Significance
Inflammation is an important arm of host defense against microbial infections, but excessive inflammation can cause human diseases. Interferon regulatory factor 5 (IRF5) is a key regulator of inflammatory responses, controlling the expression of many proinflammatory cytokines. Mutations and dysregulation of IRF5 have been linked to autoimmune and autoinflammatory diseases in humans; however, how IRF5 is activated is not well understood. We report that the kinase IKKβ, which is known to regulate the Rel/nuclear factor kappa B (NF-κB) family of transcription factors, phosphorylates IRF5 at a specific serine residue, and that this phosphorylation is critical for IRF5 activation and cytokine production. Thus, IKKβ regulates two master transcription factors, NF-κB and IRF5, which coordinately control gene expression to mediate inflammatory responses.
Keywords: IRF5, IKK, TLR, inflammation, phosphorylation
Abstract
The transcription factor interferon regulatory factor 5 (IRF5) is essential for the induction of inflammatory cytokines, but the mechanism by which IRF5 is activated is not well understood. Here we present evidence that the kinase IKKβ phosphorylates and activates IRF5 in response to stimulation in several inflammatory pathways, including those emanated from Toll-like receptors and retinoic acid-inducible gene I–like receptors. IKKβ phosphorylates mouse IRF5 at specific residues, including serine 445 (S446 in human IRF5 isoform 1), as evidenced by mass spectrometry analysis and detection with a phosphospecific antibody. Recombinant IKKβ phosphorylated IRF5 at Ser-445 in vitro, and a point mutation of this serine abolished IRF5 activation and cytokine production. Depletion or pharmacologic inhibition of IKKβ prevented IRF5 phosphorylation. These results indicate that IKKβ is an IRF5 kinase that instigates inflammation.
The interferon (IFN) regulatory factor (IRF) family of transcription factors plays a pivotal role in the development of immune cells and induction of cytokines that are important in immune and inflammatory responses (1, 2). The mammalian IRF family consists of nine members, IRF1–9 (3). Among these, IRF3 and IRF7 have been studied extensively and shown to be important for the induction of type I IFNs and other cytokines in response to various stimuli, including viral infection. For example, infection with RNA viruses leads to the activation of retinoic acid-inducible gene I (RIG-I)–like receptors (RLRs), which in turn activate the mitochondrial adaptor protein MAVS (4–8). MAVS then activates the kinase TANK-binding kinase 1 (TBK1), which phosphorylates IRF3 and IRF7, causing these transcription factors to homodimerize and enter the nucleus to turn on type I IFNs. MAVS also activates the kinase IKKβ, which activates nuclear factor kappa B (NF-κB) to induce proinflammatory cytokines. Stimulation of some Toll-like receptors (TLRs), especially those localized on the endosomal membranes, such as TLR3, 4, 7, 8, and 9, also leads to strong activation of IRF3 and IRF7 to induce type I IFNs (3, 9).
Compared with IRF3 and IRF7, much less is known about how IRF5 is activated. However, genetic studies have provided compelling evidence for an essential role of IRF5 in the production of inflammatory cytokines, such as tumor necrosis factor α (TNF-α) and interleukin 6 (IL-6), in response to TLR ligands such as lipopolysaccharides (LPS) (10). IRF5 also functions together with IRF3 and IRF7 to mediate type I IFN production in response to viral infections (11). In addition, IRF5 plays important roles in M1 macrophage polarization (12) and IgG class switching in B cells (13). Polymorphisms in the IRF5 gene have been linked to human autoimmune diseases, including systemic lupus erythematosus (14) and Sjogren syndrome (15). Thus, IRF5 is critical for regulating immune and inflammatory responses in health and disease (16).
Similar to IRF3 and IRF7, IRF5 contains a DNA-binding domain (DBD), an IRF-association domain (IAD), and a serine-rich region (SRR) at the C terminus (17, 18). The SRR is phosphorylated in response to TLR stimulation or virus infection. The crystal structure of a human IRF5 mutant, S430D, which has been proposed to mimic IRF5 phosphorylation, shows that IRF5 forms a dimer (19); however, the physiological phosphorylation sites of IRF5 have not yet been identified or validated. The kinase that mediates IRF5 phosphorylation is also unknown.
Here we report the identification of IKKβ as a kinase that phosphorylates IRF5 at several serine residues, including Ser-445 in mouse IRF5 (equivalent to Ser-446 in human IRF5 isoform 1 and Ser-462 in human IRF5 isoform 2). A point mutation of S445 to alanine abolished the ability of IRF5 to induce inflammatory cytokines. By applying mass spectrometry and developing an antibody that specifically detects IRF5 phosphorylated at S445, we validated that this serine is phosphorylated in cells stimulated by LPS or by virus infection. Depletion or pharmacologic inhibition of IKKβ prevented the phosphorylation of IRF5 and induction of inflammatory cytokines. These results demonstrate that IKKβ is an IRF5 kinase that mediates inflammatory responses.
Results
IRF5 Forms a Dimer and Is Essential for Cytokine Induction by Multiple Innate Immunity Pathways.
