Significance
RNA-directed DNA methylation (RdDM) has traditionally been associated with transposable elements, but more recent studies have shown that RdDM also targets regulatory regions of protein-coding genes. Moreover, DNA methylation in Arabidopsis appears to be more dynamic than previously believed and may correlate with transcriptional responses to environmental stresses, such as pathogens. Therefore, the identification and characterization of RdDM mechanisms is fundamental to our understanding of how biological traits are controlled at an epigenetic level. We have performed a screen based on available coexpression data and identified two previously uncharacterized genes, FRG1 and -2, involved in RdDM. This work also shows the validity of coexpression data as an approach to assign new gene functions for RdDM.
Keywords: epigenetic, plant
Abstract
DNA methylation in Arabidopsis thaliana is maintained by at least four different enzymes: DNA METHYLTRANSFERASE1 (MET1), CHROMOMETHYLASE3 (CMT3), DOMAINS REARRANGED METHYLTRANSFERASE2 (DRM2), and CHROMOMETHYLASE2 (CMT2). However, DNA methylation is established exclusively by the enzyme DRM2, which acts in the RNA-directed DNA methylation (RdDM) pathway. Some RdDM components belong to gene families and have partially redundant functions, such as the endoribonucleases DICER-LIKE 2, 3, and 4, and INVOLVED IN DE NOVO2 (IDN2) interactors IDN2-LIKE 1 and 2. Traditional mutagenesis screens usually fail to detect genes if they are redundant, as the loss of one gene can be compensated by a related gene. In an effort to circumvent this issue, we used coexpression data to identify closely related genes that are coregulated with genes in the RdDM pathway. Here we report the discovery of two redundant proteins, SNF2-RING-HELICASE–LIKE1 and -2 (FRG1 and -2) that are putative chromatin modifiers belonging to the SNF2 family of helicase-like proteins. Analysis of genome-wide bisulfite sequencing shows that simultaneous mutations of FRG1 and -2 cause defects in methylation at specific RdDM targeted loci. We also show that FRG1 physically associates with Su(var)3-9–related SUVR2, a known RdDM component, in vivo. Combined, our results identify FRG1 and FRG2 as previously unidentified components of the RdDM machinery.
Cytosine methylation is an epigenetic mark present in many eukaryotes and is involved in silencing of transposable elements and other repetitive sequences that impose threats to genome integrity. Moreover, DNA methylation in regulatory regions suppresses the expression of genes and disturbances in methylation patterns can lead to developmental defects (1).
In the model plant Arabidopsis, DNA methylation occurs at CG, CHG, and CHH sequences (H = A, T, or C) and is maintained through DNA replication by different mechanisms, depending on the sequence context: symmetric CG and CHG methylation are maintained primarily by MET1 and CMT3, respectively, whereas the asymmetric CHH methylation is maintained by either CMT2 or DRM2 (2, 3). Initial DNA methylation establishment—or de novo methylation—in any sequence context is carried out by DRM2 (4, 5). DRM2 is guided to its target loci via a complex pathway known as RNA-directed DNA methylation (RdDM). RdDM depends on the production of both small interfering RNA (siRNA), and overlapping long-noncoding transcripts. According to current knowledge, chromatin-associated proteins, including SAWADEE HOMEODOMAIN HOMOLOG1 (SHH1)/DTF1 and CLASSY1 (CLSY1), recruit and assist RNA POLYMERASE IV (Pol IV), which produces transcripts that are converted to double-stranded RNA by RNA-DEPENDENT RNA POLYMERASE2 (RDR2) and subsequently cleaved into 24-nt siRNAs by DICER-LIKE3 (DCL3). ARGONAUTE4 (AGO4) binds siRNAs and is recruited to the RNA POLYMERASE V (Pol V) complex, as well as to long noncoding transcripts (lncRNA) produced by Pol V. AGO4 ultimately interacts with DRM2, which methylates target sequences with homology to the siRNAs (6, 7). Recruitment of Pol V is mediated by methyl-CG–binding noncatalytic Su(var)3-9 histone methyltransferase homologs SUVH2 and SUVH9, and a putative chromatin remodeling complex called the DRD1-DMS3-RDM1 complex (8, 9). In addition to SUVH2 and SUVH9, SUVR2 from the same family of SET-domain proteins was also identified as an RdDM factor by a systematic analysis of DNA methylation defects in mutants of Su(var)3-9 homologs (10). However, the precise molecular function of SUVR2 in the RdDM pathway is unknown. Factors that act downstream of Pol V recruitment include the double-stranded RNA binding protein complex IDN2–IDP (11–14), SWI3B—which interacts with IDN2 and is part of the SWI/SNF chromatin remodeling complex (15)—and RRP6-like1, which is involved in stabilizing Pol V and Pol IV transcripts (16).
The majority of known RdDM components have been identified via forward genetic screens. However, redundant members of gene families are usually not recovered from classic genetic screens because of the unlikelihood of simultaneous mutation of multiple redundant genes. To identify such genes, alternative approaches, such as mass spectrometric analysis following protein-affinity purification, chemical genetics, or RNAi-based screens have been used (14, 17, 18). High-throughput expression profiling and bioinformatic tools have enabled the interrogation andcomparison of gene-expression data under various developmental and environmental conditions (19, 20). Based on coexpression data of known RdDM components, we identified the paralogous genes SNF2-RING-HELICASE–LIKE1 and -2 (FRG1 and FRG2) as two new RdDM factors. The FRG genes encode proteins containing SNF2 domains typical of ATP-dependent motor proteins in chromatin remodeling complexes, separated by a RING domain typical of E3 ubiquitin ligases. Furthermore, we show that FRG1 physically interacts with the putative histone methyltransferase SUVR2. Together with the analysis of genome-wide methylation patterns, our results indicate that FRG1/2 and SUVR2 have overlapping functions for the efficient methylation of a broad range of RdDM sites.
