Significance
Misfolded proteins are a hallmark of diverse human cardiac disorders including desmin-related cardiomyopathy and systemic amyloidosis. Defects in translational fidelity can cause neurodegeneration, however the consequences of mistranslation in other tissues, including the heart, are unknown. The fidelity of protein synthesis is largely ensured by aminoacyl-tRNA synthetases, and many tRNA synthetases contain editing domains that hydrolyze mischarged tRNAs, preventing incorporation of incorrect amino acids into proteins. Here, we show that diminished editing efficacy of the alanyl-tRNA synthetase causes misfolded protein aggregation and cell death in the mammalian heart. These results illuminate the importance of translational fidelity in cardiac homeostasis and suggest that genetic factors that disrupt the accuracy of translation may contribute to proteinopathies of the heart and other tissues.
Keywords: translational fidelity, mistranslation, protein misfolding, protein quality control, cardiomyopathy
Abstract
Misfolded proteins are an emerging hallmark of cardiac diseases. Although some misfolded proteins, such as desmin, are associated with mutations in the genes encoding these disease-associated proteins, little is known regarding more general mechanisms that contribute to the generation of misfolded proteins in the heart. Reduced translational fidelity, caused by a hypomorphic mutation in the editing domain of alanyl-tRNA synthetase (AlaRS), resulted in accumulation of misfolded proteins in specific mouse neurons. By further genetic modulation of the editing activity of AlaRS, we generated mouse models with broader phenotypes, the severity of which was directly related to the degree of compromised editing. Severe disruption of the editing activity of AlaRS caused embryonic lethality, whereas an intermediate reduction in AlaRS editing efficacy resulted in ubiquitinated protein aggregates and mitochondrial defects in cardiomyocytes that were accompanied by progressive cardiac fibrosis and dysfunction. In addition, autophagic vacuoles accumulated in mutant cardiomyocytes, suggesting that autophagy is insufficient to eliminate misfolded proteins. These findings demonstrate that the pathological consequences of diminished tRNA synthetase editing activity, and thus translational infidelity, are dependent on the cell type and the extent of editing disruption, and provide a previously unidentified mechanism underlying cardiac proteinopathy.
Proteins are the building blocks and major signaling molecules of cells, and as such, their synthesis and degradation are tightly regulated. Disruption of protein homeostasis (proteostasis) can result in the accumulation of abnormal proteins that lead to cellular pathogenesis. Like the neuroproteopathies, misfolding of specific proteins in cardiomyocytes can be caused by genetic mutations that alter the primary structure of aggregated proteins. For example, mutations in the desmin gene, which encodes a muscle-specific intermediate filament protein, or in the gene encoding αB-crystallin, a molecular chaperone for desmin, result in aggregation of these proteins and are a primary cause of hereditary cardiomyopathy (1, 2). Systemic amyloidosis, the extracellular accumulation of abnormal proteinaceous fibrils, can also occur in the heart with detrimental effects on cardiac function (3, 4). In addition, misfolded proteins and changes in the ubiquitin proteasome system or autophagy have been widely reported in failing hearts (4, 5), demonstrating the importance of protein quality control systems for cardiomyocyte homeostasis. However, whether such proteostatic changes induce pathological changes, or are consequences of the diseased state, is not clear.
In addition to genetic mutations, defective proteins can be generated by inaccurate translation. Translational fidelity is largely controlled by the precise aminoacylation of tRNAs with their cognate amino acids, a function carried out by the aminoacyl-tRNA synthetases (aaRSs). To improve substrate specificity, an editing site, which hydrolyses misactivated noncognate amino acids or mischarged tRNAs and is distinct from the aminoacylation domain, is found in approximately half of the aaRSs (6, 7). Failure of proofreading by these aaRSs may result in incorporation of the wrong amino acid into the nascent peptide, which could be detrimental to protein folding and function.
Although errors in aminoacylation are relatively low (8, 9), even small decreases in aaRS proofreading have dramatic effects on cell survival (10–12). Bacterial alanyl-tRNA synthetase (AlaRS) can misactivate glycine or serine, and Escherichia coli expressing a severe editing-deficient form (C666A) of AlaRS had increased death when grown in elevated concentrations of these amino acids (11). In mice, a point mutation (A734E) in the AlaRS editing domain in the mutant strain “sticky” caused a twofold increase in Ser-mischarged tRNAAla that resulted in formation of ubiquitinated protein aggregates in cerebellar Purkinje cells and degeneration of these neurons (10). However, because cell loss in other regions of the brain or in other tissues was not observed in the sti mutant mouse, the importance of AlaRS editing activity in other mammalian cell types remains unknown.
Here we hypothesized that further decreases in AlaRS editing function could lead to misfolded protein accumulation in additional cell types. Indeed mouse embryos homozygous for a point mutation at C723 (which corresponds to the C666 amino acid in the editing domain of bacterial AlaRS) died by midgestation. Furthermore, decreasing the amount of the sti-associated AlaRSA734E enzyme or placing the sti mutation and the more severe AlaRS (AarsC723A) mutation in trans caused extensive aggregation of misfolded proteins in mouse cardiomyocytes, eventually leading to cardiomyopathy. Using this proteome-wide mistranslation system, we demonstrate that different cell types are distinctly sensitive to errors in protein synthesis and that mistranslation can induce cardiac proteinopathy, which leads to cardiac fibrosis and decrements in heart function.