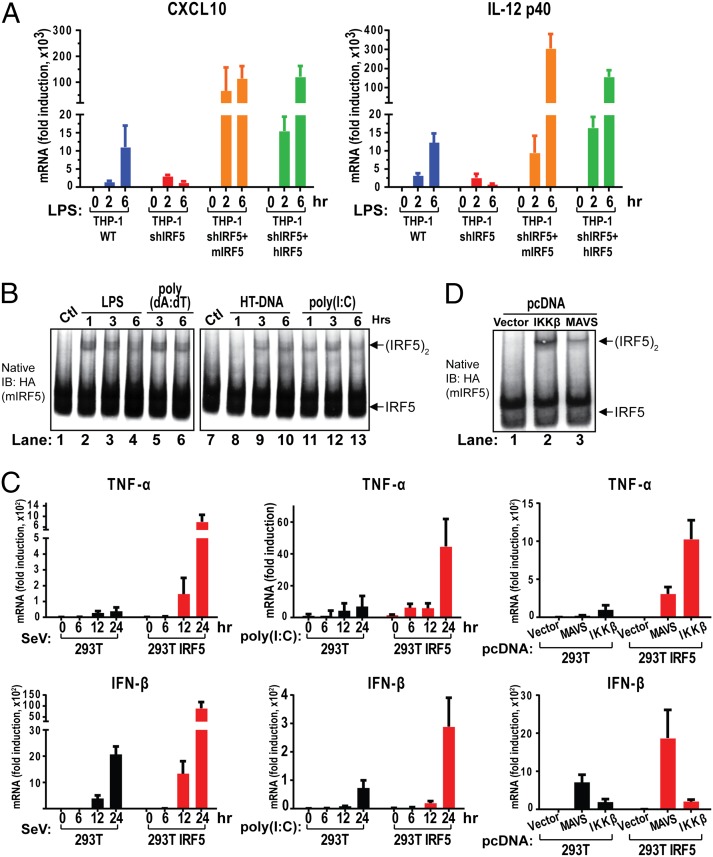
To investigate the function and active form of IRF5, we measured cytokine induction by LPS in THP1 cells depleted of endogenous IRF5 by shRNA and those reconstituted with mouse or human IRF5 (Fig. 1A and Fig. S1 A and B). The expression of chemokine (C-X-C motif) ligand 10 (CXCL10) and IL-12 (p40 subunit) was largely abolished when IRF5 was knocked down, but strongly induced when either human or mouse IRF5 was reinstalled; the higher induction levels in the IRF5 reconstituted cells were likely related to the higher levels of IRF5 (Fig. S1A). Similarly, LPS induction of IFN-β and several IFN-stimulated genes (ISGs), including IFIT3, RSAD2, and ISG15, was inhibited in the absence of IRF5 but restored when IRF5 was expressed (Fig. S1B).
Fig. 1.
IRF5 forms a dimer and mediates cytokine induction by diverse pathways. (A) Depletion of IRF5 abolishes LPS-induced cytokine production in THP-1 cells. Cells used in this experiment included WT (THP-1 WT), IRF5 knockdown (THP-1 shIRF5), IRF5 knockdown rescued with mouse IRF5 (THP-1 shIRF5+Flag-mIRF5-HA, labeled THP-1 shIRF5+mIRF5), and IRF5 knockdown rescued with human IRF5 (THP-1 shIRF5+HA-hIRF5, labeled THP-1 shIRF5+hIRF5). These cells were stimulated with 5 µg/mL LPS for the indicated time before total RNA was isolated. CXCL10 and IL-12 (p40 subunit) mRNA levels were analyzed by qRT-PCR. Unless indicated otherwise, error bars represent SDs of triplicate assays. (B) IRF5 forms dimer on activation. The THP-1 shIRF5+Flag-mIRF5-HA cell line as described in A was left untreated (control; Ctl) or stimulated by incubation with LPS (5 µg/mL) or transfection with poly(dA:dT) (2 µg/mL), HT-DNA (2 µg/mL), or poly(I:C) (2 µg/mL) for the indicated time. The formation of IRF5 dimer was analyzed by native PAGE, followed by immunoblotting with the HA antibody. (C) IRF5 promotes cytokine induction in 293T cells. WT 293T cells and cells stably expressing Flag-mIRF5-HA were stimulated with Sendai virus (SeV) or poly(I:C) (2 µg/mL) for the indicated time, followed by measurement of TNF-α and IFN-β RNA levels by qRT-PCR. (Right) The cells were transfected with empty pcDNA vector, pcDNA-Flag-MAVS (MAVS), or pcDNA-Flag-IKKβ (IKKβ) for 24 h before total RNA was isolated for analysis by qRT-PCR. (D) Overexpression of IKKβ or MAVS activates IRF5 in cells. The 293T Flag-mIRF5-HA cell line as described in C was transiently transfected with empty pcDNA vector or the vector containing Flag-MAVS or Flag-IKKβ for 24 h. Dimerization of IRF5 was analyzed by native PAGE, followed by immunoblotting with the HA antibody.
To test whether activated IRF5 forms a dimer, we stimulated THP1 cells stably expressing HA-tagged IRF5 with LPS as well as other stimuli, including poly(dA:dT) and herring testis DNA (HT-DNA), both of which are known to activate the cGAS cytosolic DNA-sensing pathway (20, 21). Poly(dA:dT) also activates the RIG-I pathway through RNA polymerase III (22, 23). We also transfected the cells with the double-stranded RNA analog poly(I:C), which is known to stimulate the RIG-I and MDA5 pathways (8). In each case, analysis by native PAGE, followed by immunoblotting showed that stimulation of the cells led to the formation of a more slowly migrating band that likely represents an IRF5 dimer, much like IRF3 dimerization following virus infection. We were not able to detect dimerization of endogenous IRF5 in THP1 cells, because the commercially available IRF5 antibody detected a strong nonspecific band at the expected IRF5 dimer position on the native gel. Thus, in the remainder of this work, we measured IRF5 activation with an IRF5 dimerization assay in THP1-HA-IRF5 stable cells or by immunoblotting with a phospho-IRF5–specific antibody (see below).