Results
RdDM Genes Are Coexpressed.
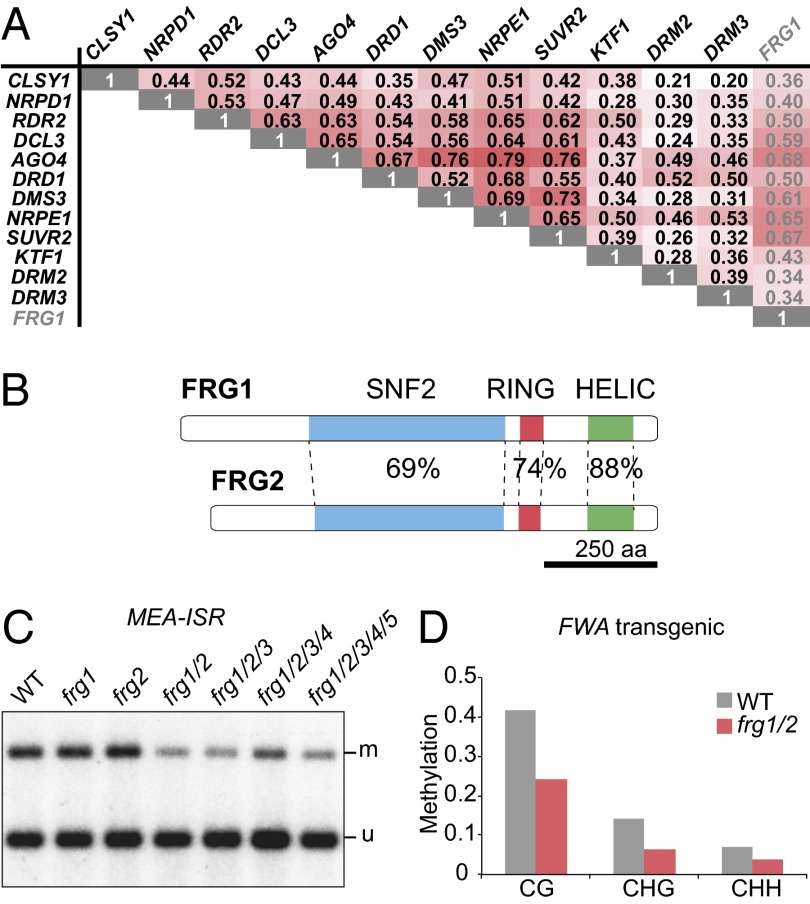
Genes within the same pathway are often coexpressed (21). To test if this is also the case for RdDM genes, we used the ATTED-II database to retrieve Pearson’s correlation coefficients (R) for pairwise comparisons of 12 genes for which expression data were available (20). The median R was 0.48, indicating that RdDM genes are in general highly coexpressed (Fig. 1A). Compared with a coexpression analysis of 100 Kyoto Encyclopedia of Genes and Genomes pathways, this value is close to the degree of coexpression observed within the proteasome pathway, which shows the highest levels of coexpression among all analyzed pathways (21). To assess which experimental conditions were causing the positive coexpression of RdDM genes, we used hierarchical clustering of the expression values for 26 different anatomical parts from ATH1 microarray databases. Common to the analyzed RdDM genes were high expression levels in the shoot apex, female flower parts, and the embryo (Fig. S1A). We searched for additional genes that were highly coexpressed with known RdDM components (R ≥ 0.55) and generated a candidate list of new RdDM components (Table S1). The top candidate was a gene encoding a protein with conserved SNF2 helicase-like domains separated by a RING domain, which we named FRG1 (Fig. 1 A and B). This candidate also fulfilled our second selection criterion, in that it has a closely related expressed paralog (22), FRG2, which is 66% identical at the protein sequence level (Fig. 1B and Fig. S1B). We thus reasoned that FRG1 and FRG2 might be functionally redundant, preventing their identification in previous genetic screens.
Fig. 1.
FRG1 is coexpressed with RdDM genes and redundantly functions with FRG2 in de novo methylation. (A) Pearson correlation coefficients for pairwise comparisons of 12 known RdDM genes and FRG1. (B) Domain architecture of FRG1 and -2. Percentages indicate identical amino acids in the corresponding domains. Scale bar indicates length of primary sequence. (C) DNA blot analysis of CHG methylation at the MEA-ISR locus. Here and subsequently, genomic DNA has been digested with the methylation sensitive enzyme MspI; the upper and lower bands correspond to methylated (m) and unmethylated (u) fractions, respectively. (D) Methylation levels of the FWA transgene in T1 from plants that have been transformed with unmethylated FWA.
FRG1 and FRG2 Are Redundantly Involved in the RdDM Pathway.