Results
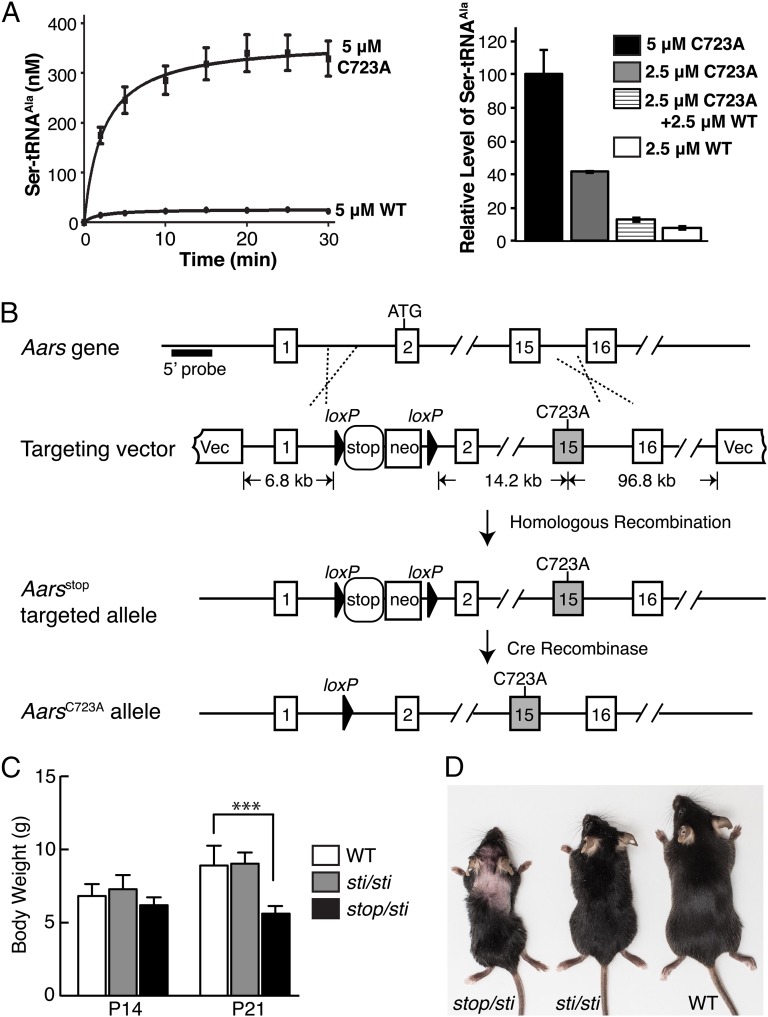
Previous results determined that Cys666 in the editing domain of bacterial AlaRS was essential for AlaRS editing (11). In agreement, mutation of Cys723 (corresponding to bacterial Cys666) to Ala in mammalian AlaRS resulted in a dramatic increase in production of mischarged Ser-tRNAAla (Fig. 1A, Left). In contrast to the twofold increase in mischarged Ser-tRNAAla generated by AlaRSA734E, the mutant enzyme found in sti mutant mice (10), in vitro misacylation assays demonstrated that the C723A enzyme produced Ser-tRNAAla at initial rates at least 15-fold higher than that produced by the wild-type enzyme.
Fig. 1.
Generation of editing-defective AlaRS mouse models. (A, Left) Misacylation of tRNAAla with serine by mouse wild-type (WT) or C723A AlaRS. (Right) Relative level of mischarged Ser-tRNAAla in the presence of WT, C723A, or equal molar ratio of WT and C723A AlaRS at 15 min. Assays were run in duplicate, and points (mean ± SD) were averaged from two experiments. (B) To generate the conditional knock-in allele of AlaRSC723A, a loxP-flanked transcriptional stop cassette was inserted in intron 1, and the TGT codon encoding cysteine at position 723 in exon 15 was replaced with a GCG alanine codon. Aars transcription remains off until the stop cassette is deleted via Cre expression. The Southern blot probe (5′ probe), the size of the left and right “arm” of the targeting vector, and the distance between the neo cassette and exon 15 are indicated. (C) Body weight of WT (n = 13), Aarssti/sti (sti/sti; n = 8), and Aarsstop/sti (stop/sti; n = 11) mice at postnatal ages (P) 14 and P21. Values represent mean ± SD; ***P < 0.0001 (one-way ANOVA). (D) Photograph of 2-mo-old WT, Aarssti/sti, and Aarsstop/sti mice. Note the dorsal alopecia characteristic of the Aarsstop/sti mouse.