To further investigate the role of IRF5 activation in inflammatory cytokine induction, we stably expressed IRF5 in HEK293T cells, which do not have detectable expression of endogenous IRF5, and then stimulated the cells by poly(I:C) transfection or infection with Sendai virus, an RNA virus known to activate the RIG-I–MAVS pathway. In both cases, the induction of TNF-α and IFN-β was strongly enhanced in 293T-IRF5 cells compared with the parental cells. Sendai virus infection was capable of inducing IFN-β in the parental 293T cells, because these cells express IRF3. Thus, TNF-α induction by cytosolic RNA or RNA viruses is critically dependent on IRF5, whereas IFN-β induction is largely dependent on IRF3 but can be further enhanced by IRF5. These results suggest that the RIG-I pathway can activate both IRF3 and IRF5. Indeed, overexpression of MAVS led to the induction of both TNF-α and IFN-β (Fig. 1C, Right), as well as the dimerization of IRF5 (Fig. 1D).
Interestingly, overexpression of IKKβ strongly induced TNF-α expression and IRF5 dimerization but only weakly induced IFN-β expression (Fig. 1 C and D). The weak induction of IFN-β by IKKβ overexpression can be explained by the fact that IRF3 is phosphorylated by TBK1 and IKKε, but not by IKKβ (24, 25). These results raise the interesting possibility that IKKβ may play an important role in IRF5 activation.
IKKβ Activates IRF5 in Vitro and Is Important for IRF5 Activation in Cells.
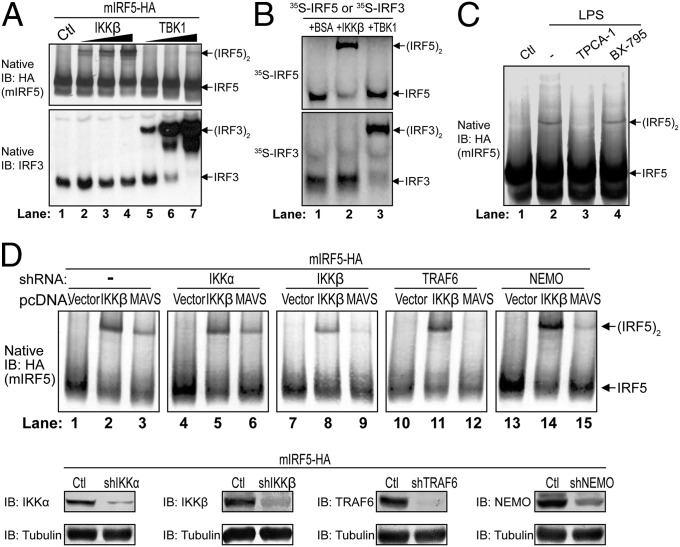
To obtain biochemical evidence for the role of IKKβ in IRF5 activation, we prepared cytosolic extracts from HEK293T cells stably expressing Flag-mIRF5-HA and incubated the extracts with recombinant IKKβ or TBK1 protein together with ATP. Native PAGE analyses of the reaction mixtures revealed that IKKβ caused the dimerization of IRF5, but not of endogenous IRF3, whereas TBK1 had the opposite effects (Fig. 2A). We also incubated in vitro-translated, 35S-labeled IRF5 or IRF3 with IKKβ or TBK1 and found that IRF5 dimerization was specifically induced by IKKβ, but not by TBK1 (Fig. 2B).
Fig. 2.
IKKβ activates IRF5 in vitro and in cells. (A and B) IKKβ activates IRF5 in vitro. (A) A cytosolic fraction (S20) from the 293T Flag-mIRF5-HA cell line was incubated with purified IKKβ or TBK1 protein in the presence of ATP. Dimerization of IRF5 or IRF3 was analyzed by native PAGE, followed by immunoblot analysis. (B) In vitro translated 35S-IRF5 or 35S-IRF3 protein was incubated with BSA, IKKβ, or TBK1 in the presence of ATP. Dimerization of IRF5 or IRF3 was analyzed by native PAGE, followed by autoradiography. Ctl, control cytosolic fraction without kinase. (C) IKKβ inhibitor blocks IRF5 activation by LPS. THP-1 shIRF5 cells stably reconstituted with Flag-mIRF5-HA were treated with IKKβ inhibitor (TPCA-1; 20 µM) or TBK1 inhibitor (BX-795; 10 µM) for 2 h before stimulation with LPS (5 µg/mL) for 2 h. IRF5 activation was analyzed by native PAGE and immunoblotting. Ctl, DMSO control. (D) Knockdown of IKKβ or TRAF6 abolishes IRF5 activation by MAVS. IKKα, IKKβ, TRAF6, or NEMO was stably knocked down in 293T Flag-mIRF5-HA cells using lentiviral shRNA as indicated. These cells were transfected with empty pcDNA vector, pcDNA-Flag-MAVS, or pcDNA-Flag-IKKβ for 24 h. (Upper) Activation of IRF5 was analyzed by native PAGE and immunoblot analysis. (Lower) Knockdown efficiency for each gene was analyzed by immunoblot analysis.