To test if FRG1 and FRG2 are indeed required for DNA methylation, we first measured the extent of non-CG methylation at the MEDEA-Intergenic Subtelomeric Repeat (MEA-ISR) locus in frg1 and frg2 single- and double-mutants, as well as higher-order mutants with other FRG paralogs, including FRG3, FRG4, and FRG5 (Fig. S1 B and C). This process was performed by digestion with a methylation-sensitive enzyme and subsequent DNA blot analysis, a well-established and sensitive assay for RdDM (23). Although neither frg1 nor frg2 mutants alone showed a defect, frg1 frg2 double-mutants showed reduced (but not completely eliminated) methylation of MEA-ISR (Fig. 1C and Fig. S2A). Additional mutations in FRG3, -4, and -5 did not lead to further reduction in MEA-ISR methylation, indicating that only FRG1 and FRG2 have redundant functions in RdDM and that the other family members are not involved in this pathway (Fig. 1C). According to the ATH1 microarray data, FRG1 and FRG2 are expressed at intermediate levels throughout plant development (Fig. S1D), but FRG2 expression does not significantly correlate with FRG1 and other RdDM genes.
FWA is another locus that is normally methylated in Arabidopsis. In vegetative tissue, hypomethylation of CG sites leads to ectopic expression and a dominant late-flowering phenotype (24). Unmethylated FWA copies that are introduced into wild-type plants by Agrobacterium-mediated transformation become efficiently methylated and silenced, whereas RdDM mutants fail to efficiently methylate and silence transgenic FWA (5). To test if the FRG proteins are involved in de novo methylation of FWA, we transformed frg1 frg2 double-mutants with an unmethylated FWA construct. We observed a reduction in CG, CHG, and CHH methylation compared with transformed wild-type plants (Fig. 1D). However, this reduction was moderate (Fig. 1D) and was associated with only a very small increase in flowering time, as measured by the number of leaves per plant at the onset of flowering (Fig. S2B). Thus, consistent with the only partial reduction of RdDM at MEA-ISR, the frg1 frg2 double-mutants show only a partial reduction of RdDM mediated FWA de novo methylation.
frg1 frg2 Double-Mutants Show a Partial Decrease in RdDM.
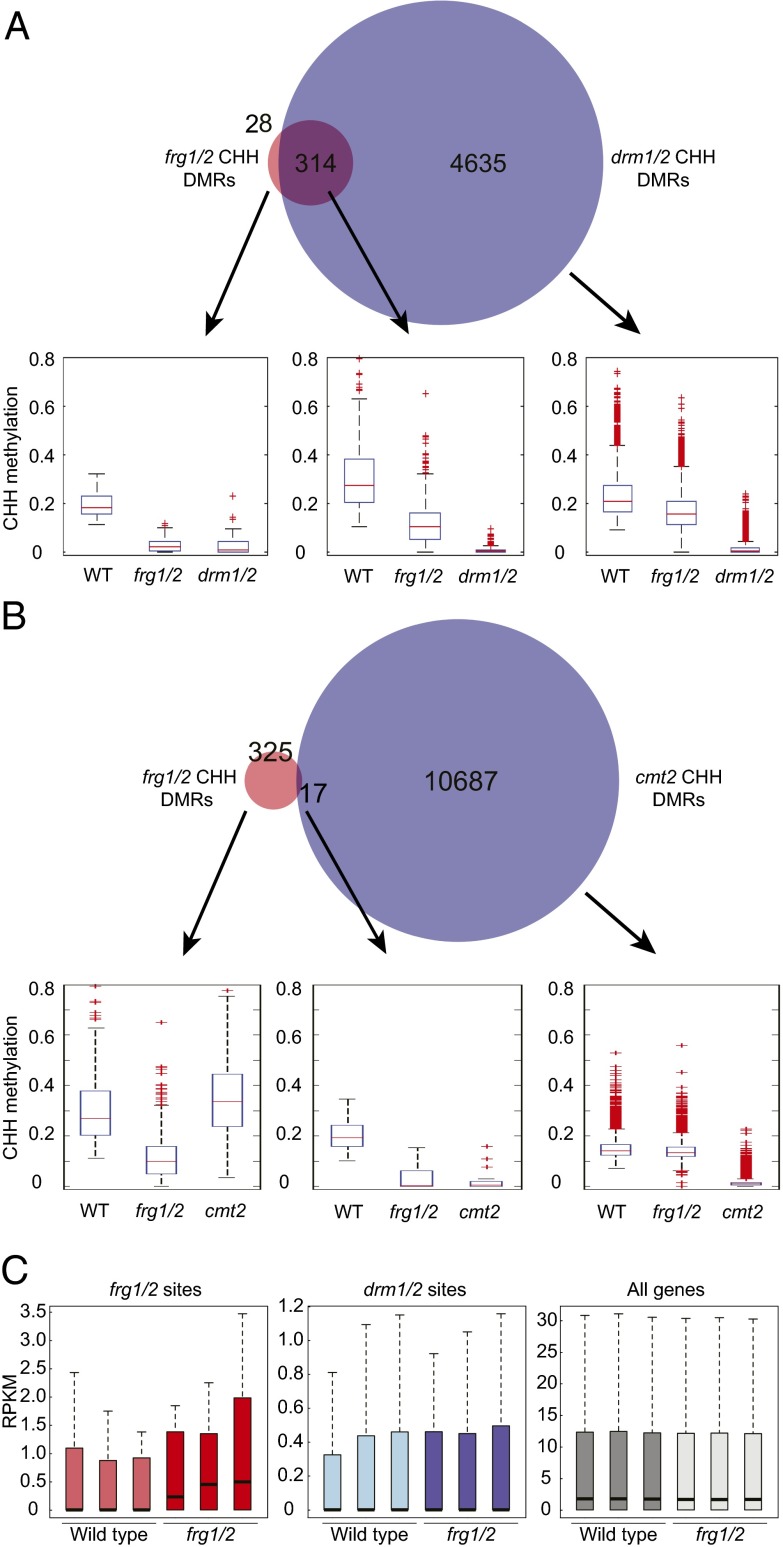
To further define the role of FRG1 and FRG2 in RdDM, we analyzed genome-wide DNA methylation patterns in rosette leaves of 3-wk-old plants at single base resolution by whole-genome bisulfite sequencing and defined differentially methylated regions (DMRs) with reduced DNA methylation levels in the frg1 frg2 double-mutant compared with wild-type (hypo-DMRs). We identified 342 frg1/2 hypo-DMRs in the CHH context and compared them to the 4,635 and 10,687 CHH hypo-DMRs previously identified in drm1 drm2 double-mutants (DRM1 encodes a lowly expressed paralog of DRM2) and cmt2 mutants, respectively (3, 10). Of frg1/2 hypo-DMRs, 93% overlapped with drm1/drm2 hypo-DMRs (Fig. 2A), whereas only 5% of frg1/2 hypo-DMRs overlapped with cmt2 hypo DMRs (Fig. 2B) in the CHH context, strongly indicating that DNA methylation defects in frg1 frg2 mostly occur at RdDM sites. The distribution of DNA methylation levels at drm1/2 or cmt2 hypo-DMRs showed that CHH methylation is moderately reduced in frg1 frg2 double-mutants over drm1/2 sites, but not over cmt2 sites, confirming that FRG1 and FRG2 are specifically involved in RdDM (Fig. 2 A and B and Fig. S2C). The relatively small number of frg1/2 DMRs compared with drm1/2 DMRs indicates that FRG1 and FRG2 are mostly required for DNA methylation at a subset of RdDM target sites. However, CHH methylation levels in frg1 frg2 double-mutants were slightly reduced at drm1/2 CHH hypo-DMRs that did not overlap with frg1/2 DMRs (Fig. 2A). This finding indicates that regions with weakly reduced DNA methylation in frg1 frg2 double-mutants were not identified by the stringent parameters for calling DMRs, and that the function of FRG1 and FRG2 is not restricted to the defined frg1/2 hypo-DMRs.
