Significance
Peptides are valuable leads for drug development, offering advantages over other molecular classes. Specifically, they can bind potently and selectively to drug targets, including protein–protein interactions that are too challenging for small-molecule therapeutics. However, peptides are poor drugs because of their low in vivo stability and poor oral bioavailability. We propose a strategy for improving the oral bioavailability of peptides by identifying appropriate amides for chemical modification using temperature coefficients measured by NMR. The modified peptides have improved solvation properties, making them more membrane permeable. This approach for identifying sites for modification is a rapid method for guiding peptide drug design.
Keywords: cyclic peptide, permeability, N-methylation
Abstract
Enhancing the oral bioavailability of peptide drug leads is a major challenge in drug design. As such, methods to address this challenge are highly sought after by the pharmaceutical industry. Here, we propose a strategy to identify appropriate amides for N-methylation using temperature coefficients measured by NMR to identify exposed amides in cyclic peptides. N-methylation effectively caps these amides, modifying the overall solvation properties of the peptides and making them more membrane permeable. The approach for identifying sites for N-methylation is a rapid alternative to the elucidation of 3D structures of peptide drug leads, which has been a commonly used structure-guided approach in the past. Five leucine-rich peptide scaffolds are reported with selectively designed N-methylated derivatives. In vitro membrane permeability was assessed by parallel artificial membrane permeability assay and Caco-2 assay. The most promising N-methylated peptide was then tested in vivo. Here we report a novel peptide (15), which displayed an oral bioavailability of 33% in a rat model, thus validating the design approach. We show that this approach can also be used to explain the notable increase in oral bioavailability of a somatostatin analog.
Peptides are potentially valuable compounds for drug development, offering many advantages over other molecular classes (1–4). Specifically, their ability to mimic endogenous bioactive molecules allows them to bind potently and selectively to “difficult” drug targets, including protein–protein interactions that are too challenging for small-molecule therapeutics. However, the widespread use of peptides in the clinic has been slow in coming, in large part because of their generally low stability in vivo, high clearance, and poor oral bioavailability.
The low oral bioavailability of peptides is attributed to a disparity between their physicochemical properties and those traditionally expected for “drug-likeness” (5, 6), leading to a perception that peptides are good drug leads but poor drugs. However, this perception is being challenged by a growing number of peptides that seem to be stable (7) and well absorbed within the gastrointestinal tract (8) and examples in which cyclic peptides have shown orally delivered bioactivity in animal disease models, including inflammatory pain (9) and neuropathic pain (10), prompting us to devise new rules for predicting pharmacokinetic properties of this compound class. Arguably the most famous example of a peptide with poor drug-likeness but reasonable oral bioavailability is cyclosporin A, widely used as the immunosuppressant drug cyclosporine (11). Two structural features of cyclosporin A in particular are thought to contribute to its oral bioavailability, namely its macrocyclic architecture and backbone N-methylation.
Cyclization imparts increased rigidity to a parent peptide, which not only improves its stability against proteolytic degradation but also directs it into specific conformations that might be favorable for membrane permeability. In a recent study, the relative rates of diffusion of cyclic hexapeptide stereoisomers across a membrane were traced back to intramolecular hydrogen bond networks, steric protection of backbone amides, and the relative stabilities of aqueous and membrane-associated conformations (12). That study corroborated earlier findings, showing that cyclic peptides have improved membrane permeability over their linear counterparts (13). A common theme among these studies is that cyclization seems to be an important contributing factor toward oral bioavailability.
Backbone N-methylation is another structural feature that seems to be correlated with improved membrane permeability (14, 15) and can, in some cases (16–18), be important for enhancing the bioactivity and selectivity of peptides. For example, N-methylation of Arg-Gly-Asp (16), an integrin ligand (18), and the melanocortin family of peptides (17) resulted in cyclic peptide analogs with improved selectivity for their target receptors and enhanced or comparable bioactivities. Interestingly, N-methylation of a cyclic somatostatin peptide improved its oral bioavailability without significantly modifying its biological activity and selectivity (19). Recently, Ovadia et al. (20) demonstrated that the number of N-methyl groups can have a significant effect on the cell permeability of a series of cyclic hexapeptides. Structures of cyclosporin A (21) and other N-methylated macrocyclic compounds [e.g., aureobasidin A (22) and the destruxins (23)] suggest that N-methylation can affect conformation and hydrogen bonding potential, which might in turn affect membrane permeability.