To determine the in vivo effects of this dramatic loss in AlaRS editing activity, the C723A mutation was introduced into the mouse Aars gene via homologous recombination (Fig. 1B). To achieve conditional expression of the knock-in allele, a loxP-flanked transcriptional stop cassette (13, 14) was also inserted in intron 1 of this gene. Constitutive expression of AarsC723A was induced by crossing Aarsstop/+ mice to the EIIa-Cre strain to remove the stop cassette (15). The resulting AarsC723A/+ offspring were viable and indistinguishable from wild-type littermates. We hypothesized that wild-type AlaRS in the AarsC723/+ mouse could edit in trans the Ser-tRNAAla produced by C723A AlaRS. An in vitro mixing of C723A AlaRS with wild-type AlaRS confirmed this expectation (Fig. 1A, Right).
Still, careful examination of brains from aged AarsC723A/+ mice revealed mild loss of cerebellar Purkinje cells (Fig. S1). This minor neuronal loss could result from the ability of eukaryotic translation elongation factor 1 alpha to capture some of the mischarged tRNA and prevent its clearance by wild-type AlaRS (10). In comparison, Purkinje cell loss is not observed in aged Aarssti/+ mice, and Aarssti/sti mice have early onset and severe Purkinje cell degeneration (10). Moreover, no abnormalities were observed in brain regions other than the cerebellum or in other organs (SI Materials and Methods) from 10- to 18-mo-old AarsC723A/+ mice (Fig. S1). Together these observations suggest that due to the further reduction in AlaRS editing activity, the effects of the AarsC723A mutation are more detrimental than the Aarssti mutation in vivo. Consistent with this notion, no AarsC723A/C723A offspring were obtained from AarsC723A/+ intercrosses, demonstrating that homozygous expression of this mutation was lethal (Table S1). In addition, no viable E8.5/E9.5 AarsC723A/C723A embryos were observed, and 26% (n = 9/34) of decidua at these time points contained embryos that were too reabsorbed for genotyping, suggesting that the homozygous embryos died at an earlier stage of development (Table S1). These results, compared with our previous findings of neuron-specific defects in the sti mutant mouse (10), indicate that the AarsC723A mutation likely causes a higher rate of translational errors in vivo than the sti mutation and that phenotypes caused by deficiencies in AlaRS editing are dependent on the extent of the residual editing activity of the mutant enzyme.
To create a mouse model with an editing deficiency between that of AarsC723A/C723A and Aarssti/sti mice, we generated Aarsstop/sti compound heterozygotes. These mice were viable and present at weaning age in the expected Mendelian ratios. Although initially indistinguishable from wild-type littermates, at 3 wk of age Aarsstop/sti mice were smaller than wild-type or Aarssti/sti mice (Fig. 1C). In addition, larger regions of alopecia were consistently observed in Aarsstop/sti than in Aarssti/sti mice (Fig. 1D). The gene dosage effect of the Aarssti mutation reinforces the notion that cells differ in their sensitivity to disruptions in translational fidelity.
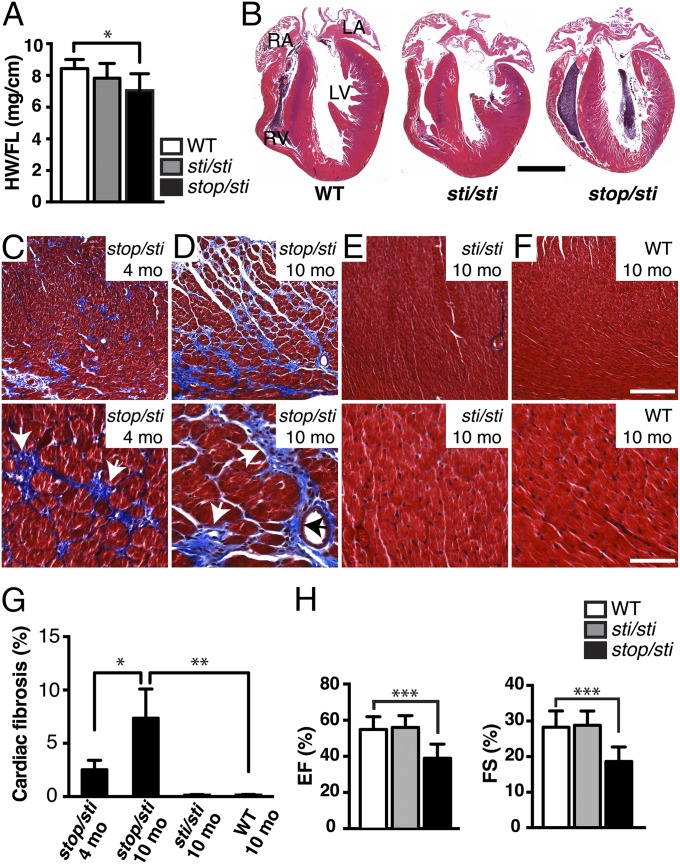
Histological analysis of the Aarsstop/sti brain demonstrated that the temporal and spatial pattern of Purkinje cell degeneration was very similar to that observed in Aarssti/sti mice (Fig. S1). However, gross morphological examination of other tissues from 10-mo-old mice revealed that the heart of Aarsstop/sti mice was smaller than those from age-matched Aarssti/sti or wild-type mice (Fig. 2 A and B). Although histological analysis of other organs did not reveal abnormalities, myofiber disarray was observed in the ventricular myocardium of hearts from Aarsstop/sti mice. Masson’s trichrome and picrosirius red staining of sections from 4- and 10-mo-old Aarsstop/sti mice revealed extensive fibrosis, which typically forms where cardiomyocytes have degenerated. Both interstitial and perivascular fibrosis was observed in the left and right ventricular walls and in the interventricular septum (IVS) of affected hearts (Fig. 2 C and D and Fig. S2). Moreover, the development of cardiac fibrosis in the Aarsstop/sti mouse appeared to be age-related: At 1 mo of age, no apparent fibrosis was observed in heart sections; at 4 mo of age, ∼2.5% of the heart tissue was fibrotic; and by 10 mo of age, fibrotic areas increased to about 7.5% of the Aarsstop/sti heart (Fig. 2G). In contrast, no fibrotic lesions were observed in 10-mo-old Aarssti/sti or wild-type mice (Fig. 2 E–G).