Fig. 2.
frg1/2 double mutants show reduced DNA methylation at RdDM sites. (A and B) Overlap between frg1/2 and drm1/2 (A) or cmt2 (B) CHH hypo-DMRs. Box plots show CHH methylation levels in the respective DMR fractions. For all box plots shown here and subsequently: horizontal line, median; edges of boxes, 25th (Bottom) and 75th (Top) percentiles; error bars, minimum and maximum points within 1.5× interquartile range; red dots, outliers. (C) mRNA abundance in reads per kilobase per million reads at frg1/2 CHH hypo DMRs (Left), drm1/2 CHH hypo DMRs (Center), and all genes (Right) in wild-type and frg1 frg2 double-mutants. Three biological replicates per genotype are shown.
To test if loss of DNA methylation in frg1 frg2 double-mutants is associated with changes in gene expression, we analyzed genome-wide mRNA transcript levels by RNA-seq. Overall, gene-expression levels did not differ strongly in wild-type and frg1 frg2 double-mutants, indicating that FRG1 and FRG2 are not generally involved in transcription (Fig. 2C, Right). Analysis of differential expression in frg1 frg2 double-mutants compared with wild-type showed 55 genes that were more than twofold up- and 136 genes that were more than twofold down-regulated. Many of these up-regulated genes were annotated as transposable elements (n = 8) or unknown protein/pseudogene/other RNA (n = 19) (Dataset S1). In agreement with the absence of major transcriptional changes, frg1 frg2 double-mutants did not display morphological or developmental differences compared with wild-type plants. Transcript levels at RdDM sites were low in wild-type, as well as frg1 frg2 double-mutants (Fig. 2C, Center). However, a slight overall increase over frg1/2 DMR sites in frg1 frg2 double-mutants was observed and is indicative of derepression at some sites that have lost DNA methylation in frg1 frg2 double-mutants (Fig. 2C, Left). Together with the DNA methylation defect, these results show that FRG1 and FRG2 are required for efficient transcriptional silencing at a subset of RdDM sites.
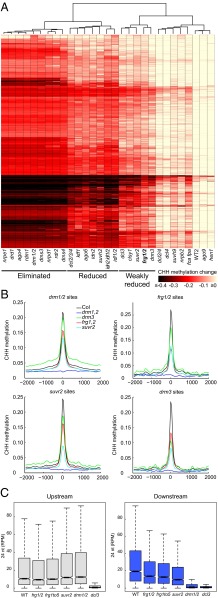
Previous analysis of CHH methylation patterns revealed that different classes of RdDM mutants show different levels of methylation defects and, accordingly, the different mutants were categorized as “eliminated,” “reduced,” and “weakly reduced” (10). Hierarchical clustering of CHH methylation patterns at DRM1/DRM2-dependent sites placed the frg1 frg2 double-mutant into the “weakly reduced” class, clustering together with the RdDM mutants drm3, suvr2, dcl3, and clsy1 (Fig. 3A). As in the other weakly reduced mutants, CHH methylation in the frg1 frg2 double-mutant was reduced over a broad range of DRM1/DRM2-dependent sites, and some sites were more affected than others. Consistently, frg1 frg2, suvr2, and drm3 mutants showed slight to moderate reductions in average CHH methylation over the CHH hypo-DMRs of each of these mutants, as well as the drm1 drm2 double-mutant (Fig. 3B). We sought to further test whether the frg1 frg2 double-mutant affects methylation at a unique subset of loci, or whether it was typical of other weak RdDM mutants, by comparing the CHH hypo-DMRs of frg1 frg2 double-mutants with those of previously characterized weak mutants (10). The overlap between DMRs ranged from 30% of frg1/2 CHH hypo-DMRs with drm3 to 87% with ktf1, but in general the overlap appeared to be larger with stronger mutants (i.e., mutants with more DMRs) (Fig. S3). Moreover, in all pairwise comparisons, frg1/2 methylation levels were slightly reduced in the DMRs fraction that did not overlap with frg1/2 DMRs and, conversely, the compared mutants showed slightly decreased methylation in frg1/2-specific DMRs (Fig. S3). Therefore, the weak RdDM mutants, including frg1/2, have broadly overlapping genomic regions with weakly reduced methylation levels, but also some specific loci that show stronger methylation defects in the different genotypes.