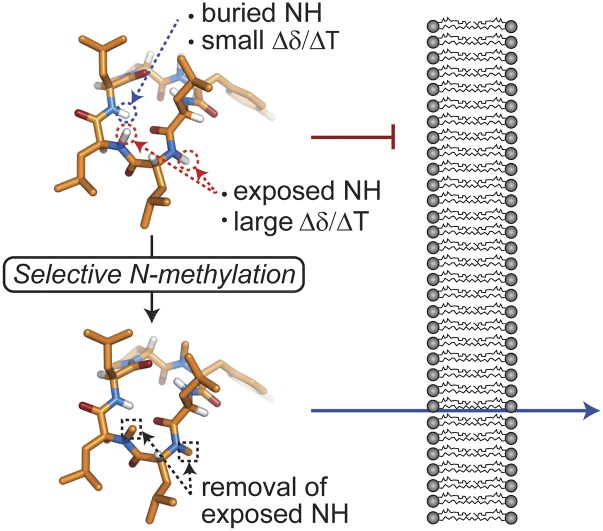
On the basis that the internal hydrogen bonding potential of peptides is a key determinant of passive membrane permeability, the hypothesis is that N-methylation of the most exposed NH groups of a cyclic peptide might improve its membrane permeability. This is consistent with a recent study showing that on-resin modification of leucine-rich cyclic peptides via nonspecific N-methylation can result in small-molecule-like oral bioavailability (24). One approach to test this hypothesis was to use structural information to identify exposed NH groups that can be specifically targeted for N-methylation. Accurate determination of the atomic positions of the NH groups of a peptide requires elucidation of the cyclic peptide structure, which can be time consuming, particularly in a drug design environment. However, we reasoned that complete determination of a cyclic peptide structure is not necessary; rather, approaches to rapidly probe the hydrogen bonding network (i.e., amide temperature coefficients and hydrogen–deuterium exchange determined from NMR spectroscopy experiments) might provide a means of rapidly identifying the most exposed NH groups, which can then be selectively N-methylated. Furthermore, we postulated that the resulting N-methylated derivatives would have improved membrane permeability relative to the nonmethylated parent compounds (Fig. 1).
Fig. 1.
Schematic illustration of the principle of using selective N-methylation of small cyclic peptides to improve membrane permeability. Exposed amides of cyclic peptides, as shown in red circles in the top peptide, are thought to reduce or prevent membrane permeability. Selective N-methylation of the amides (shown in boxes on the bottom peptide) can increase membrane permeability of the peptide by masking the exposed NH moieties.
Here we report the use of cyclic hexapeptide scaffolds to investigate how amide temperature coefficients and hydrogen–deuterium exchange experiments can be used to study the interplay between N-methylation, conformation, and permeability. We emphasize that although both amide temperature coefficient experiments and hydrogen–deuterium exchange have been used previously to provide supplementary information in structural studies of proteins, their potential as structural markers of peptide permeability has not been studied. We propose a structure-informed method for directing the design of cyclic peptides with promising pharmacokinetic properties. This method has been applied to a series of cyclic hexapeptide stereoisomers to improve their membrane permeability. We validated this approach for the design of orally bioavailable peptides with in vitro permeability and in vivo pharmacokinetics studies.
Results
Synthesis of Cyclic Hexapeptides and Selective N-Methylation.
The scaffolds selected for investigation of N-methylation on peptide permeability and oral bioavailability were cyclic hexapeptides with the general sequence, cyclo(Leu-Leu-Leu-Leu-Pro-Tyr), and are detailed in Fig. 2A and SI Appendix. Synthesis of the linear peptides was carried out using Fmoc solid-phase peptide synthesis. The incorporation of N-methylated residues is challenging because it requires robust conditions to couple subsequent residues, owing to the more sterically hindered methyl amide that has been introduced. To overcome these difficult couplings, the coupling reagent HCTU, O-(1H-6-chlorobenzotriazol-1-y1)-N,N,N′,N′-tetramethyluronium hexafluorophosphate, was substituted for the more reactive HATU, O-(7-azabenzotriazol-1-y1)-N,N,N′,N′-tetramethyluronium hexafluorophosphate, and the coupling time and the excess of the coupling amino acids were increased, yielding linear protected peptides of 90–95% purity for almost every N-methylated peptide. The choice of linear sequence for cyclization was also found to be crucial for successful assembly of N-methylated peptides; it was desirable to avoid an N-methylated residue at the N terminus of the peptide, which was found to reduce the yield of the cyclization step.
Fig. 2.
Synthesis of nonmethylated and N-methylated cyclic peptides. (A) Cyclic leucine-rich peptides synthesized for this study. All of the peptides share a common sequence unless indicated (*Pro instead of Leu residue). The chirality of each residue is indicated, and N-methylated residues are represented by shading. (B) Synthesis of these peptides was carried out from the side chain protected linear peptides in DMF (dimethylformamide) with HATU/DIPEA (N,N-diisopropylethylamine). After cyclization peptides were purified by RP-HPLC.
Amide Temperature Coefficients and Hydrogen–Deuterium Exchange Studies.
We measured the amide NMR chemical shift temperature coefficients (ΔδNH/ΔT) for a series of nonmethylated hexapeptide stereoisomers (1–5) to determine whether these parameters could be used to monitor the conformational behavior of the backbone amides. It has been proposed that backbone amides having very negative ΔδNH/ΔT (i.e., <−4.6 ppb/K) are water-exposed, whereas those having less-negative or slightly positive ΔδNH/ΔT (i.e., ≥−4.6 ppb/K) are considered to be involved in intramolecular hydrogen bonds (25).