Fig. 2.
Cardiac abnormalites in Aarsstop/sti mice. (A) Ratio of the heart weight to femur length (HW/FL) from 10-mo-old WT (n = 11), Aarssti/sti (sti/sti; n = 6), and Aarsstop/sti (stop/sti; n = 8) mice. Values represent mean ± SD; *P < 0.01 (one-way ANOVA). (B) Hematoxylin and eosin staining of longitudinal sections of hearts from 10-mo-old mice with the indicated genotypes. (Scale bar, 2 mm.) (C–F) Masson’s trichrome stain of heart sections from 4- (C) and 10-mo-old (D) Aarsstop/sti and 10-mo-old Aarssti/sti (E) and WT (F) mice. Higher magnification photos of a representative area of each image are shown in the Lower panel. Note the high levels of interstitial fibrosis (blue; white arrows) and perivascular fibrosis (blue; black arrows) in Aarsstop/sti but not Aarssti/sti or WT hearts. (Scale bars, 200 μm and 60 μm for the Upper and Lower panel, respectively.) (G) Quantification of fibrotic areas (as a percentage of total area). Values are mean ± SD; *P < 0.01 (unpaired t test) and **P < 0.001 (one-way ANOVA); n = 4 mice for each data point. (H) Ejection fraction (EF%; Left graph) and fractional shortening (FS%; Right graph) of 10-mo-old WT (n = 15), Aarssti/sti (n = 6), and Aarsstop/sti (n = 9) mice. Values are mean ± SD; ***P < 0.0001 (one-way ANOVA).
In an alternative effort to diminish the editing activity of the AlaRS enzyme in vivo, we generated AarsC723A/sti mice. Similar to Aarsstop/sti mice, these mice were smaller than Aarssti/sti mice, with larger regions of dorsal alopecia. In addition, widespread myocardial fibrosis was present in the hearts of 4-mo-old AarsC723A/sti mice (Fig. S3), confirming that editing activity in between that of AlaRSA734E or AlaRSC723A results in death of cardiomyocytes.
Fibrotic cardiac muscle is stiffer and less compliant than normal heart muscle and is commonly associated with cardiac failure (16). To determine if fibrosis affected the contractile function of the Aarsstop/sti heart, echocardiography was performed on 10-mo-old Aarsstop/sti, Aarssti/sti, and wild-type mice (Fig. S4 and Table S2). Echocardiogram parameters revealed that the Aarsstop/sti mutant heart exhibited a slight, but significant, reduction in the thickness of the left ventricular posterior wall. Although not significant, the Aarsstop/sti septum also tended to be thinner than that of wild-type mice (IVSs). Consistent with the observed pathology, less systolic thickening of the left ventricle was seen in the Aarsstop/sti heart, indicating myocardial dysfunction. Furthermore, ventricular output, as measured by the ejection fraction and fractional shortening of the left ventricle, was significantly reduced in Aarsstop/sti but not Aarssti/sti or AarsC723A/+ mice (Fig. 2H and Fig. S1).
Electrophysiological function of Aarsstop/sti and Aarssti/sti hearts was further evaluated by electrocardiogram telemetry in 10-mo-old mice. Electrocardiographic characteristics were similar between different genotypes, and ventricular arrhythmias were not observed in any mice (Fig. S5).