Fig. 3.
frg1/2 double-mutants belong to the “weakly reduced” class of RdDM mutants and are defective in the downstream RdDM pathway. (A) Heatmap of CHH methylation changes within drm1/2 hypo-DMRs clustered by absolute change in CHH methylation levels (rows) and genotype (columns). The color scale is capped for values ≤−0.4 and ≥0. (B) Average distribution of CHH methylation levels over hypo-DMRs of drm1/2, frg1/2, suvr2, or drm3 mutants. The x axis indicates distance from the DMR midpoints in base pairs. (C) Abundance of 24-nt siRNAs in reads per million at “upstream” and “downstream” siRNA clusters, referring to “pol-iv only” and “shh1/drm2/pol-v” clusters, respectively, as previously defined (25).
FRG1 and FRG2 Act in the Downstream RdDM Pathway.
Previously, genome-wide clusters of 24-nt siRNAs were defined that depended only on Pol IV (upstream), or additionally required SHH1, Pol V, and DRM2 (downstream) (25). To further investigate the role of FRG1 and FRG2 in the RdDM pathway, we performed small RNA-seq to profile frg1 frg2 double-, frg quintuple-, as well as suvr2 mutants. We also included the upstream mutant dcl3, as well as downstream drm1 drm2 double-mutants as controls. As expected, 24-nt siRNAs were lost in dcl3 at both upstream and downstream clusters, whereas drm1/2 only affected downstream clusters (Fig. 3C). The abundance of 24-nt siRNAs in frg1 frg2 double-, frg quintuple-, and suvr2 mutants compared with wild-type was not reduced at upstream clusters and was moderately reduced at downstream clusters, with suvr2 showing a slightly stronger effect (Fig. 3C). These data confirm previous analysis of siRNAs in suvr2 by Northern blots (10), and suggest that FRG1 and FRG2, as well as SUVR2, act in the downstream portion of the RdDM pathway.
FRG1 Physically Interacts with SUVR2.
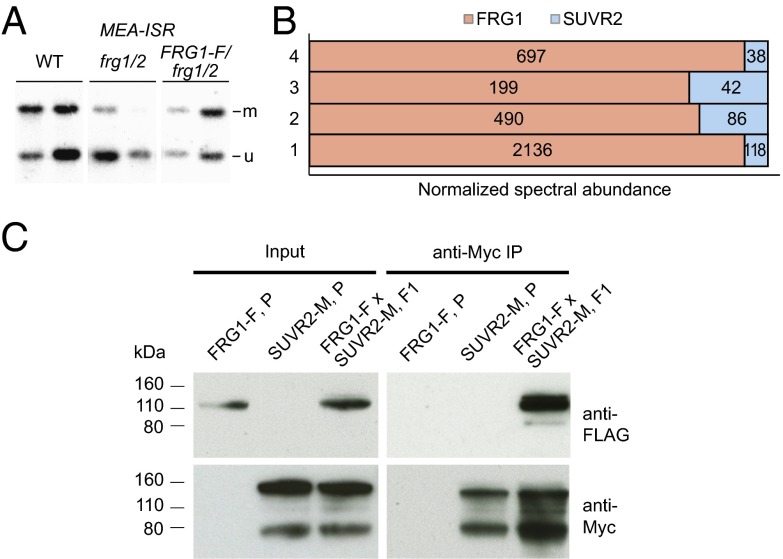
To gain insight into the molecular function of FRG1, we sought to identify interacting proteins using immunoprecipitation (IP) experiments. We generated a construct containing the FRG1 gene driven by its endogenous promoter with a 3′ epitope tag encoding three FLAG peptides followed by a biotin ligase recognition peptide (BLRP). The construct was introduced into frg1 frg2 double-mutants and complementation of the DNA methylation defect by the tagged fusion protein was confirmed by MEA-ISR Southern blots (Fig. 4A). Subsequently, we performed large-scale IP experiments followed by mass spectrometry to identify proteins that copurify with FRG1. The only protein that was enriched in all four independent IP experiments in samples from the tagged FRG1 line compared with the untagged wild-type control was SUVR2 (Table S2). The normalized spectral abundance of SUVR2 peptides was 5–20% compared with FRG1, indicating that either the interaction was weak/transient or only a fraction of the Flag-tagged FRG1 interacted with SUVR2 (Fig. 4B). To confirm the FRG1–SUVR2 interaction, we crossed the FLAG-tagged FRG1 line with a line that expresses a complementing SUVR2 protein fused to a 9xMyc tag at the C terminus (Fig. S4) and demonstrated their interaction in F1 plants by co-IP (Fig. 4C). In conclusion, FRG1 physically interacts with a known component of the RdDM pathway, SUVR2.
Fig. 4.
