Significance
Mechanical signaling plays many roles in cell physiology, including stem cell differentiation and reprogramming. To better understand the roles of mechanical forces in stem cells, we created genetically coded probes for actin that enables the direct measurement of forces in actin in living cells. This is the first force probe created for oligomeric proteins. We reprogrammed HEK-293 and Madin-Darby canine kidney cells into stem-like cells by culturing them on a soft substrate without using transcription factors. The mechanical properties of the microenvironment, and thus the local forces, promote cell reprogramming. Surprisingly, the cells showed close association of stemness to high tension in actin. Given the universal presence of actin in animal cells, the actin force probes have broad applications in biology.
Keywords: actin, force probe, stem cell, reprogramming, cell mechanics
Abstract
Cell mechanics plays a role in stem cell reprogramming and differentiation. To understand this process better, we created a genetically encoded optical probe, named actin–cpstFRET–actin (AcpA), to report forces in actin in living cells in real time. We showed that stemness was associated with increased force in actin. We reprogrammed HEK-293 cells into stem-like cells using no transcription factors but simply by softening the substrate. However, Madin-Darby canine kidney (MDCK) cell reprogramming required, in addition to a soft substrate, Harvey rat sarcoma viral oncogene homolog expression. Replating the stem-like cells on glass led to redifferentiation and reduced force in actin. The actin force probe was a FRET sensor, called cpstFRET (circularly permuted stretch sensitive FRET), flanked by g-actin subunits. The labeled actin expressed efficiently in HEK, MDCK, 3T3, and bovine aortic endothelial cells and in multiple stable cell lines created from those cells. The viability of the cell lines demonstrated that labeled actin did not significantly affect cell physiology. The labeled actin distribution was similar to that observed with GFP-tagged actin. We also examined the stress in the actin cross-linker actinin. Actinin force was not always correlated with actin force, emphasizing the need for addressing protein specificity when discussing forces. Because actin is a primary structural protein in animal cells, understanding its force distribution is central to understanding animal cell physiology and the many linked reactions such as stress-induced gene expression. This new probe permits measuring actin forces in a wide range of experiments on preparations ranging from isolated proteins to transgenic animals.
Cells have only three sources of free energy—chemical, electrical, and mechanical potential (1)—and life is driven by the flow of energy between them. The flow of mechanical energy has not been well studied in living cells outside of muscle, yet all cells are subject to endogenous and exogenous mechanical forces. The lack of progress in measuring these forces (stress) has primarily been due to a lack of suitable probes, but that has now been remedied with the development of genetically coded FRET-based force sensors (2). The literature shows many macroscopic effects of mechanical stress on cellular processes including cell motility, embryogenesis, stem cell replication and differentiation (3, 4), bone and muscle homeostasis, gene expression, protein folding (5), and membrane potential (6). However, these studies lacked the ability to measure stress in specific structural proteins, and the analyses have tended to treat cytoskeletal stresses as uniform, which we now know is not true (2, 7). We, and other groups (8), have shown that the distribution of forces is nonuniform in both time and space and protein specificity (9).
The force sensors consist of a FRET pair (typically CFP/YFP) connected by a protein linker, a biological analog of a mechanical spring (2, 8, 10). We have used a variety of different linkers and some that match the compliance of the host protein (7, 11, 12). All of the published work on force probes has required a host protein that is linear, so the probes can be coded into the DNA of the host protein. However, several key fibrous structural proteins, such as actin and tubulin, have multimeric structures, so a new approach was required to insert the sensors. Here we present, to our knowledge, the first probe to measure force in a polymeric protein, f-actin, that fulfills many structural and motor functions in cells. For example, f-actin forms filaments that bridge chromatin domains in the nucleus to the cell membrane and the extracellular matrix, providing a path for the well-known effects of mechanical stress on gene expression (13), stem cell differentiation, and mobility (14, 15).
Our actin probe, named actin–cpstFRET (circularly permuted stretch sensitive FRET)–actin (AcpA), consists of a dipole orientation-based FRET sensor called cpstFRET flanked with β-actin monomers. The sensor consists of a pair of tandemly connected circularly permutated CFP/YFPs (cpCerulean/cpVenus; Fig. 1, Inset) (7). Mechanical loading twists the dipole orientation from near parallel to diagonal, decreasing FRET efficiency. The angular sensor has a much wider dynamic range and higher sensitivity than the sensors with linear linkers that use the distance dependence of FRET (7). The sensor shows ∼80% energy transfer at rest and reductions as low as 20% with physiologically relevant forces (7). Perhaps because the sensor somewhat resembles the structure of actin dimers, the actin probe AcpA expresses well and labels f-actin efficiently, presumably at random locations. For imaging purposes, we found that, as previously published, polarized FRET (16–18) reduces bleed-through corrections and provides increased contrast.