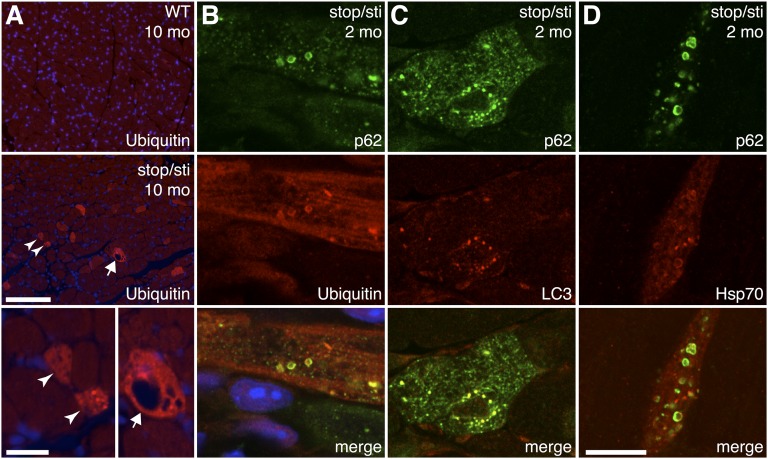
Previously we showed that accumulation of misfolded proteins in Aarssti/sti Purkinje cells accompanied their degeneration (10). To determine whether similar mechanisms occurred in Aarsstop/sti cardiomyocytes, ubiquitin immunofluorescence was performed on heart sections of 10-mo-old mutant and wild-type mice (Fig. 3A). In the wild-type heart, ubiquitin signals were present in the cytoplasm and in the nucleus, as previously reported (17). However, ubiquitin-positive puncta characteristic of protein aggregates were not detected in the wild-type heart. In contrast, ubiquitin puncta were present in ∼2.4% of cardiomyocytes in Aarsstop/sti hearts (n = 4). In addition, large vacuoles and a reduction in fiber diameter were often observed in aggregate-containing cardiomyocytes, consistent with cellular atrophy and cell death (Fig. 3A).
Fig. 3.
Formation of misfolded protein aggregates in Aarsstop/sti mutant cardiomyocytes. (A) Immunofluorescence with antibodies to ubiquitin on heart sections from 10-mo-old WT and Aarsstop/sti mice. Nuclei are counterstained with Hoechst dye (blue). Higher magnification photos of Aarsstop/sti cardiomyocytes are shown in the Lower panel. Arrowheads indicate myofibers with intense ubiquitin signals and reduced fiber diameter. An arrow indicates a cardiomyocyte with elevated ubiquitin and vacuoles. (B–D) High-magnification confocal microscopic images showing colocalization of p62 with ubiquitin (B), LC3 (C), and Hsp70 (D) within the cytoplasm of representative cardiomyocytes of 2-mo-old Aarsstop/sti mice. [Scale bars, (A, Upper and Middle panels) 100 μm, (A, Lower panel) 20 μm, and (B–D) 10 μm.]
To determine the onset of proteinopathy in mutant cardiomyocytes, ubiquitin immunofluorescence was performed on hearts from younger Aarsstop/sti mice. At 1 mo of age, most cardiomyocytes in Aarsstop/sti hearts appeared normal, with cells containing ubiquitinated aggregates only occasionally observed. However, the number of aggregate-containing cells drastically increased by 2 mo of age, constituting ∼1.8% of total cells in the Aarsstop/sti heart. In comparison, cardiomyocytes of Aarssti/sti mice appeared normal, and ubiquitin-positive inclusions were rarely observed at any given age. Our observations that protein aggregates occurred in prefibrotic Aarsstop/sti hearts and that the number of cardiomyocytes with aggregates increased with age suggest that misfolded proteins induce cardiomyocyte death.
In addition to the ubiquitin–proteasome protein degradation pathway, aggregates of ubiquitinated proteins can be degraded by autophagy. Thus, autophagy is often up-regulated as an adaptive response to the increased production of misfolded proteins. Correspondingly, suppression of autophagy in diseased hearts leads to cardiomyopathy (5). The autophagic adapter protein p62/sequestosome 1 noncovalently interacts with ubiquitinated proteins through its C-terminal domain and facilitates their degradation via autophagy (18, 19). Recent studies demonstrate that p62 is often associated with aggresomes in desmin-associated cardiomyopathies (5, 20). Coimmunofluorescence using antibodies to p62 and ubiquitin in 2-mo-old Aarsstop/sti hearts demonstrated that the majority of ubiquitinated aggregates also contained p62 (Fig. 3B). Ubiquitin and p62-positive puncta were also observed in cardiomyocytes of AarsC723A/sti mice, further demonstrating the convergence of the two mouse models (Fig. S3).
Immunofluorescence with antibodies to microtubule-associated protein 1 light chain 3 (LC3), a marker of autophagosomes, on sections of Aarsstop/sti hearts also showed colocalization of LC3 and protein aggregates (Fig. 3C). In addition, coimmunofluorescence with antibodies to stress-inducible Hsp70, a molecular chaperone often colocalized with intracellular inclusions (21, 22), revealed colocalization of p62-positive aggregates with Hsp70, further confirming that these structures contain misfolded or unfolded proteins (Fig. 3D). Lastly, because AlaRS editing defects have the potential to cause mistranslation of any protein containing alanine, we determined if protein aggregates in Aarsstop/sti hearts contained desmin, previously shown to form aggregates in the genetic desminopathy disorders (23). In both wild-type and Aarsstop/sti hearts, desmin was detected at the sarcomeric Z-disk (Fig. S6). However, no colocalization of p62 and desmin was observed in the Aarsstop/sti heart, suggesting desmin is not a major component of protein aggregates.