In vivo interaction of FRG1 and SUVR2. (A) DNA blot showing that FLAG-tagged FRG1 complements the DNA methylation defect of frg1/2 at the MEA-ISR locus. Two biological replicates per genotype are shown. (B) Abundance of FRG1 and SUVR2 peptides in mass spectrometric analyses of four independent FRG1-FLAG IP experiments. The combined relative abundances of FRG1 and SUVR2 peptides are set to 100% on the x axis. (C) Co-IP assay for interaction of FRG1-FLAG with Myc-SUVR2. Input shows the expression of the tagged proteins in parental lines (P) and in the offspring from the crossed parental lines (F1). Myc-SUVR2 was immunopurified (IP) with anti-Myc antibody and the presence of copurified FRG1-FLAG was analyzed by Western blot using anti-FLAG antibody.
Discussion
Using an RNA coexpression approach, this study identifies FRG1 and FRG2 as redundant genes required for efficient RdDM. The observation that frg1 frg2 double-mutants show only a partial reduction of RdDM, and that this phenotype is not enhanced in higher-order mutant combinations with other FRG paralogs, suggests that FRG1 and FRG2 are acting at a step in the RdDM pathway that is needed for efficient methylation, but is not completely required for functioning of the RdDM pathway at the majority of RdDM target sites. However, a subset of RdDM sites seems particularly prone to loss of methylation in frg1 frg2 double, as well as other weakly reduced mutants. Although the precise molecular function of FRG1 and FRG2 is unknown, it is tempting to speculate that they may act as regulators of efficient RdDM at particular RdDM sites, and may therefore be involved in dynamic control of methylation during stress or through development.
Similar to two other components of the RdDM pathway, CLSY1 and DRD1 (defective in RNA-directed DNA methylation 1), FRG1 and FRG2 belong to a superfamily of helicase-like SWI2/SNF2 ATP-dependent chromatin remodeler-related proteins (26–28). CLSY1 and DRD1 interact with Pol IV and Pol V, respectively, and might be required to provide a chromatin environment favorable for recruitment or transcriptional activity of these heterochromatic RNA polymerases. Recently, SWI3B, a component of the ATP-dependent chromatin remodeling complex SWI/SNF, was described as an IDN2 interactor that is guided by lncRNAs to position nucleosomes at RdDM target sites (15). The 24-nt siRNA profile of frg1 frg2 double-mutants did not show any changes at siRNA clusters that depend on Pol IV but not on genes downstream of siRNA production, which argues against an involvement of FRG1 and FRG2 in Pol IV transcription. Therefore, it is more likely, that FRG1 and FRG2 might function at or downstream of lncRNA production, or might function in the DRM2 targeting step of the pathway.
Phylogenetic analyses of the SNF2 superfamily in Arabidopsis place the five FRG paralogs in the family of yeast Rad5- and Rad16-related genes (22). Within this family, they show the closest homology to yeast DIS1/RIS1/ULS1, a RING-type E3 ligase that mediates ubiquitylation of small ubiquitin-like modifier-conjugated substrates (29, 30) (Fig. S1). Yeast DIS1 was initially identified in a screen for genes that interfere with the silencing of the cryptic mating-type loci and it was proposed that it facilitates the accessibility of heterochromatin during mating-type switching (31). The presence of both a RING and a SNF2 helicase domain suggests that FRG1 and FRG2 likely have dual functions: (i) ATP-driven alteration of the chromatin environment at RdDM sites and (ii) regulation of RdDM via ubiquitylation or ubiquitin-like posttranslational modification. These possibilities provide the basis for future functional characterization of FRG1 and FRG2 and an anticipation of further insights into the dynamic regulation of RdDM. A connection between ubiquitin and RdDM is provided by the requirement of histone H2B de-ubiquitylation for CHH methylation by the ubiquitin protease UBP26 (32). H2B monoubiquitylation promotes H3K4 methylation (33) and is regarded as an active histone mark that counteracts repressive H3K9 methylation. Therefore, hypothetical H2B ubiquitylation by FRG1/2 for the purpose of methylating DNA seems counterintuitive, but detailed analysis of the involved histone variants and dynamics might reveal new insights into the role of ubiquitin or ubiquitin-like modification in this context.
FRG1 was found to physically interact with another known component of the RdDM pathway, SUVR2. In addition, like frg1 frg2 double-mutants, suvr2 mutants only partially compromise the RdDM pathway. SUVR2 encodes a Su(var)3-9 homolog, although it has been previously reported that recombinant SUVR2 does not methylate histones in vitro (34). SUVR2 may thus require interactions with other proteins or posttranslational modifications for its histone methylation activity, or may act on nonhistone substrates. Intriguingly, SUVR2 has been very recently shown to bind noncovalently to ubiquitin through its WIYLD domain (35), although the natural ubiquitylated substrate for SUVR2 binding is unknown. Because FRG1 and FRG2 encode putative ubiquitin E3 ligases, it therefore seems possible that SUVR2 might bind to substrates that are ubiquitylated by FRG1 and FRG2. Future identification of FRG and SUVR2 substrates should shed light on these hypotheses, and aid in the further molecular characterization of the RdDM pathway.
Materials and Methods
Plant Materials.
All plants used in this study were in the Columbia-0 ecotype and grown under long-day conditions. We used the following mutant lines: frg1-1 (SALK_027637), frg1-2 (SALK_063135), frg2-1 (SALK_057016), frg3-1 (SALK_071872), frg4-1 (SAIL_735_G06), frg5-1 (SALK_022785), and suvr2-1 (SAIL_832_E07). Unless stated differently, frg1 indicates the frg1-1 allele.
RdDM Coexpression List.