Values of ΔδNH/ΔT were measured for peptides 1–5 in six different solvent compositions (30%, 90%, and 100% vol/vol acetonitrile; 30% and 100% vol/vol trifluroethanol; and 100% vol/vol chloroform), as shown in Table 1. Aside from demonstrating the utility of ΔδNH/ΔT measurements in different solvents that may be useful for peptide solubilization, the different solvents may also be useful for mimicking specific solvent environments that are associated with the peptides as they pass from an aqueous environment through a membrane. In solvents that contain some proportion of water, the values of the ΔδNH/ΔT were generally consistent with large negative ΔδNH/ΔT remaining relatively invariant regardless of the concentration of the cosolvent. As an example, ΔδNH/ΔT of Leu-3 in 5 was −8.29, −8.18, and −7.32 ppb/K in 30% (vol/vol) acetonitrile, 90% (vol/vol) acetonitrile, and 30% (vol/vol) trifluroethanol, respectively. Because of the similarity in ΔδNH/ΔT values obtained from different aqueous solvent mixtures, we chose to focus on the 30% (vol/vol) acetonitrile solvent system to explore the potential of amide temperature coefficients as structural markers of peptide permeability.
Table 1.
NMR amide temperature coefficients of peptides 1–5 in different solvents
| ΔδHN/ΔT (ppb/K) | |||||||
| Acetonitrile (polar aprotic) | Trifluroethanol (polar protic) | Chloroform (nonpolar) | |||||
| Residue | 30%* | 90%* | 100% | 30%* | 100%* | 100% | |
| Cyclic peptide 1 | |||||||
| 1 | Leu | −6.13 | −7.18 | −1.53 | −7.30 | −5.24 | −0.87 |
| 2 | d-Leu | −8.02 | −9.34 | −6.98 | −10.29 | −6.19 | −1.93 |
| 3 | d-Leu | −1.94 | −2.22 | −2.50 | −1.91 | −2.51 | −0.94 |
| 4 | d-Leu | −5.01 | −3.97 | −0.05 | −4.85 | −3.08 | −0.17 |
| 5 | Pro† | ||||||
| 6 | Tyr | −2.42 | −2.63 | −2.18 | −2.17 | −2.26 | −1.50 |
| Cyclic peptide 2 | |||||||
| 1 | Leu | −8.70 | −8.53 | −5.49 | −8.56 | −3.28 | −9.83 |
| 2 | Leu | −7.89 | −4.51 | −2.69 | −5.11 | −3.95 | −6.08 |
| 3 | d-Leu | −0.72 | −0.23 | −0.76 | −0.79 | −1.86 | −4.00 |
| 4 | d-Pro† | ||||||
| 5 | Pro† | ||||||
| 6 | Tyr | −2.53 | −2.21 | −2.11 | −2.43 | −0.64 | −3.37 |
| Cyclic peptide 3 (conformation 1) | |||||||
| 1 | d-Leu | −6.55 | −11.00 | −1.49 | −7.90 | −0.04 | −0.40 |
| 2 | d-Leu | −6.09 | −7.50 | −3.80 | −6.33 | −5.66 | −1.74 |
| 3 | d-Leu | −2.21 | −2.00 | −4.03 | −2.08 | −3.52 | −6.70 |
| 4 | Leu | −7.41 | −8.00 | −1.19 | −6.85 | −1.62 | −1.50 |
| 5 | Pro† | ||||||
| 6 | Tyr | −0.94 | −0.20 | −4.42 | −0.90 | −2.11 | −1.44 |
| Cyclic peptide 3 (conformation 2) | |||||||
| 1 | d-Leu | −1.96 | −1.90 | −1.49 | −1.13 | −0.04 | −0.40 |
| 2 | d-Leu | −5.29 | −5.30 | −3.80 | −5.48 | −5.66 | −1.74 |
| 3 | d-Leu | −6.63 | −5.10 | −4.03 | −6.14 | −3.52 | −6.70 |
| 4 | Leu | −1.95 | −1.20 | −1.19 | −1.60 | −1.62 | −1.50 |
| 5 | Pro† | ||||||
| 6 | Tyr | −8.57 | −6.90 | −4.42 | −4.19 | −2.11 | −1.44 |
| Cyclic peptide 4 | |||||||
| 1 | d-Leu | −6.91 | −5.68 | −0.05 | −6.90 | 0.26 | −1.33 |
| 2 | d-Leu | −6.76 | −9.55 | −3.37 | −6.36 | −5.89 | −1.63 |
| 3 | Leu | −2.37 | −3.02 | −1.45 | −3.05 | −2.87 | −0.70 |
| 4 | d-Leu | −8.57 | −7.17 | −0.45 | −7.90 | −6.53 | −0.14 |
| 5 | Pro† | ||||||
| 6 | Tyr | −4.07 | −3.08 | −0.41 | −2.57 | −1.53 | −1.83 |
| Cyclic peptide 5 | |||||||
| 1 | Leu | −0.93 | −1.25 | −0.26 | −0.88 | −1.44 | −1.09 |
| 2 | d-Leu | −7.74 | −7.12 | −0.07 | −5.94 | −2.04 | −2.30 |
| 3 | Leu | −8.29 | −8.18 | −2.00 | −7.32 | −7.57 | −2.34 |
| 4 | Leu | −5.69 | −6.63 | −2.99 | −3.41 | −2.40 | −0.40 |
| 5 | d-Pro† | ||||||
| 6 | Tyr | −8.01 | −8.23 | −2.24 | −7.47 | −7.14 | −2.01 |
Organic solvent mixtures with H2O (vol/vol).
No NH proton present in Pro.