To test which kinase is important for IRF5 activation in cells, we treated THP1 cells stably expressing Flag-mIRF5-HA with the IKKβ inhibitor TPCA-1 or TBK1 inhibitor BX-795, and then stimulated the cells with LPS. TPCA1, but not BX-795, inhibited IRF5 dimerization, suggesting that IKKβ is responsible for the LPS-induced dimerization of IRF5.
To further examine the role of IKKs and other signaling molecules in IRF5 activation, we used shRNA to stably knock down the expression of IKKα, IKKβ, TNF receptor-associated factor 6, or NF-κB essential modulator (NEMO) in HEK293T cells stably expressing Flag-mIRF5-HA. These cells were transfected with IKKβ or MAVS, followed by analysis of IRF5 dimerization by native PAGE. The results show that IKKβ, TRAF6, and NEMO, but not IKKα, were required for IRF5 dimerization induced by MAVS. IKKβ knockdown partially inhibited IRF5 dimerization induced by IKKβ overexpression, presumably because the shRNA only partially reduced the IKKβ level. Knocking down other proteins, including IKKα, TRAF6, and NEMO, had little effect on IRF5 activation by IKKβ. Taken together, these results suggest that TRAF6, NEMO, and IKKβ mediate IRF5 activation by MAVS.
Phosphorylation of IRF5 at Ser-445 by IKKβ Is Important for Cytokine Induction.
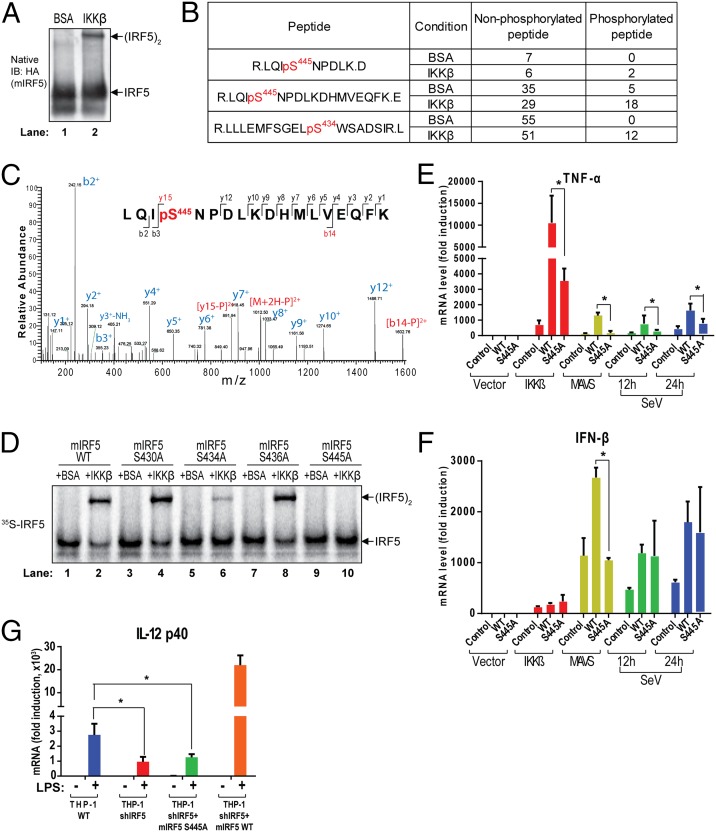
To map the phosphorylation site(s) of IRF5, we incubated Flag-mIRF5-HA, which was partially purified from HEK293T cells stably expressing the protein, with IKKβ or with BSA (as a control) in the presence of ATP and Mg2+ at 30 °C for 1 h. IKKβ, but not BSA, caused IRF5 dimerization in this reaction (Fig. 3A). The IRF5 protein from these reaction mixtures was further purified and analyzed by mass spectrometry, which revealed that peptides containing phosphorylated Ser-445 and Ser-434 of mIRF5 were greatly enriched in the reactions that contained IKKβ, whereas the total counts of mIRF5 peptides were similar in both reactions (Fig. 3 B and C and Fig. S2 A–D). In addition, we also detected mIRF5 peptides containing phosphorylation at Ser-430 and 436 (Fig. S2E).
Fig. 3.
Mapping and functional analysis of IRF5 phosphorylation sites. (A) IKKβ activates IRF5 in vitro. IRF5 partially purified from 293T Flag-mIRF5-HA cells was incubated with IKKβ or BSA in the presence of ATP. Activation of IRF5 was analyzed by native PAGE and immunoblot analysis. (B) IKKβ phosphorylates IRF5 at Ser-445 and Ser-434. IRF5 in reaction mixtures described in A was purified with a Flag antibody and then analyzed by tandem mass spectrometry. The sequences of the peptides and number of nonphosphorylated and phosphorylated peptides in each condition are shown. (C) Representative tandem mass spectrum (MS2) after HCD fragmentation of the ion with m/z = 1061.50 (z = 2+) indicating phosphorylation at S445. “b” and “y” ions with or without neutral loss are labeled in blue. Diagnostic ions for phosphorylation are highlighted in red. (D) Serine 445 is essential for IRF5 activation by IKKβ in vitro. WT or mutant 35S-IRF5 proteins were translated in vitro and incubated with IKKβ or BSA in the presence of ATP. Dimerization of IRF5 was analyzed by native PAGE, followed by autoradiography. (E–G) Serine 445 of IRF5 is required for cytokine induction in cells. (E and F) 293T cell lines stably expressing WT or S445A IRF5 were transfected with expression vectors for IKKβ or MAVS for 24 h, or infected with Sendai virus for the indicated time. Total RNA was isolated for the measurement of TNF-α and IFNβ RNA levels by qRT-PCR. (G) WT (THP-1 WT), IRF5 knockdown (THP-1 shIRF5), and IRF5 knockdown and rescued with WT or S445A mouse IRF5 (THP-1 shIRF5+mIRF5 WT or THP-1 shIRF5+mIRF5 S445A) THP-1 cell lines were stimulated with 5 µg/mL LPS for 6 h before total RNA was isolated. IL-12 p40 mRNA levels were analyzed by qRT-PCR. *P < 0.05, a statistically significant difference.