We used 16 known RdDM genes individually as input (genes A). Then we selected for the genes showing coexpression (based in Pearson coefficient) ≥ 0.55 (gene B). As a second step, we ranked those genes and sorted them according to two different parameters. The first parameter was occurrence, or how often gene B appears coexpressed with genes A; the second parameter was the magnitude of the Pearson correlation coefficient for one particular gene A to another gene B. Finally, we chose genes that are members of a gene family, and selected those genes for which phylogenetic analyses showed very close homologous gene pairs.
Phylogenetic Analysis.
We retrieved protein sequences from the FRG1 family from The Arabidopsis Information Resource (TAIR). The alignment of protein sequences was done with CLUSTALW2 (www.ebi.ac.uk/Tools/msa/clustalw2/). The CLUSTALW2 tree data output was then loaded into Phylodendron tree printer (iubio.bio.indiana.edu/treeapp/treeprint-form.html).
Generation of Gateway Entry and Destination Clones.
We amplified genomic fragments containing the promoter and genomic sequences of FRG1 from the BAC MZE19 using the primers FRG1C-tagging Forward (5′-CACCTCGTTCGTTGTCTGATTTGTTG-3′) and FRG1C-tagging Reverse (5′-GACCATAAACAGATACTTGAGATCG-3′). We cloned the corresponding PCR product into pENTR/d-TOPO vector (Invitrogen). We then introduced C-terminal 3xFLAG-BLRP tag into an AscI site in the pENTR/d-TOPO vector and then digested the resulting pENTR/d with the restriction enzyme MluI. Subsequently, we recombined the restriction fragment into a modified Gateway destination vector based on the pEarleyGate vectors described in ref. 36.
DNA Gel Blotting.
MEA-ISR PCR products for probing were generated with primers MEA-ISR Forward (5′-AAACCTTTCGTAAGCTACAGCCACTTTGTT-3′) and MEA-ISR Reverse 5′-TCGGATTGGTTCTTCCTACCTCTTTACCTT-3′).
Bisulfite Sequencing and Analysis.
Bisulfite sequencing and analysis were conducted as previously described (37).
Affinity Purification.
Approximately 10 g of flower tissue from transgenic FRG1-Flag-BLRP, or Columbia (as a negative control) were ground in liquid nitrogen, and resuspended in 50 mL of IP buffer [50 mM Tris pH7.6, 150 mM NaCl, 5 mM MgCl2, 5% (vol/vol) glycerol, 0.1% Nonidet P-40, 2.8 mM β-mercaptoethanol, 1 μg/mL pepstatin, 1 mM PMSF, and 1× protease inhibitor mixture tablet (Roche, 14696200)]. The cleared lysate was incubated at 4 °C for 2 h with 200 µL of anti-FLAG M2 magnetic beads (Sigma). The immunoprecipitate was washed once with 40 mL and three times with 1 mL of IP buffer, followed by two washes with 1 mL IP buffer lacking detergent. Proteins were released from the beads by two consecutive elutions in 400 µL 3xFlag peptide (150 ng/µL) for 20 min at room temperature.
Mass Spectrometric Analysis.
Mass spectrometric analyses were conducted as described in ref. 9.
Co-IP Assays.
IP was performed with c-Myc 9E10 agarose (50% slurry; Santa Cruz Biotechnology). Western blotting was performed with anti-FLAG M2 antibody-HRP conjugate (Sigma; A8592) and c-Myc 9E10 mouse monoclonal antibody (Santa Cruz Biotechnology; sc-40).
Whole Genome Bisulfite Sequencing.
Genomic DNA was extracted from 1 g of 3-wk-old plant aerial tissue using DNeasy Plant Mini Kit (Qiagen). Libraries for bisulfite sequencing were generated and sequenced as described in ref. 10. Read statistics are listed in Table S3. Data for mutants other than the frg1/2 were obtained from GSE39901. Data were analyzed exactly as previously described (10). Briefly, identical reads were removed and unique reads were mapped to the Arabidopsis genome (TAIR10) using the BSseeker (38). Methylation levels were calculated as #C/(#C+#T). DMRs were identified by comparing frg1/2 to three published wild-type replicates (10). Methylated cytosines and unmethylated cytosines were compared in 100-bp bins using Fisher’s Exact Test. DMRs were called by using cut-offs of Benjamini–Hochberg-corrected false-discovery rate < 0.01 and absolute methylation differences of 10% for CHH, and selecting bins with at least four cytosines covered by at least four reads in the wild-type replicates. Finally, DMRs within 200 bp were merged.
mRNA Sequencing.
Total RNA was isolated with TRIzol (Ambion) from 0.1 g of 14-d-old seedlings grown at continuous light and 21 °C on Murashige and Skoog medium. Individual libraries from three biological replicates each were generated according to the manufacturer’s instructions (Illumina). All libraries were sequenced with the HiSEq. 2000 platform according to the manufacturer’s instructions (Illumina) at 50-bp length. Reads were mapped to the TAIR10 genome with TopHat (39) using defaults settings, except that only uniquely mapping reads and up to one mismatch were allowed and intron length was set to 40–1,000. Read statistics are listed in Table S3. Reads per kilobase of transcript per million reads mapped values were calculated from read overlaps with gene exons, frg1/2 DMRs and previously published drm1/2 DMRs (10), using the trimmed mean of M-values normalization method (40) and only including DMRs that contained at least one read in at least one sample. Differential gene expression was analyzed with DESeq. (41).
smRNA Sequencing.