To confirm that ΔδNH/ΔT could be used to predict whether the amide of interest is solvent-exposed or involved in an intramolecular hydrogen bond, we measured the hydrogen–deuterium exchange rates for the amides of 1–5 and correlated them to ΔδNH/ΔT values, as shown in SI Appendix, Fig. S1. As anticipated, amides having very negative ΔδNH/ΔT (i.e., <−4.6 ppb/K) generally displayed more rapid exchange rates, whereas those having less-negative ΔδNH/ΔT (i.e., ≥−4.6 ppb/K) generally displayed slower exchange rates. Two amides with ΔδNH/ΔT more negative than −4.6 ppb/K displayed relatively slow exchange rates: Leu-4 of 3 and Leu-4 of 5. Overall, however, the results suggested that ΔδNH/ΔT can be used here to identify the level of solvent exposure for amides or probe the hydrogen-bond network of cyclic peptides.
Amide Temperature Coefficients and N-Methylation.
We subsequently investigated whether ΔδNH/ΔT measurements could be used to monitor the interplay between N-methylation, hydrogen bonding, and membrane permeability. We conducted a directed “N-methyl scan” of 5, a nonmethylated cyclic hexapeptide that can be N-methylated at three specific locations (i.e., Leu-2, Leu-3, and Tyr-6) to produce 12, a peptide reported to have noteworthy oral bioavailability (24). The peptides 6–11 were synthesized and represent N-methyl intermediates of 12 relative to 5.
Fig. 3A shows the ΔδNH/ΔT for these peptides. The changes in the ΔδNH/ΔT values observed for each residue as N-methyl groups were progressively added to 5 may reflect conformational restraints imposed by N-methylation. Importantly, the three residues of 5 that showed the most negative ΔδNH/ΔT (i.e., Leu-2, Leu-3, and Tyr-6) also correspond to those that require N-methylation to improve membrane permeability, as highlighted in Fig. 3A. Strikingly, as these residues were progressively N-methylated, the remaining residues that needed to be N-methylated to produce 12 consistently displayed ΔδNH/ΔT with large negative values. Similarly, residues of 5 that were not solvent-exposed and did not need to be N-methylated to produce 12 consistently displayed ΔδNH/ΔT with less-negative values. As a specific example, the N-methyl intermediate 8, which contained one N-methyl group on the amide of Tyr-6, displayed ΔδNH/ΔT of −7.76 and −7.34 ppb/K for Leu-2 and Leu-3, respectively, and −0.42 and −3.84 ppb/K for Leu-1 and Leu-4, respectively. Furthermore, a relationship between the N-methylation position and membrane permeability was observed, with the permeability of 5 increasing as it was progressively modified.
Fig. 3.
Amide temperature coefficient measurements for N-methylated peptides. (A) The table shows the progression from the nonmethylated 5 to the N-methylated analog 12. Amide temperature coefficients (in ppb/K) are shown for the amides in each peptide; residues with very negative ΔδNH/ΔT are shaded red, and N-methylated residues are indicated in orange. Apparent permeability (Papp; 10−6 cm/s) is also shown. (B) The temperature coefficient-directed N-methylation approach is pictorially represented. Exposed amides requiring N-methylation are shown in red, and the buried/shielded amides are indicated in blue text.
To examine the effect of N-methylation of amides that displayed less-negative ΔδNH/ΔT values, we synthesized 13, which is N-methylated at Leu-1. Indeed, 13 did not show improved permeability compared with 5. Additionally, we synthesized 14, which contains two N-methyl groups—one at Leu-1 and another at Tyr-6. Compared with the other peptides (i.e., 9, 10, and 11) that have two N-methyl groups at residues that displayed the most negative ΔδNH/ΔT, 14 has reduced permeability.
Overall, these results allowed us to propose a strategy for improving the membrane permeability (and possibly oral bioavailability) of peptides. As illustrated in Fig. 3B, ΔδNH/ΔT can be used to identify amides that can be N-methylated for the purposes of improving membrane permeability; the resulting “optimal” or “near-optimal” peptide only exhibits small ΔδNH/ΔT values. We propose that it is possible to start with a nonmethylated peptide (or a partially N-methylated peptide) and identify exposed residues for N-methylation to increase membrane permeability.
Permeability of Designed N-Methylation Products.
To test this method for improving the membrane permeability of peptides according to ΔδNH/ΔT measurements, we designed and synthesized N-methylated analogs of the nonmethylated compounds 1–4 based on their measured ΔδNH/ΔT. The nonmethylated peptides and their designed products, 15–18, were tested in parallel artificial membrane permeability assay (PAMPA) and Caco-2 permeability assay, which are commonly used as predictors of permeability. As shown in Fig. 4 A and B, the designed peptides generally displayed higher membrane permeability than their nonmethylated counterparts. For example, using the ΔδNH/ΔT-guided approach, the percent permeability of 1 in the PAMPA assay was improved from ∼6% to 17% in 15, and its permeability in the Caco-2 assay was improved from 1 × 10−6 cm/s to 21 × 10−6 cm/s. Fig. 4C shows that the backbone amides of Leu-1 and d-Leu-2 in 1 that displayed the most negative ΔδNH/ΔT values were subsequently N-methylated to produce 15; the nonmethylated amides of 15 generally displayed small ΔδNH/ΔT values.
Fig. 4.