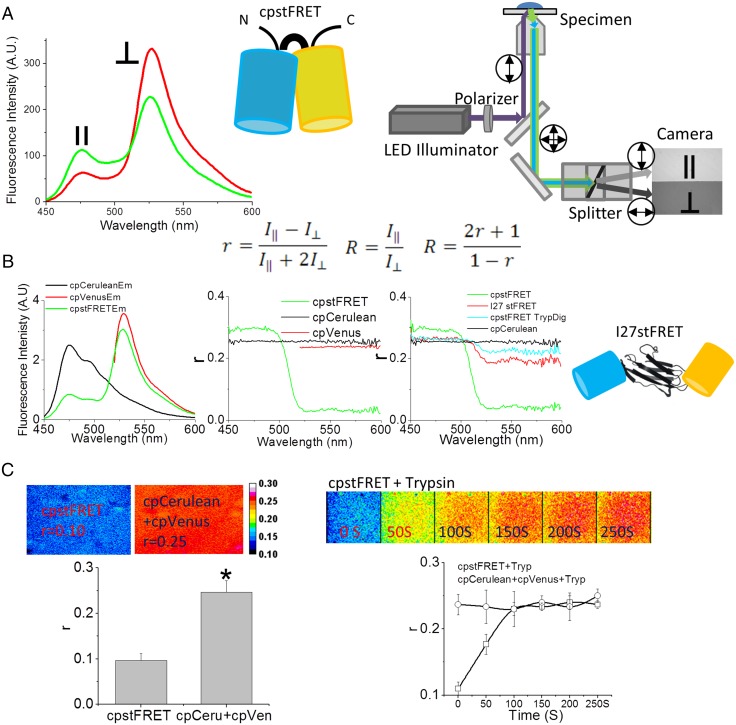
Fig. 1.
Fluorescence anisotropy is a sensitive method to detect FRET changes in cpstFRET. (A) Schematic diagram of the cpstFRET force sensor and the microscope setup. (Left) cpstFRET anisotropy scanned by a spectrofluorimeter; ‖, emission parallel to excitation; ⊥, emission perpendicular to excitation. (Middle) The cyan barrel represents cpCerulean, and the yellow barrel represents cpVenus. (Right) The wide-field fluorescence microscope setup for anisotropy measurements. Double-headed arrows indicate the polarization in the light path. (Equation Inset) R, anisotropy FRET ratio; r, anisotropy. (B) Anisotropy measured by spectrofluorimeter. (Left) The emission spectra of a purified protein solution of cpstFRET (green), cpVenus (red), and cpCerulean (black). (Middle) The emission spectral anisotropy of the three protein solutions. (Right) The anisotropy of cpCerulean (black), cpstFRET (green), cpstFRET after cleavage of the linker by trypsin for 20 s (cyan), and I27stFRET (red). Schematic diagram shows I27stFRET, with I27 as the linker between cpVenus and cpCerulean. (C) cpstFRET protein solution anisotropy measured in the microscope. (Left) FRET anisotropy, r, images of free-floating dilute probe cpstFRET (r = 0.1) and cpCerulean and cpVenus in a 1:1 ratio with no visible FRET (r = 0.25). (Right) R images of cpstFRET protein with linker cleaved by trypsin up to 250 s. All images were processed and pseudocolored by the 16-color map of ImageJ. The calibration bar was set from 0.08 to 0.30.
The actin probe appeared nontoxic, as we were able to establish multiple stable cell lines expressing the probe. Furthermore, the anatomy of stable cell lines and the founders was similar. We expressed the actin probes in HEK, Madin-Darby canine kidney (MDCK), 3T3, and bovine aortic endothelial (BAEC) cells and compared the actin distributions to the cells expressing Actin–GFP. Actin–GFP is a widely accepted standard for mapping actin, and functional studies and histology showed our labeled actin distribution was similar. We observed dynamic changes in the force in actin upon applying reversible, physiologically relevant, mechanical, and pharmaceutical perturbations including reprogramming. We were easily able to reprogram our stable cell lines into stem-like cells by softening the substrate (19). Mechanical cues such as matrix stiffness, surface topology, and cell shape are known to play critical roles in stem cell self-renewal and linage differentiation (3, 4). Counter to our intuition, we found that reprogramming increased tension in f-actin relative to the parent. The increased tension was reversible upon replating the cells on coverslips, suggesting that increased force in actin may be essential to reprogramming and retaining stemness. The actin probe has broad applicability in biology, as actin is so common and it permits the cross-correlation of actin forces with biochemical and electrical activities in living cells.