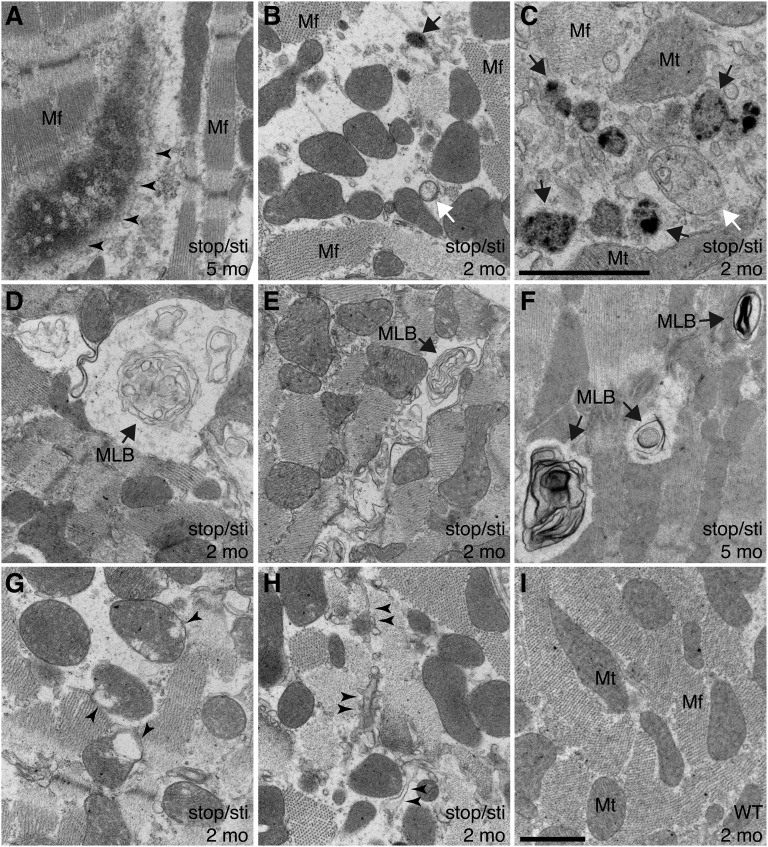
We further performed transmission electron microscopy that revealed multiple pathological changes in Aarsstop/sti cardiomyocytes. Consistent with our immunofluorescence observations, deposition of protein aggregates that disrupt myofibrillar structure was observed in mutant, but not wild-type, cardiomyocytes (Fig. 4A). In addition, many mutant cardiomyocytes had abnormal accumulation of autophagic vacuoles of different stages, indicative of elevated autophagy (Fig. 4 B and C). Multilamellar bodies indicative of impaired lysosomal degradation of autophagic cargos (24) were apparent in mutant, but not wild-type, cells (Fig. 4 D–F), suggesting that autophagic clearance is likely compromised in Aarsstop/sti mutant cells. In addition to accumulation of autophagic structures, we observed abnormal mitochondria and disruption of the sarcoplasmic reticulum in mutant cardiomyocytes (Fig. 4 B, G, and H). These pathological changes may reflect the toxic effects of misfolded proteins and may contribute to degeneration of cardiomyocytes.
Fig. 4.
Ultrastructural changes in Aarsstop/sti cardiomyocytes. (A–I) Representative transmission electron microscope images of hearts from 2-mo- (B–E, G, and H) and 5-mo-old (A and F) Aarsstop/sti mice and 2-mo-old WT (I) mice. (A) Formation of large protein aggregates (arrowheads) that disrupt myofibril (Mf) structures. (B and C) Accumulation of autophagic vacuoles (white arrow, early autophagosome; black arrow, autolysosome) accompanied by loss of Mf structures. (D–F) Multilamellar bodies (MLBs, arrows), products of active autophagy and a hallmark of insufficient lysosomal activity, are also present. (G) Defective mitochondria (Mt) with disrupted cristae (arrowheads). (H) Disrupted sarcoplasmic reticulum (arrowheads) in Aarsstop/sti cardiomyocytes. [Scale bars, (A, B, and D–I) 1 μm and (C) 1 μm.]
Discussion
The effect of mistranslation on the cellular homeostasis of complex higher organisms remains poorly understood. To explore the pathological impact of increased translational errors in vivo, we genetically decreased the editing function of the mouse AlaRS enzyme. This decrease in editing in turn affects global translation by increasing the incorporation of noncognate amino acids into nascent proteins. Our “fine-tuning” of the editing efficiency of AlaRS demonstrated that the severity of the resulting phenotypes directly correlated with the residual editing activity of the enzyme. The originally identified Aarssti (A734E) mutation is the mildest allele in our series. Homozygosity for this mutation selectively induces protein aggregation and apoptosis in cerebellar Purkinje cells (10). In contrast, embryos homozygous for the AarsC723A allele, which causes dramatic increases in mischarged Ser-tRNAAla, died before midgestation, suggesting that editing activity is essential for mammalian development. Interestingly, mice that were heterozygous for the AarsC723A mutation were largely phenotypically normal, with only loss of a minor population of Purkinje cells in aged mice. This finding indicates that AlaRS expressed from the wild-type allele is sufficient in trans to hydrolyze the Ser-tRNAAla generated by the mutant enzyme. Further genetic regulation of the editing efficacy of the Aarssti allele by either lowering the level of the enzyme (as in Aarsstop/sti mice) or placing the Aarssti allele over the AarsC723A mutation resulted in phenotypes intermediate to those in Aarssti/sti and AarsC723A/C723A mice, including loss of Purkinje cells and widespread protein aggregate formation in cardiomyocytes and subsequent death of these cells. These results suggest that the response to mistranslation varies among cells and that protein aggregates may be induced in additional cell types upon further decreases in translational fidelity.