Membrane permeability of cyclic peptides. (A) PAMPA results for peptides 1–5 and their corresponding N-methylated peptides, 15–18 and 12, respectively. Atenolol and quinidine were used as controls of known and varied permeability. (B) Caco-2 assay results, represented as apparent permeability (Papp; 10−6 cm/s), for the same peptides. Errors are shown as SD of six replicates. (C) The temperature coefficient-directed N-methylation approach that was applied to produce 15 from 1 is pictorially represented.
In Vitro and in Vivo Pharmacokinetic Studies.
To confirm whether the in vitro permeability of the designed peptides correlated with absorption in vivo and detectable plasma concentrations of the cyclic peptide, the in vivo pharmacokinetics of 15 in male Wistar rats was measured by examining plasma concentrations after i.v. (1 mg/kg, n = 4) and oral (10 mg/kg, n = 4) administration. Analysis of pharmacokinetics of 15 (SI Appendix, Table S1) revealed a fraction of absorption (F%) of 33% ± 2.9% (Fig. 5), which shows that 15 had permeability across rat intestines. The F% of 15 is an improvement (P < 0.05) over our measured F% of the reference peptide 12 (24), for which we determined a value of 20% ± 6.6%. In combination with the in vitro permeability measurements, this data suggest that the strategic N-methylations made to peptide 1 to produce 15 resulted in a peptide with improved membrane permeability potential in both model assays and the whole animal.
Fig. 5.
Relative plasma concentrations assessed by LCMS up to 8 h after administration of 15 at 10 mg/kg p.o. (olive oil) vs. 1 mg/kg i.v. (DMSO) in Wistar rats. The absolute oral bioavailability (F) of 15 was determined to be 33% ± 2.9% (n = 4; error in SD; pharmacokinetic data in SI Appendix, Table S2).
Solution NMR Studies.
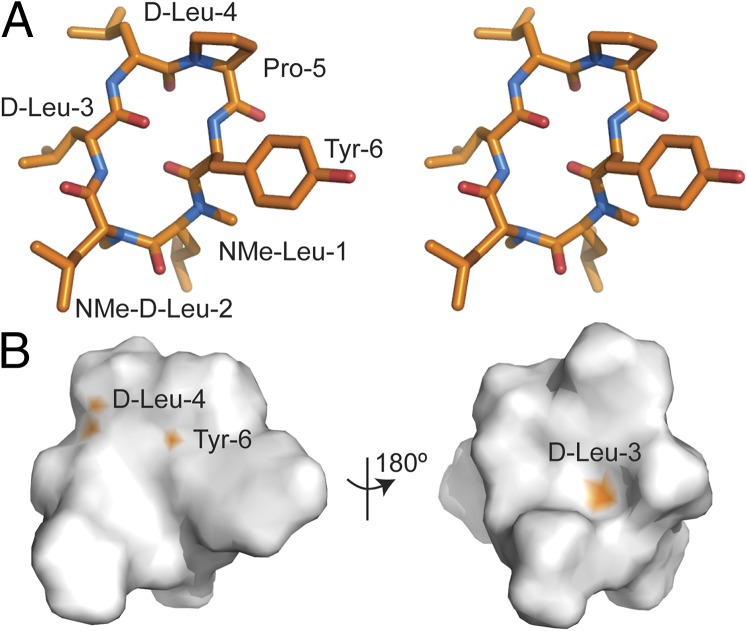
To provide structural insight into the high oral bioavailability of 15, we elucidated its solution structure by NMR spectroscopy, as shown in Fig. 6 and SI Appendix, Figs. S3 and S4. The amides of d-Leu-3 and Tyr-6, which displayed small ΔδNH/ΔT values (i.e., −3.34 ppb/K and −4.13 ppb/K), are involved in intramolecular hydrogen bonds with the carbonyls of Tyr-6 and d-Leu-3, respectively. The N-methyl groups of d-Leu-1 and d-Leu-2 protrude away from the core of the structure, pointing toward the solvent. Interestingly, the amide of d-Leu-4, which has a ΔδNH/ΔT of −1.17 ppb/K, is not involved in an intramolecular hydrogen bond but instead was shielded from the solvent by the side-chains of two neighboring leucine residues (i.e., d-Leu-3 and d-Leu-4).
Fig. 6.
Solution structure and surface representation of 15. (A) Stereo view of the tertiary structure of 15 elucidated by NMR spectroscopy. (B) Surface representation of 15. The solvent accessible area of the nonmethylated amides are colored in orange.
Application to a Therapeutic Peptide: Veber-Hirschmann Peptide.
We have so far demonstrated the utility of ΔδNH/ΔT on a set of model peptides that have no reported biological function. To test whether this approach could be used to improve the oral bioavailability of a peptide with bioactivity, we examined the N-methylated Veber-Hirschmann peptide (SI Appendix, Fig. S2), which is selective toward the somatostatin receptor subtypes sst2 and sst5 and has a reported oral bioavailability of 9.9% (19). We synthesized the nonmethylated analog and measured its ΔδNH/ΔT in 30% vol/vol acetonitrile for each constituent residue. The ΔδNH/ΔT values that we obtained are consistent with those previously measured for the nonmethylated analog in 100% vol/vol DMSO (26). As shown in SI Appendix, Fig. S2, four residues displayed very negative ΔδNH/ΔT values (i.e., d-Trp-1, Lys-2, Phe-4, and Phe-6); three of these residues (i.e., d-Trp-1, Lys-2, and Phe-4) correspond to the residues that were modified to improve oral bioavailability while maintaining bioactivity in a previous study (19).