To test which serine residues are important for IRF5 activation by IKKβ, we mutated each serine residue identified above to alanine, in vitro translated the mutant proteins in the presence of [35S]methionine, and used the proteins in reactions that contained IKKβ or BSA (Fig. 3D). Among the mutants tested, the S445A mutation completely inhibited, and S434A mutation partially inhibited, IRF5 dimerization, whereas the other mutations had little inhibitory effect. Interestingly, Ser-434 and Ser-445 are the most conserved residues among IRF5 proteins from different species and are homologous to Ser-385 and Ser-396, respectively, of human IRF3 (Fig. S2D), known critical phosphorylation sites essential for type I IFN induction (26).
A previous study showed that a S480A mutation in human IRF5 (equivalent to S439A of mouse IRF5) impaired its ability to induce IFN-α (18). When this serine was mutated to aspartic acid, (S430D in the version of human IRF5 used in the study), IRF5 formed a dimer whose crystal structure was solved (19). Therefore, we mutated this residue (S439A in mouse IRF5) as well as other serine residues (S430A and S445A) and transfected them into HEK293T cells together with IKKβ or MAVS. IRF5-S445A failed to dimerize in response to stimulation by IKKβ or MAVS, whereas the S430A and S439A mutations had no effect (Fig. S3A). Immunoblot analysis showed that the IRF5 serine mutants were expressed at similar levels to that of WT IRF5 (Fig. S3A, Lower). The S445A mutation abrogated the ability of IRF5 to stimulate the induction of TNF-α in response to IKKβ, MAVS, or Sendai virus infection (Fig. 3E), whereas mutations at other serine residues did not have significant inhibitory effects (Fig. S3B).
We also tested the IRF5 S445D mutant and found that this mutation largely inhibited IRF5 dimerization (Fig. S3C) and abrogated the ability of IRF5 to boost TNF-α induction by IKKβ (Fig. S3D). Thus, the S445D mutation does not appear to mimic the effect of phosphorylation. The S445A mutation also partially inhibited IFN-β induction by MAVS, but did not significantly affect IFN-β induction by Sendai virus (Fig. 3F), presumably because IRF3 plays a dominant role in IFN-β induction in response to Sendai virus infection.
As shown previously, IKKβ only weakly induced IFN-β in a manner independent of IRF5, again consistent with a dominant role of IRF3 in IFN-β induction (Fig. 3F). To determine the role of IRF5 phosphorylation in TLR signaling, we established a THP-1 stable cell line depleted of endogenous IRF5 and reconstituted with WT IRF5 or the S445A mutant. The S445A mutation largely abrogated the ability of IRF5 to induce IL-12 in response to LPS stimulation (Fig. 3G). Taken together, these results suggest that IKKβ phosphorylates mIRF5 at Ser-445, and that this phosphorylation is important for inflammatory cytokine induction.
Detection of IRF5 Phosphorylation at Ser-445 with a Phosphospecific Antibody.
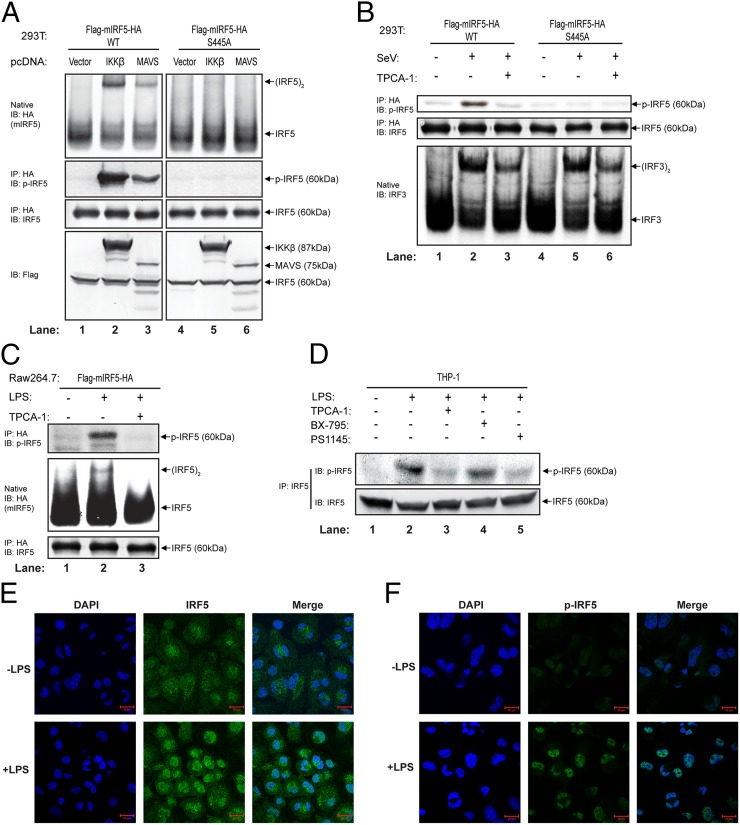
To further investigate IRF5 phosphorylation in cells, we developed an antibody that recognizes IRF5 phosphorylated at Ser-445 by immunizing rabbits with a synthetic phosphopeptide (IRLQIpS445NPDLC) corresponding to amino acids 440–450 of mouse IRF5 (identical to residues 441–451 of human IRF5). To test the specificity of this antibody, we transfected 293T cells stably expressing WT or the S445A mutant of Flag-mIRF5-HA with an expression vector encoding IKKβ or MAVS, both of which stimulated dimerization of WT, but not S445A, IRF5. Immunoprecipitation with the HA antibody followed by immunoblotting with the pIRF5 antibody showed that the antibody selectively detected WT, but not S445A IRF5, after stimulation (Fig. 4A), confirming that this antibody is specific for IRF5 phosphorylated at Ser-445.