Results
Anisotropy Measurements of FRET in Stress Probes.
FRET efficiency depends on both the distance and the dipole angular orientation between donor and acceptor. In most FRET experiments, the donor/CFP and acceptor /YFP spectral emission overlap, and that requires cross-talk corrections. However, as proposed by Piston’s group, fluorescence anisotropy provides a straightforward way to minimize those errors (16); FRET emission is more depolarized than donor or acceptor emission, as the dipole orientations are not the same and the dual lifetimes allow more Brownian motion. The measurement of polarized FRET uses polarized excitation and paired orthogonally polarized emission for the acceptor. This ratio is typically parameterized as fluorescence anisotropy or polarization (16, 17).
To verify the correlation of fluorescence anisotropy and traditional FRET efficiency, we used purified cpstFRET protein solutions and examined them in a spectrofluorimeter (Fig. 1, Inset) and in a microscope (Fig. 1A). In the spectrofluorimeter, we recorded orthogonal and parallel emission spectra using vertically polarized donor excitation (Fig. 1 A, Left). In the microscope, equipped with a dual-band filter set, and a polarized dual-view splitter, we recorded the perpendicular and parallel polarized signals simultaneously (Fig. 1 A, Right). The anisotropy r and the FRET ratio R were calculated using the equations shown in the Inset of Fig. 1.
We scanned the protein solution spectra of cpstFRET, cpVenus, and cpCerulean using the spectrofluorimeter. Fig. 1 B, Left shows their emission spectra from 450–600 nm. The Middle panel shows the anisotropy r. cpVenus and cpCerulean showed high r values between 0.23 and 0.24 across the spectra, corresponding to a high polarization of emission and little Brownian motion during the fluorescence lifetime. For cpstFRET, r was high (0.27) for cpCerulean donor emission (between 450 and 500 nm) and low (0.05) for the FRET from acceptor emission, 525–600 nm—increased anisotropy of the quenched cpCerulean and low anisotropy of FRET. To test the correlation of anisotropy to FRET, we cleaved the sensor linker with trypsin and measured r. We also measured changes with a probe called I27stFRET, which uses the I27 domain of titin as a physiological linker, and that had 10–15% efficiency (Fig. 1B, Right). With no link, the fluorophores float randomly and eliminate FRET. Twenty seconds of trypsin digestion cleaved cpstFRET into donors and acceptors and eliminated FRET. The anisotropy r increased from 0.05 to 0.23 over 525–600 nm because the fluorescence came from the directly excited donor. Between 450 and 500 nm, r decreased from 0.27 to 0.24 due to the elimination of the quenching of cpCerulean, which increased the lifetime. I27stFRET had r = 0.20 between 525 and 600 nm.
In the microscope, cpstFRET had r = 0.10, and a 1:1 mixture of donor and acceptor (essentially zero FRET) also gave r = 0.25 (Fig. 1 C, Left), matching the data from the spectrofluorimeter. As with the spectrofluorimeter, trypsin digestion of cpstFRET led to a rapid increase of r of the FRET channel from 0.10 to 0.25 as FRET efficiency decreased from 80 to 0. Trypsin had no effect on a mixture of unlinked donors and acceptors (Fig. 1C, Right). Thus, as previously published, anisotropy is a sensitive method to measure changes in FRET efficiency.
AcpA Probe Reports Force in Actin.
To estimate the intrinsic stress sensitivity of the probe, we used DNA springs (20) to push apart the two fluorophores of the sensor. We covalently linked 60 mer of single-stranded DNA between the donor and acceptor GFPs using the natural peripheral cysteines. Single-stranded DNA is like a floppy string with a short persistence length (∼1 nm) and thus exerts minimal force on the probe. Adding the complementary strand created dsDNA that is much stiffer (persistence length, ∼50 nm) to push apart the donor and acceptor with forces of 7–20 pN (7, 21), and this caused a significant reduction in FRET. Although we do know roughly the absolute sensitivity of the probe, optical images have a voxel resolution of ∼1 µm3 that averages over many proteins, labeled and unlabeled, that may be under different levels of stress. The most important results from these probes are the gradients of force in time and space and the factors that modulate these forces.