Discussion
Poor oral bioavailability is a key limitation in the development of peptide therapeutics. In this study we were interested in investigating a method to help accelerate the design of membrane permeable peptide drug leads. We used a series of cyclic hexapeptide stereoisomers as a model system for studying the effect of N-methylation on membrane permeability. On the basis of a systematic analysis of ΔδNH/ΔT measurements acquired using NMR spectroscopy for nonmethylated and N-methylated peptides, we propose a structure-informed approach that potentially can be applied as a general method for improving the oral bioavailability of drug leads. Specifically, we found that ΔδNH/ΔT can be used to identify amides (of a nonmethylated or partially N-methylated peptide) that are suitable for selective N-methylation, which can lead to a cyclic peptide with passive membrane permeability. We validated this approach on a set of five cyclic hexapeptides and showed that we can achieve an oral bioavailability of 33%. We further demonstrated that this approach can be used to explain the improvement in membrane permeability of peptide drug leads by examining the Veber-Hirschmann peptide.
The approach is based on the observation that the amide chemical shift is dependent on temperature. The main reason for this effect is related to the length of a hydrogen bond between the amide proton and a nearby (intramolecular or intermolecular) hydrogen bond acceptor (27), which is strongly influenced by thermal motions induced by temperature increases. We found that amide temperature coefficients can be used to identify solvent-exposed amides for the cyclic peptides used in this study. This is not surprising: temperature coefficients have previously been used to determine hydrogen bonded and solvent-shielded amide protons in linear peptides and proteins (28). In general, we observed that amides with very negative temperature coefficients (i.e., <−4.6 ppb/K) displayed fast hydrogen–deuterium exchange rates, whereas amides having less-negative temperature coefficients (i.e., ≥−4.6 ppb/K) displayed slower exchange rates. It should be noted that many different methods for assigning this cutoff value have been proposed (25, 29, 30), suggesting that temperature coefficients that are close to the value of −4.6 ppb/K need to be interpreted with some caution. This finding is consistent with an earlier report using NMR amide temperature coefficients to probe the hydrogen bond network of cyclic disulfide-rich cyclic peptides (31).
We note that hydrogen–deuterium exchange rates can also be used as an indicator of hydrogen-bonding/solvent-exposure, and thus be used to identify amides for N-methylation in a manner similar to how amide temperature coefficients are used in this study. We focus on amide temperature coefficients in this study because they are easily measured in different solvent systems (i.e., composition and pH) compared with hydrogen–deuterium exchange rates, which can be tedious to measure owing to the need for solvent exchange and careful attention to the kinetics of the measurements.
In this approach, we use N-methylation of solvent accessible amides to confer solvation properties to peptides that are favorable for membrane permeability. We postulate that certain residues are better candidates for N-methylation, which can be identified by NMR amide temperature coefficient measurements. One of our designed peptides, 15, showed an increased permeability in vitro as well as a high oral bioavailability of 33%. It is clear that 15 is highly permeable not only in in vitro permeability assays but also within the gastrointestinal tract of a living rat, thus validating the design strategy.
The solution structure of 15, an N-methylated peptide that we designed, confirmed that the N-methyl groups are positioned at surface-exposed locations, whereas the remaining unmodified amides are involved in hydrogen bonds, with the exception of one amide, which is protected from the solvent by two leucine side-chains. This is consistent with the hypothesis that the sequestration of both amide carbonyl and amide groups leads to improved membrane permeability. However, a caveat of this method is that N-methylation of solvent-exposed amides may not always improve membrane permeability. For example, in a study by Kessler and coworkers (18), seven analogs of the cyclic hexapeptide cyclo(GRGDfL) with different solvent-exposed amides N-methylated were designed according to the bioactive conformation; however, the analogs did not show improved transport across the membrane. In these cases, other factors (such as sequence composition and backbone conformation) may be limiting their permeability. In a separate study, Kessler and coworkers (32) proposed that specific backbone conformations are crucial for regulating membrane permeability after extensive conformational studies of two highly permeable cyclic alanine-rich peptides with different N-methylation patterns. Nevertheless, the approach used here is still a rapid means of improving the solvation properties of peptides, and therefore useful within the drug design and optimization process.
It should be noted that N-methylation can in some cases significantly affect biological activity by either directly modifying interactions of the backbone with its target or changing the peptide conformation. However, we postulate that N-methylation of exposed amides is less disruptive to the peptide conformation compared with N-methylation of buried amides; therefore, biological activity is more likely to be retained. Indeed, it is possible for N-methylation to improve the membrane permeability of a drug lead without having a significant effect on its biological activity. For example, multiple N-methylation of a somatostatin analog improved its oral bioavailability without modifying its biological activity and selectivity (19). We showed that amide temperature coefficients could be used to rapidly identify the residues of the somatostatin analog that could be N-methylated, suggesting that the approach can be applied to peptides with therapeutic potential.
In summary, we have demonstrated that NMR amide temperature coefficients can be used as a structural marker to direct the design of membrane permeable peptides. By studying the relationship between conformation and permeability, we investigated an approach to identify solvent-exposed amides that could then be modified by N-methylation (or, in fact, with other chemical modifications). Furthermore, although the focus of this investigation was on using amide temperature coefficients as the structural marker, other measures, such as hydrogen–deuterium exchange rates, can also be used alone or in conjunction with amide temperature coefficients to direct peptide design. This strategy is advantageous over the traditional structure-directed design because it provides comparatively rapid information on the nature of all amides and potential sites for modification. We further demonstrated that this strategy can be applied to peptides with therapeutic potential.