Fig. 4.
IKKβ-dependent phosphorylation of IRF5 at Serine 445 in response to virus infection and LPS stimulation. (A) 293T cells stably expressing WT or S445A Flag-mIRF5-HA were transfected with expression vectors for IKKβ or MAVS for 24 h. (Upper) Aliquots of the cell extracts were analyzed for IRF5 dimerization by native PAGE, whereas other aliquots were immunoprecipitated with the HA antibody, followed by immunoblotting with an antibody against IRF5 or phosphorylated IRF5 at Ser-445. Expression of IKKβ and MAVS was examined by immunoblotting with the Flag antibody (Lower). (B) 293T cell lines as described above were treated with or without 20 µM TPCA-1 for 2 h before being infected with Sendai virus for 24 h. (Upper) IRF5 was immunoprecipitated with an HA antibody, followed by immunoblotting with an antibody against IRF5 or phosphorylated IRF5. Dimerization of IRF3 was detected by native PAGE and immunoblot analysis (Lower). (C) Raw 264.7 cell stably expressing Flag-mIRF5-HA was treated with or without TPCA-1 (20 µM) for 2 h before being stimulated with LPS (5 µg/mL) for 2 h. IRF5 was immunoprecipitated with an HA antibody, followed by immunoblotting with an antibody against IRF5 or phosphorylated IRF5. Dimerization of IRF5 was detected by immunoblotting of cytosolic extracts. (D) THP-1 cells were treated with or without the IKKβ inhibitors (TPCA-1 and PS1145) or TBK1 inhibitor (BX-795) for 2 h before stimulation with LPS (5 µg/mL) for 2 h. Phosphorylated IRF5 was immunoprecipitated with an IRF5 antibody, followed by immunoblotting with the same antibody or the phospho-IRF5 (S445) antibody. (E and F) Phosphorylated IRF5 accumulates in the nucleus. Differentiated THP-1 cells were stimulated with LPS for 2 h. Nuclear translocation and phosphorylation of IRF5 were monitored by confocal immunofluorescence using antibodies against IRF5 (E) or p-IRF5 (F).
To determine whether IRF5 is phosphorylated at Ser-445 in response to physiological stimuli, we infected 293T cells stably expressing WT or S445A Flag-mIRF5-HA with Sendai virus. Immunoblotting with the p-IRF5 antibody confirmed that WT, but not S445A IRF5, was phosphorylated in the virus-infected cells, and that this phosphorylation was abolished by the IKK inhibitor TPCA1 (Fig. 4B, Upper). Sendai virus-induced dimerization of endogenous IRF3 was not affected by overexpression of WT or S445A IRF5 and was only partially inhibited by TPCA1 (Fig. 4B, Lower). LPS stimulation of the macrophage cell line Raw264.7 stably expressing Flag-mIRF5-HA also led to IKK-dependent phosphorylation of IRF5 at Ser-445 (Fig. 4C).
To test wheteher endogenous IRF5 is phosphorylated at Ser-445, we stimulated THP1 cells with LPS and then immunoprecipitated IRF5 with an IRF5 antibody, followed by immunoblotting with the p-IRF5 antibody (Fig. 4D). We also tested the effect of several kinase inhibitors on IRF5 phosphorylation and found that only IKKβ inhibitors (TPCA-1 and PS1145), and not TBK1 inhibitor (BX-795), could inhibit the phosphorylation of IRF5 at Ser-445 in response to LPS (Fig. 4D). Finally, we performed immunofluorescence analyses in THP1 cells using IRF5 and p-IRF5 antibodies. Consistent with previous reports (27), IRF5 translocated into the nucleus in response to LPS stimulation (Fig. 4E). Importantly, p-IRF5 signal was barely detectable in the absence of stimulation, and LPS stimulation led to accumulation of p-IRF5 in the nucleus (Fig. 4F). These experiments demonstrate that LPS stimulates the phosphorylation of endogenous IRF5 at Ser-445 and its subsequent translocation to the nucleus.
Discussion
In this report, we present evidence that IKKβ is an IRF5 kinase and identify Ser-445 of mouse IRF5 (Ser-446 of human IRF5) as a critical phosphorylation site essential for IRF5 to induce cytokines. We have developed an antibody specific for IRF5 phosphorylated at Ser-445, and used this antibody to demonstrate that IRF5 is phosphorylated at Ser-445 in an IKKβ-dependent manner in response to LPS stimulation or Sendai virus infection. Our results suggest that IKKβ plays a crucial role in activating both NF-κB and IRF5, two master regulators of proinflammatory cytokines.