Our results define a common mechanism of protein aggregation in cardiomyocytes and neurons. The underlying sensitivity of these two cell types to mistranslation may lie in the postmitotic nature of these cells, in which protein aggregates are not diluted by cell division (25). Moreover, hydrolysis of misacylated tRNA may be subject to additional regulation. Indeed, Ser-tRNAAla can also be hydrolyzed by a free-standing editing domain protein, Aarsd1 (AlaXP) (26, 27), which could compensate for editing deficiencies in AlaRS. Lastly, the differential sensitivity of cells to translation errors may be due to the varying capacity of protein quality control systems in different cell types.
Although decreased translation fidelity causes protein misfolding and aggregation in both cardiomyocytes and Purkinje cells, the downstream pathways that lead to cell death in the two cell types may be quite different. Purkinje cells in sticky mutant mice undergo apoptosis (10), whereas cardiomyocytes in the Aarsstop/sti mouse were negative for cleaved caspase 3 and TUNEL labeling. In addition, abnormal mitochondria with disorganized cristae that may be a contributing factor in cell death were observed in Aarsstop/sti cardiomyocytes. Interestingly, a point mutation in the editing domain of human mitochondrial AlaRS (AARS2) has been associated with infantile mitochondrial cardiomyopathy (28), suggesting increased errors in either cytoplasmic or mitochondrial protein synthesis can lead to cell death in the heart.
In summary, our data show that a global reduction in translational fidelity, rather than disruption of a specific protein, can induce defects in proteostasis in numerous cell types in the mouse. Indeed, fidelity of protein synthesis is not only subjected to genetic changes that modulate translation machinery, it can also be affected by environmental factors such as oxidative stress. The cysteine 182 residue that is critical for editing activity of threonyl-tRNA synthetase in E. coli can be oxidized by reactive oxygen species, and thus, overall translational fidelity is impaired as a result (29). In mammalian cells, various environmental factors that trigger oxidative stress are known to increase Met-misacylation of tRNAs (30). It is thus possible that proteinopathy can occur in the brain, heart, or even other tissues as a combination of genetic and/or environmental factors that compromise translational fidelity.
Materials and Methods
Misacylation Assays.
Misacylation assays were performed with mouse wild-type or mutant C723A AlaRS, 10 μM 3H-l-serine, and 10 μM human tRNAAla following methods described in ref. 31. Overexpression and purification of human tRNAAla was performed as previously described (10). Details are provided in SI Materials and Methods.
Mice.
All animal protocols were approved by The Jackson Laboratory Animal Care and Use Committee. Homologous recombination of the Aarsstop allele was performed in C57BL/6J-Tyrc-2J ES cells. Aarssti/sti and Tg(EIIa-cre)C5379Lmgd/J mice were maintained on a C57BL/6 genetic background (10, 15). Generation of the knock-in mouse and genotyping information are described in SI Materials and Methods.
Echocardiography and Electrocardiography.
Cardiac function, heart dimensions, and electrical activity of the heart were evaluated by echocardiography and electrocardiography on anesthetized mice. Details are provided in SI Materials and Methods.
Histology.
Hematoxylin and eosin, Masson’s trichrome, and picrosirius red staining was performed on 10% (vol/vol) neutral buffered formalin (NBF)-fixed tissue sections. Details are described in SI Materials and Methods.
Immunofluorescence.
The 10% NBF-fixed, paraffin-embedded heart sections were deparaffinated, rehydrated, and microwaved in 0.01 M citrate buffer (pH 6.0) three times for 2 min each. Tissue sections were then blocked with 4% (wt/vol) goat serum in phosphate-buffered saline with Tween-20 (PBST) for 30 min, incubated with primary antibodies overnight at 4 °C, washed in PBST, and incubated with secondary antibodies for 1 h. Details about antibodies are described in SI Materials and Methods.
Electron Microscopy.
Mice were transcardially perfused with 2% (wt/vol) paraformaldehyde and 2% (wt/vol) glutaraldehyde. Tissues were prepared for analysis using standard procedures.
Statistics.
Results are mean ± SD. Statistical significance was determined by unpaired t tests or one-way ANOVA followed by Dunnett’s multiple-comparisons test. P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We are grateful to Jennifer Cook and Krystal-Leigh Baker for technical assistance, Litao Sun for tRNAAla, Jennifer Ryan for ECG and echocardiography recordings, Mark Lessard for help with imaging and quantification, Pete Finger for electron microscopy assistance, and Doug McMinimy for mouse photos. We also thank Dr. Karen Svenson for helpful discussions, Dr. Greg Cox for comments on the manuscript, and The Jackson Laboratory sequencing, histology, and microinjection services for their contributions. These studies were supported by National Institutes of Health Grant NS042613 (to S.L.A.), and services used in this study were supported by Cancer Center Core Grant CA34196 (The Jackson Laboratory). Support was also provided by aTyr Pharma, Inc., The National Foundation for Cancer Research, and National Cancer Institute Grant CA92577. J.S.S. was supported by a fellowship from the American Health Assistance Foundation. S.L.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420196111/-/DCSupplemental.
