;12(4):685–695. [Google Scholar]
- 7.Titman D. Ecological competition between algae: Experimental confirmation of resource-based competition theory. Science. 1976;192(4238):463–465. doi: 10.1126/science.192.4238.463. [DOI] [PubMed] [Google Scholar]
- 8.Michaelis L, Menten ML. The kenetics of the inversion effect. Biochem Z. 1913;49:333–369. [Google Scholar]
- 9.Edwards K, Thomas M, Klausmeier CA, Litchman E. Allometric scaling and taxonomic variation in nutrient utilization traits and maximum growth rate of phytoplankton. Limnol Oceanogr. 2012;57(2):554–566. [Google Scholar]
- 10.Björkman K, Duhamel S, Karl DM. Microbial group specific uptake kinetics of inorganic phosphate and adenosine-5′-triphosphate (ATP) in the north pacific subtropical gyre. Front Microbiol. 2012;3:189. doi: 10.3389/fmicb.2012.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orchard ED, Ammerman JW, Lomas MW, Dyhrman ST. Dissolved inorganic and organic phosphorus uptake in the Sargasso Sea: Variability in Trichodesmium and the microbial community. Limnol Oceanogr. 2010;55(3):1390–1399. [Google Scholar]
- 12.Sohm JA, Capone DG. Phosphorus dynamics of the tropical and subtropical North Atlantic: Trichodesmium spp. versus bulk plankton. Mar Ecol Prog Ser. 2006;317:21–28. [Google Scholar]
- 13.Martiny AC, Huang Y, Li W. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol. 2009;11(6):1340–1347. doi: 10.1111/j.1462-2920.2009.01860.x. [DOI] [PubMed] [Google Scholar]
- 14.Martiny AC, Coleman ML, Chisholm SW. Phosphate acquisition genes in Prochlorococcus ecotypes: Evidence for genome-wide adaptation. Proc Natl Acad Sci USA. 2006;103(33):12552–12557. doi: 10.1073/pnas.0601301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mather RL, et al. Phosphorus cycling in the North and South Atlantic Ocean subtropical gyres. Nat Geosci. 2008;1(7):439–443. [Google Scholar]
- 16.Chisholm SW. In: Primary Productivity and Biogeochemical Cycles in the Sea. Falkowski PG, Woodhead AD, editors. Plenum; New York: 1992. pp. 213–237. [Google Scholar]
- 17.Martiny AC, Kathuria S, Berube PM. Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. Proc Natl Acad Sci USA. 2009;106(26):10787–10792. doi: 10.1073/pnas.0902532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scanlan DJ, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73(2):249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhee G-Y. A continuous culture study of phosphate uptake, growth rate and polyphosphate in Scenedesmus sp. J Phycol. 1973;9(4):495–506. [Google Scholar]
- 20.Goldman JC, Glibert PM. In: Nitrogen in the Marine Environment. Carpenter EJ, Capone DG, editors. Academic; New York: 1983. pp. 233–273. [Google Scholar]
- 21.Moore LR, Goericke R, Chisholm SW. Comparative physiology of Synechococcus and Prochlorococcus: Influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar Ecol Prog Ser. 1995;116(1):259–275. [Google Scholar]
- 22.Shalapyonok A, Olson RJ, Shalapyonok LS. Ultradian growth in Prochlorococcus spp. Appl Environ Microbiol. 1998;64(3):1066–1069. doi: 10.1128/aem.64.3.1066-1069.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ammerman JW, Hood RR, Case DA, Cotner JB. Phosphorus deficiency in the Atlantic: An emerging paradigm in oceanography. Eos Trans AGU. 2003;84(18):165–170. [Google Scholar]
- 24.Kane A, et al. Improving the parameters of a global ocean biogeochemical model via variational assimilation of in situ data at five time series stations. J Geophys Res. 2011;116:C06011. [Google Scholar]
- 25.Parekh P, Follows MJ, Boyle EA. Decoupling of iron and phosphate in the global ocean. Global Biogeochem Cycles. 2005;19:GB2020. [Google Scholar]
- 26.Polovina JJ, Howell EA, Abecassis M. Ocean’s least productive waters are expanding. Geophys Res Lett. 2008;35:L03618. [Google Scholar]
- 27.Flombaum P, et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA. 2013;110(24):9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casey JR, et al. Phytoplankton taxon-specific orthophosphate (Pi) and ATP utilization in the western subtropical North Atlantic. Aquat Microb Ecol. 2009;58(1):31–44. [Google Scholar]
- 29.Larsen A, Tanaka T, Zubkov MV, Thingstad TF. P-affinity measurements of specific osmotroph populations using cell-sorting flow cytometry. Limnol Oceanogr. 2008;6:355–363. [Google Scholar]
- 30.Tovar-Sanchez A, et al. A trace metal clean reagent to remove surface-bound iron from marine phytoplankton. Mar Chem. 2003;82(1):91–99. [Google Scholar]
- 31.Casey JR, Aucan JP, Goldberg SR, Lomas MW. Changes in partitioning of carbon amongst photosynthetic pico- and nano-plankton groups in the Sargasso Sea in response to changes in the North Atlantic Oscillation. Deep Sea Res Part II Top Stud Oceanogr. 2013;93:58–70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.