IKKβ is activated by a variety of stimulatory agents, including inflammatory cytokines and microbial pathogens that activate different pattern recognition receptors (28, 29). Consistent with the pleiotropic functions of IKKβ, we found that IRF5 is activated by multiple pathways, including those that engage TLRs and cytosolic DNA and RNA sensors. Not all stimuli that activate IKKβ are capable of activating IRF5, however; for example, we found that TNF-α treatment or MyD88 overexpression, both known to strongly stimulate IKKβ, could not activate IRF5 (Fig. S4). Thus, IRF5 activation requires other signals in addition to IKKβ. A similar scenario was recently reported in the cytosolic DNA-sensing pathway, which uses the adaptor protein STING to not only activate TBK1, but also recruit IRF3, thereby specifying the phosphorylation of IRF3 by TBK1 (30). It is possible that similar adaptor proteins may be engaged by TLR and other pathways to recruit IRF5 for phosphorylation by IKKβ.
Through mass spectrometry, we identified several serine residues on mIRF5 that are phosphorylated by IKKβ, including Ser-430, 434, 436, and 445. Our functional analyses showed that Ser-445, and to a lesser extent Ser-434, is required for IRF5 dimerization, whereas mutations of other serine residues had no effect. These results differ from those of a previous report showing that Ser-436 and Ser-439 (equivalent to Ser-477 and Ser-480 in the human IRF5 used in the previous study) were important for IFN-α induction (18). Importantly, Ser-434 and 445 of mIRF5 are homologous to Ser-385 and 396 of human IRF3, and they reside in a highly conserved region (Fig. S2D) (26). The p-IRF5 antibody that we developed clearly detected the phosphorylation of IRF5 at Ser-445 in cells stimulated with LPS or infected with Sendai virus, consistent with the phosphorylation of IRF3 at Ser-396 in response to RNA virus infection. Collectively, our results demonstrate that Ser-445 is phosphorylated by IKKβ in cells in response to stimulation, and that this phosphorylation is critical for IRF5 activation.
It is interesting that despite homologous domain structures and considerable sequence similarities between IRF5 and IRF3, these proteins are phosphorylated by distinct but homologous kinases, IKKβ and TBK1, respectively. It has been reported that IKKα is responsible for the phosphorylation of IRF7 in response to stimulation of endosomal TLRs, such as TLR7 and TLR9 (31). Thus, IKK and IKK-like kinases may be largely responsible for the activation of IRFs, and further work is needed to identify the kinase specific for each IRF. Future research should also explore the biochemical basis for the specificity of IRF phosphorylation by a cognate IKK or IKK-like kinase. In the case of IRF5, which is essential for the production of inflammatory cytokines and has been closely linked to human autoimmune diseases (16), the work reported here, which includes the discovery of IKKβ as an IRF5 kinase, identification of Ser-445 of mIRF5 (Ser-446 of human IRF5) as a critical phosphorylation site, and development of antibody that recognizes phosphorylated IRF5 at Ser-445, should facilitate further research on the mechanism of IRF5 activation and its role in human diseases.
Materials and Methods
Antibodies and Other Reagents.
The following antibodies were used in this study: IRF3, IKKα, TRAF6, NEMO (Santa Cruz Biotechnology), phospho-IKKα/β, phospho-TBK1, IκBα, phospho-IκBα (Cell Signaling), Flag antibody (M2), Tubulin, M2-conjugated agarose, and anti-HA-conjugated agarose (Sigma-Aldrich), HA (Thermo Scientific), and IRF5 (Abcam). The antibody against phosphor-Ser445 IRF5 was generated by immunizing rabbits with a synthetic peptide (IRLQIpS445NPDLC). LPS, HT-DNA, poly(dA:dT), and poly(I:C) were obtained from Sigma-Aldrich. Plasmid and DNA or RNA ligands were transfected into cells using Lipofectamine 2000 (Life Technologies). The kinase inhibitors were dissolved in DMSO and used at the following final concentrations: TBK1 inhibitor (BX795; Selleckchem), 10 μM; IKKβ inhibitor (TPCA-1; Sigma-Aldrich), 20 μM; IKKβ inhibitor (PS1145; Sigma-Aldrich), 20μM. GST-IKKβ and GST-TBK1 recombinant proteins were expressed and purified from Sf9 cells.
Expression Constructs.
For expression in mammalian cells, cDNA encoding N-terminal Flag- or HA-tagged mouse IRF5 S430A, IRF5 S434A, IRF5 S436A, and IRF5 S439A were cloned into pcDNA3. HA-tagged mouse IRF5 WT, IRF5 S445A and human IRF5 WT were cloned into pcDNA3 and pTY-EF1a-GFP-IRES-hygroR lentiviral vectors. Mutants were constructed with the QuikChange Site-Directed Mutagenesis Kit (Stratagene).
Partial Purification of IRF5 for in Vitro Reaction.
Because IRF5 spontaneously forms dimer when the protein is affinity-purified with a purification tag (e.g., Flag, GST), we attempted to partially purify IRF5 from the 293T FG-mIRF5-HA stable cell line. Cytosolic extracts from these cells were first fractionated using a HiTrap Heparin HP column (GE Healthcare). Fractions containing IRF5, as detected by immunoblot analysis, were concentrated and buffer- exchanged three times with hypotonic buffer (20 mM Tris⋅HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2) using Amicon Ultra 0.5-mL centrifugal filters (Millipore). The partially purified IRF5 was used for in vitro assays.