References
- 1.Goldfarb LG, Dalakas MC. Tragedy in a heartbeat: Malfunctioning desmin causes skeletal and cardiac muscle disease. J Clin Invest. 2009;119(7):1806–1813. doi: 10.1172/JCI38027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLendon PM, Robbins J. Desmin-related cardiomyopathy: An unfolding story. Am J Physiol Heart Circ Physiol. 2011;301(4):H1220–H1228. doi: 10.1152/ajpheart.00601.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quarta CC, Kruger JL, Falk RH. Cardiac amyloidosis. Circulation. 2012;126(12):e178–e182. doi: 10.1161/CIRCULATIONAHA.111.069195. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Robbins J. Heart failure and protein quality control. Circ Res. 2006;99(12):1315–1328. doi: 10.1161/01.RES.0000252342.61447.a2. [DOI] [PubMed] [Google Scholar]
- 5.Zheng Q, Su H, Ranek MJ, Wang X. Autophagy and p62 in cardiac proteinopathy. Circ Res. 2011;109(3):296–308. doi: 10.1161/CIRCRESAHA.111.244707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 7.Schimmel P. Development of tRNA synthetases and connection to genetic code and disease. Protein Sci. 2008;17(10):1643–1652. doi: 10.1110/ps.037242.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loftfield RB, Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem J. 1972;128(5):1353–1356. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jakubowski H, Goldman E. Editing of errors in selection of amino acids for protein synthesis. Microbiol Rev. 1992;56(3):412–429. doi: 10.1128/mr.56.3.412-429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443(7107):50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 11.Beebe K, Ribas De Pouplana L, Schimmel P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 2003;22(3):668–675. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol. 2006;13(10):1091–1100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 14.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93(12):5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber KT, Brilla CG, Janicki JS. Myocardial fibrosis: Functional significance and regulatory factors. Cardiovasc Res. 1993;27(3):341–348. doi: 10.1093/cvr/27.3.341. [DOI] [PubMed] [Google Scholar]
- 17.Hilenski LL, Terracio L, Haas AL, Borg TK. Immunolocalization of ubiquitin conjugates at Z-bands and intercalated discs of rat cardiomyocytes in vitro and in vivo. J Histochem Cytochem. 1992;40(7):1037–1042. doi: 10.1177/40.7.1318894. [DOI] [PubMed] [Google Scholar]
- 18.Vadlamudi RK, Joung I, Strominger JL, Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J Biol Chem. 1996;271(34):20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 19.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 20.Olivé M, et al. Expression of mutant ubiquitin (UBB+1) and p62 in myotilinopathies and desminopathies. Neuropathol Appl Neurobiol. 2008;34(1):76–87. doi: 10.1111/j.1365-2990.2007.00864.x. [DOI] [PubMed] [Google Scholar]
- 21.Warrick JM, et al. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23(4):425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 22.Waelter S, et al. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell. 2001;12(5):1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clemen CS, Herrmann H, Strelkov SV, Schröder R. Desminopathies: Pathology and mechanisms. Acta Neuropathol. 2013;125(1):47–75. doi: 10.1007/s00401-012-1057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hariri M, et al. Biogenesis of multilamellar bodies via autophagy. Mol Biol Cell. 2000;11(1):255–268. doi: 10.1091/mbc.11.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soonpaa MH, Field LJ. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am J Physiol. 1997;272(1 Pt 2):H220–H226. doi: 10.1152/ajpheart.1997.272.1.H220. [DOI] [PubMed] [Google Scholar]
- 26.Nawaz MH, Merriman E, Yang XL, Schimmel P. p23H implicated as cis/trans regulator of AlaXp-directed editing for mammalian cell homeostasis. Proc Natl Acad Sci USA. 2011;108(7):2723–2728. doi: 10.1073/pnas.1019400108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chong YE, Yang XL, Schimmel P. Natural homolog of tRNA synthetase editing domain rescues conditional lethality caused by mistranslation. J Biol Chem. 2008;283(44):30073–30078. doi: 10.1074/jbc.M805943200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Götz A, et al. Exome sequencing identifies mitochondrial alanyl-tRNA synthetase mutations in infantile mitochondrial cardiomyopathy. Am J Hum Genet. 2011;88(5):635–642. doi: 10.1016/j.ajhg.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ling J, Söll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci USA. 2010;107(9):4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Netzer N, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462(7272):522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beebe K, Waas W, Druzina Z, Guo M, Schimmel P. A universal plate format for increased throughput of assays that monitor multiple aminoacyl transfer RNA synthetase activities. Anal Biochem. 2007;368(1):111–121. doi: 10.1016/j.ab.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.