Total RNA was isolated with TRIzol (Ambion) from 0.1-g flowers. siRNAs were purified as described in ref. 3. Individual TruSeq small RNA libraries from three biological replicates each were generated according to the manufacturer’s instructions (Illumina), except that 13 cycles of PCR amplification were used. All libraries were sequenced with the HiSeq. 2000 platform according to the manufacturer’s instructions (Illumina) at 50-bp length. Read statistics are listed in Table S3. First adapter sequences were removed, and then reads were mapped to the TAIR10 genome using Bowtie (38) by allowing no mismatches and only keeping reads that uniquely map to the genome. Small RNA counts were normalized to the size of each small RNA library by dividing to the number of reads to the number of total uniquely mapping reads of 18–34 bp in size. For the analysis of normalized reads in “upstream” and “downstream” clusters, the “pol-iv only” and “shh1/drm2/pol-v” clusters from ref. 25 were used, respectively.
Accession Numbers.
All sequencing data are available at National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible via GEO Series accession no. GSE62801.
Supplementary Material
Acknowledgments
We thank Mahnaz Akhavan for her support in high-throughput sequencing at the University of California, Los Angeles, Broad Stem Cell Research Center BioSequencing Core Facility; and Julia Chambers for technical assistance. Work in the S.E.J. laboratory was supported by NIH Grant GM60398; work in the J.A.W. laboratory was supported by NIH Grant GM089778; M.G was supported by a European Molecular Biology Organization ALTF 986-2011 Long-Term Fellowship; H.S. is a Howard Hughes Medical Institute Fellow of the Damon Runyon Cancer Research Foundation (DRG-2194-14); S.F. was a Special Fellow of the Leukemia & Lymphoma Society; M.V.C.G. was supported by US Public Health Service National Research Service Award GM07104 and a University of California, Los Angeles, Dissertation Year Fellowship; and S.E.J. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Database deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE62801).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420515111/-/DCSupplemental.
References
- 1.Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293(5532):1070–1074. doi: 10.1126/science.293.5532.1070. [DOI] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21(1):64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao X, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13(24):2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 5.Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12(13):1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 6.Matzke MA, Mosher RA. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15(6):394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 7.Zhong X, et al. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell. 2014;157(5):1050–1060. doi: 10.1016/j.cell.2014.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson LM, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507(7490):124–128. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Law JA, et al. A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr Biol. 2010;20(10):951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152(1-2):352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang CJ, et al. IDN2 and its paralogs form a complex required for RNA-directed DNA methylation. PLoS Genet. 2012;8(5):e1002693. doi: 10.1371/journal.pgen.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie M, Ren G, Costa-Nunes P, Pontes O, Yu B. A subgroup of SGS3-like proteins act redundantly in RNA-directed DNA methylation. Nucleic Acids Res. 2012;40(10):4422–4431. doi: 10.1093/nar/gks034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ausin I, Mockler TC, Chory J, Jacobsen SE. IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat Struct Mol Biol. 2009;16(12):1325–1327. doi: 10.1038/nsmb.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ausin I, et al. INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(22):8374–8381. doi: 10.1073/pnas.1206638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Y, Rowley MJ, Böhmdorfer G, Wierzbicki AT. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol Cell. 2013;49(2):298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, et al. An Rrp6-like protein positively regulates noncoding RNA levels and DNA methylation in Arabidopsis. Mol Cell. 2014;54(3):418–430. doi: 10.1016/j.molcel.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gille S, Hänsel U, Ziemann M, Pauly M. Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc Natl Acad Sci USA. 2009;106(34):14699–14704. doi: 10.1073/pnas.0905434106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alonso JM, Ecker JR. Moving forward in reverse: Genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet. 2006;7(7):524–536. doi: 10.1038/nrg1893. [DOI] [PubMed] [Google Scholar]
- 19.Manfield IW, et al. Arabidopsis Co-expression Tool (ACT): Web server tools for microarray-based gene expression analysis. Nucleic Acids Res. 2006;34(web server issue):W504–W509. doi: 10.1093/nar/gkl204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obayashi T, et al. ATTED-II: A database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35(database issue):D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams EJ, Bowles DJ. Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 2004;14(6):1060–1067. doi: 10.1101/gr.2131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaked H, Avivi-Ragolsky N, Levy AA. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics. 2006;173(2):985–994. doi: 10.1534/genetics.105.051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao X, Jacobsen SE. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA. 2002;99(suppl 4):16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soppe WJ, et al. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol Cell. 2000;6(4):791–802. doi: 10.1016/s1097-2765(05)00090-0. [DOI] [PubMed] [Google Scholar]
- 25.Law JA, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498(7454):385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith LM, et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19(5):1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 2011;7(7):e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanno T, et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14(9):801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 29.Knizewski L, Ginalski K, Jerzmanowski A. Snf2 proteins in plants: Gene silencing and beyond. Trends Plant Sci. 2008;13(10):557–565. doi: 10.1016/j.tplants.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Uzunova K, et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem. 2007;282(47):34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Buchman AR. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol Cell Biol. 1997;17(9):5461–5472. doi: 10.1128/mcb.17.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sridhar VV, et al. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447(7145):735–738. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 33.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418(6893):104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 34.Thorstensen T, et al. The Arabidopsis SUVR4 protein is a nucleolar histone methyltransferase with preference for monomethylated H3K9. Nucleic Acids Res. 2006;34(19):5461–5470. doi: 10.1093/nar/gkl687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahman MA, et al. The arabidopsis histone methyltransferase SUVR4 binds ubiquitin via a domain with a four-helix bundle structure. Biochemistry. 2014;53(13):2091–2100. doi: 10.1021/bi401436h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45(4):616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 37.Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006;4(11):e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.