Methods and Materials
General.
A detailed description of the methods and materials is provided in SI Appendix.
Peptide Synthesis.
Cyclic peptides were synthesized using Fmoc solid-phase peptide synthesis on 2-chlorotrityl chloride resin. Peptides were cleaved from the resin using 1% trifluoroacetic acid in dichloromethane. Head-to-tail cyclization was performed in DMF with 3 equivalent HATU and 6 equivalent DIPEA. Following removal of the solvent, side-chain protecting groups were removed in 94:3:3 TFA/TIPS (triisopropylsilane)/water. After cyclization, the peptides were purified by RP-HPLC. Purity of fractions was assessed using ESI-MS and analytical HPLC. A detailed description is provided in SI Appendix.
NMR Experiments.
All spectra were recorded on a Bruker ARX 500 MHz spectrometer. Chemical shifts were internally referenced to DSS at 0 ppm. Amide temperature coefficient measurements, hydrogen–deuterium exchange studies, and structure determination were carried out as described previously (31). A detailed description is provided in SI Appendix.
PAMPA and Caco-2 Permeability Studies.
A 96-well system preloaded with artificial lipid membrane was used (BD Biosciences); additional details are provided in SI Appendix. Caco-2 cells were cultured and seeded as described in SI Appendix. Caco-2 cell monolayers were used for experimentation 6 or 21 d after seeding. Acceptor and donor well concentrations were measured by LC-MS.
In Vivo Pharmacokinetic Studies.
Male Wistar rats (250 ± 20 g) were bred at, and obtained from, the Australia Animal Resource Centre (Canning Vale, WA). Rats were surgically implanted with a jugular vein catheter and fasted overnight, as previously described (33). Dosing, sample collection, and quantification were carried out as described in SI Appendix. All experiments were approved by the animal ethics committee of The University of Queensland.
Supplementary Material
Acknowledgments
We thank Olivier Cheneval and Phillip Walsh for help with peptide synthesis. Caco-2 cells were kindly provided by Prof. Istvan Toth. This work was supported by Australian Research Council Linkage Grant LP110200213, co-funded by Pfizer and a Queensland Government Smart Futures Co-investment Grant. C.K.W. was supported by a National Health and Medical Research Council (NHMRC) Early Career Research Fellowship (546578). D.P.F. and D.J.C. are NHMRC Senior Principal Research Fellows (APP1027369, APP1026501).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417611111/-/DCSupplemental.
References
- 1.Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81(1):136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 2.Vlieghe P, Lisowski V, Martinez J, Khrestchatisky M. Synthetic therapeutic peptides: Science and market. Drug Discov Today. 2010;15(1-2):40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Northfield SE, et al. Disulfide-rich macrocyclic peptides as templates in drug design. Eur J Med Chem. 2014;77:248–257. doi: 10.1016/j.ejmech.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Baeriswyl V, Heinis C. Polycyclic peptide therapeutics. ChemMedChem. 2013;8(3):377–384. doi: 10.1002/cmdc.201200513. [DOI] [PubMed] [Google Scholar]
- 5.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1-3):3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 6.Veber DF, et al. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 7.Busby RW, et al. Linaclotide, through activation of guanylate cyclase C, acts locally in the gastrointestinal tract to elicit enhanced intestinal secretion and transit. Eur J Pharmacol. 2010;649(1-3):328–335. doi: 10.1016/j.ejphar.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Bock JE, Gavenonis J, Kritzer JA. Getting in shape: Controlling peptide bioactivity and bioavailability using conformational constraints. ACS Chem Biol. 2013;8(3):488–499. doi: 10.1021/cb300515u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong CT, et al. Orally active peptidic bradykinin B1 receptor antagonists engineered from a cyclotide scaffold for inflammatory pain treatment. Angew Chem Int Ed Engl. 2012;51(23):5620–5624. doi: 10.1002/anie.201200984. [DOI] [PubMed] [Google Scholar]
- 10.Clark RJ, et al. The engineering of an orally active conotoxin for the treatment of neuropathic pain. Angew Chem Int Ed Engl. 2010;49(37):6545–6548. doi: 10.1002/anie.201000620. [DOI] [PubMed] [Google Scholar]
- 11.Borel JF, Feurer C, Gubler HU, Stähelin H. Biological effects of cyclosporin A: A new antilymphocytic agent. Agents Actions. 1976;6(4):468–475. doi: 10.1007/BF01973261. [DOI] [PubMed] [Google Scholar]
- 12.Rezai T, et al. Conformational flexibility, internal hydrogen bonding, and passive membrane permeability: Successful in silico prediction of the relative permeabilities of cyclic peptides. J Am Chem Soc. 2006;128(43):14073–14080. doi: 10.1021/ja063076p. [DOI] [PubMed] [Google Scholar]