Purification of IRF5 for Mapping Phosphorylation Sites.
To determine the phosphorylation site(s) induced by IKKβ, reaction mixture (60 µL) containing 20 mM Hepes-KOH (pH 7.0), 2 mM ATP, 5 mM MgCl2, 40 µL of partially purified Flag-mIRF5 from the 293T stable cell line, and 2 µg Flag-IKKβ or BSA was incubated at 30 °C for 1 h, followed by incubation with M2-conjugated agarose at 4 °C for 4 h. The beads were washed three times with lysis buffer containing 150 mM NaCl and 1% Triton X-100. Bound proteins were then eluted by boiling in 2× Laemmli Sample Buffer before SDS/PAGE and silver staining. Gel slices from each lane were excised and digested with trypsin in situ. Digested samples were subjected to mass spectrometry using Q Exactive, and raw data were analyzed using Mascot (Matrix Science).
Confocal Microscopy.
THP-1 cells (4 × 105) were seeded and differentiated with 50 nM phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) for 48 h and then cultured for another 48 h by replacing the PMA-containing media with fresh media without PMA. The differentiated cells were left unstimulated or stimulated with LPS for 2 h. The cells were immunostained with IRF5 antibody (Abcam; ab21689) or phosphospecific IRF5 antibody. The images were acquired and processed with the Zeiss LSM 700 confocal laser scanning microscope system.
Note Added in Proof.
Cohen and coworkers have independently identified IKKβ as an IRF5 kinase (32). The phosphorylation site Ser-462 of human IRF5 isoform 2 in their paper is equivalent to Ser-446 of human IRF5 isoform 1 in our paper.
Supplementary Material
Acknowledgments
We thank Drs. Lijun Sun and Siqi Liu for assistance with protein purification and phospho-specific antibody production. This work was supported by grants from the National Institutes of Health (R01 AI093967) and the Cancer Prevention and Research Institute of Texas (CPRIT; RP120718). J.R. was supported by a CPRIT predoctoral training fellowship (RP140110). Z.J.C. is an Investigator at the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 17348.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1418516111/-/DCSupplemental.
References
- 1.Honda K, Taniguchi T. IRFs: Master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6(9):644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 2.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 3.Ikushima H, Negishi H, Taniguchi T. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb Symp Quant Biol. 2013;78:105–116. doi: 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- 4.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 5.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437(7062):1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 6.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 7.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19(6):727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 9.Noppert SJ, Fitzgerald KA, Hertzog PJ. The role of type I interferons in TLR responses. Immunol Cell Biol. 2007;85(6):446–457. doi: 10.1038/sj.icb.7100099. [DOI] [PubMed] [Google Scholar]
- 10.Takaoka A, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434(7030):243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 11.Lazear HM, et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 2013;9(1):e1003118. doi: 10.1371/journal.ppat.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krausgruber T, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12(3):231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 13.Lien C, et al. Critical role of IRF-5 in regulation of B-cell differentiation. Proc Natl Acad Sci USA. 2010;107(10):4664–4668. doi: 10.1073/pnas.0911193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham RR, et al. Argentine and Spanish Collaborative Groups A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38(5):550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 15.Miceli-Richard C, et al. Association of an IRF5 gene functional polymorphism with Sjögren’s syndrome. Arthritis Rheum. 2007;56(12):3989–3994. doi: 10.1002/art.23142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazzari E, Jefferies CA. IRF5-mediated signaling and implications for SLE. Clin Immunol. 2014;153(2):343–352. doi: 10.1016/j.clim.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Barnes BJ, Moore PA, Pitha PM. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J Biol Chem. 2001;276(26):23382–23390. doi: 10.1074/jbc.M101216200. [DOI] [PubMed] [Google Scholar]
- 18.Barnes BJ, Kellum MJ, Field AE, Pitha PM. Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol Cell Biol. 2002;22(16):5721–5740. doi: 10.1128/MCB.22.16.5721-5740.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W, et al. Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat Struct Mol Biol. 2008;15(11):1213–1220. doi: 10.1038/nsmb.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4(5):491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 25.Sharma S, et al. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300(5622):1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 26.Hiscott J, Lin R, Nakhaei P, Paz S. MasterCARD: A priceless link to innate immunity. Trends Mol Med. 2006;12(2):53–56. doi: 10.1016/j.molmed.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Schoenemeyer A, et al. The interferon regulatory factor, IRF5, is a central mediator of Toll-like receptor 7 signaling. J Biol Chem. 2005;280(17):17005–17012. doi: 10.1074/jbc.M412584200. [DOI] [PubMed] [Google Scholar]
- 28.Israël A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb Perspect Biol. 2010;2(3):a000158. doi: 10.1101/cshperspect.a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu F, Xia Y, Parker AS, Verma IM. IKK biology. Immunol Rev. 2012;246(1):239–253. doi: 10.1111/j.1600-065X.2012.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5(214):ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoshino K, et al. IkappaB kinase-alpha is critical for interferon-alpha production induced by Toll-like receptors 7 and 9. Nature. 2006;440(7086):949–953. doi: 10.1038/nature04641. [DOI] [PubMed] [Google Scholar]
- 32.Lopez-Pelaez M, et al. Protein kinase IKKβ-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc Natl Acad Sci USA. 2014;111:17432–17437. doi: 10.1073/pnas.1418399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.