- 13.Okumu FW, Pauletti GM, Vander Velde DG, Siahaan TJ, Borchardt RT. Effect of restricted conformational flexibility on the permeation of model hexapeptides across Caco-2 cell monolayers. Pharm Res. 1997;14(2):169–175. doi: 10.1023/a:1012092409216. [DOI] [PubMed] [Google Scholar]
- 14.Pauletti GM, et al. Structural requirements for intestinal absorption of peptide drugs. J Control Release. 1996;41(1-2):3–17. [Google Scholar]
- 15.Chatterjee J, Gilon C, Hoffman A, Kessler H. N-methylation of peptides: A new perspective in medicinal chemistry. Acc Chem Res. 2008;41(10):1331–1342. doi: 10.1021/ar8000603. [DOI] [PubMed] [Google Scholar]
- 16.Dechantsreiter MA, et al. N-Methylated cyclic RGD peptides as highly active and selective alpha(V)beta(3) integrin antagonists. J Med Chem. 1999;42(16):3033–3040. doi: 10.1021/jm970832g. [DOI] [PubMed] [Google Scholar]
- 17.Doedens L, et al. Multiple N-methylation of MT-II backbone amide bonds leads to melanocortin receptor subtype hMC1R selectivity: Pharmacological and conformational studies. J Am Chem Soc. 2010;132(23):8115–8128. doi: 10.1021/ja101428m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatterjee J, et al. Multiple N-methylation by a designed approach enhances receptor selectivity. J Med Chem. 2007;50(24):5878–5881. doi: 10.1021/jm701044r. [DOI] [PubMed] [Google Scholar]
- 19.Biron E, et al. Improving oral bioavailability of peptides by multiple N-methylation: Somatostatin analogues. Angew Chem Int Ed Engl. 2008;47(14):2595–2599. doi: 10.1002/anie.200705797. [DOI] [PubMed] [Google Scholar]
- 20.Ovadia O, et al. The effect of multiple N-methylation on intestinal permeability of cyclic hexapeptides. Mol Pharm. 2011;8(2):479–487. doi: 10.1021/mp1003306. [DOI] [PubMed] [Google Scholar]
- 21.Loosli HR, et al. Peptide conformations. Part 31. The conformation of cyclosporin A in the crystal and in solution. Helv Chim Acta. 1985;68(3):682–704. [Google Scholar]
- 22.In Y, Ishida T, Takesako K. Unique molecular conformation of aureobasidin A, a highly amide N-methylated cyclic depsipeptide with potent antifungal activity: X-ray crystal structure and molecular modeling studies. J Pept Res. 1999;53(5):492–500. doi: 10.1034/j.1399-3011.1999.00046.x. [DOI] [PubMed] [Google Scholar]
- 23.Steiner JR, Barnes CL. Crystal and molecular structure of Destruxin B. Int J Pept Protein Res. 1988;31(2):212–219. doi: 10.1111/j.1399-3011.1988.tb00025.x. [DOI] [PubMed] [Google Scholar]
- 24.White TR, et al. On-resin N-methylation of cyclic peptides for discovery of orally bioavailable scaffolds. Nat Chem Biol. 2011;7(11):810–817. doi: 10.1038/nchembio.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cierpicki T, Otlewski J. Amide proton temperature coefficients as hydrogen bond indicators in proteins. J Biomol NMR. 2001;21(3):249–261. doi: 10.1023/a:1012911329730. [DOI] [PubMed] [Google Scholar]
- 26.Kessler H, et al. Peptide conformations. 28. Relayed heteronuclear correlation spectroscopy and conformational analysis of cyclic hexapeptides containing the active sequence of somatostatin. J Am Chem Soc. 1983;105(23):6944–6952. [Google Scholar]
- 27.Wagner G, Pardi A, Wuthrich K. Hydrogen bond length and proteon NMR chemical shifts in proteins. J Am Chem Soc. 1983;105(18):5948–5949. [Google Scholar]
- 28.Hruby VJ. Conformations of peptides in solution as determined by NMR spectroscopy and other physical methods. In: Weinstein B, editor. Chemistry and Biochemistry of Amino Acids, Peptides and Proteins. Marcel Dekker; New York: 1974. pp. 1–188. [Google Scholar]
- 29.Andersen NH, et al. Extracting information from the temperature gradients of polypeptide NH chemical shifts. 1. The importance of conformational averaging. J Am Chem Soc. 1997;119(36):8547–8561. [Google Scholar]
- 30.Baxter NJ, Williamson MP. Temperature dependence of 1H chemical shifts in proteins. J Biomol NMR. 1997;9(4):359–369. doi: 10.1023/a:1018334207887. [DOI] [PubMed] [Google Scholar]
- 31.Wang CK, et al. Combined X-ray and NMR analysis of the stability of the cyclotide cystine knot fold that underpins its insecticidal activity and potential use as a drug scaffold. J Biol Chem. 2009;284(16):10672–10683. doi: 10.1074/jbc.M900021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beck JG, et al. Intestinal permeability of cyclic peptides: Common key backbone motifs identified. J Am Chem Soc. 2012;134(29):12125–12133. doi: 10.1021/ja303200d. [DOI] [PubMed] [Google Scholar]
- 33.Lohman RJ, et al. An antagonist of human protease activated receptor-2 attenuates PAR2 signaling, macrophage activation, mast cell degranulation, and collagen-induced arthritis in rats. FASEB J. 2012;26(7):2877–2887. doi: 10.1096/fj.11-201004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.