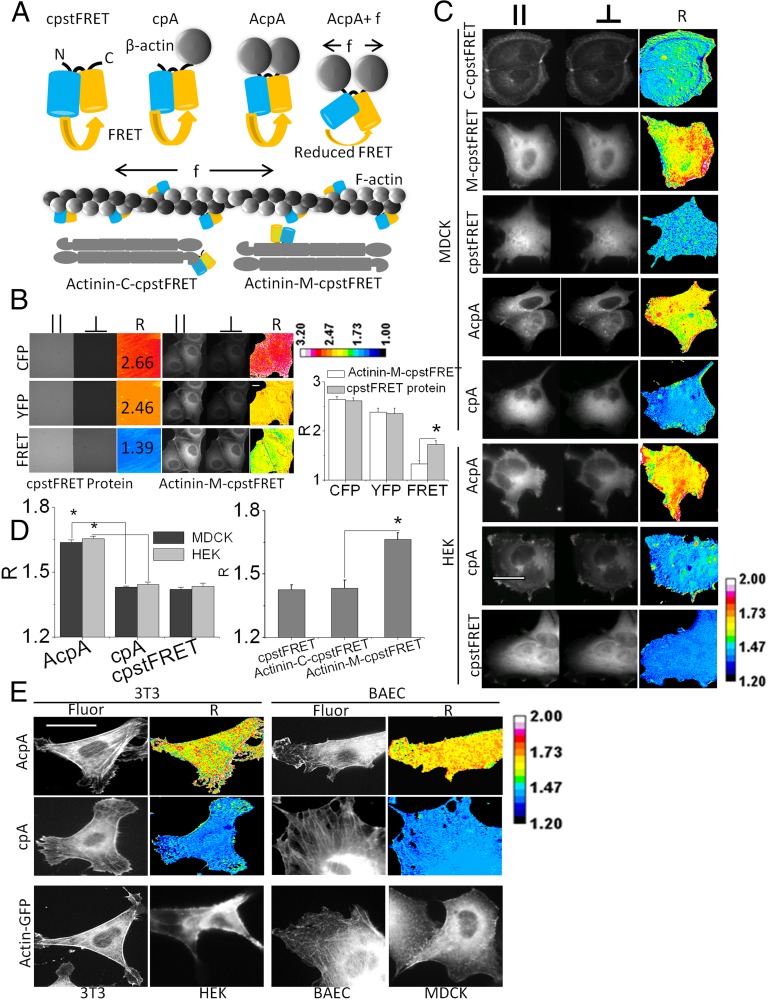
To create chimeric actin (Fig. 2A), we flanked cpstFRET with β-actin to allow it to integrate naturally into microfilaments, and we named the construct AcpA. As a force-free control to test for possible environmental effects on FRET aside from those induced by forces, we created a version of cpA that had only had one actin monomer, and this left the other end of the probe dangling free and the probe devoid of tension. To generalize our results, we created covalent cpstFRET actinin, an actin cross-linker that forms antiparallel homodimers and cross-links parallel microfilaments (22). We inserted cpstFRET between spectrin repeat domains 3 and 4 toward the middle of actinin and created what we called actinin–M–cpstFRET. We also created the force-free version with C-terminal tagged actinin: actinin–C–cpstFRET (Fig. 2A). Once expressed in cells, the actin probe could incorporate into f-actin filaments, bridge adjacent filaments, or bind to other actin binding proteins, but it appeared to us that the majority incorporated into f-actin.
Fig. 2.
Actin–cpstFRET–Actin and actinin–M–cpstFRET report force in actin and the actin cross-linker actinin. (A) Schematic diagram of actin–cpstFRET and actinin–cpstFRET chimeric constructs. cpA, cpstFRET–β-actin with one actin molecule attached to the C-terminal of cpstFRET; AcpA, Actin–cpstFRET–Actin with one actin molecule attached on each end of cpstFRET. f indicates force, which expands the structure of cpstFRET and leads to a reduction of FRET. The f-actin cartoon only presents what we expect to be the dominant configuration of probe incorporation and does not intend to exclude incorporation into cross-links. Actinin–C–cpstFRET/C–cpstFRET represents C-terminal–tagged actinin (force-free); Actinin–M–cpstFRET/M–cpstFRET represents actinin with the sensor incorporated in the middle of the host. (B) Comparison of the anisotropy ratio R of purified cpstFRET protein in solution and in the actinin host in MDCK cells. R is calculated from parallel and perpendicular polarizations using the equations shown in Fig. 1. The R images are presented in a 16-pseudocolor map with a range from 1.0 to 3.2. (Scale bar, 20 μm.) Histograms show R from cpstFRET protein and actinin–M–cpstFRET, n ≥ 3. P < 0.05 by Student t test. (C) cpstFRET reporting constitutive tension in actin and actinin in HEK and MDCK cells. MDCK cells expressed Actinin–M–cpstFRET, Actinin–C–cpstFRET, cpstFRET, AcpA, and cpA. HEK expressed cpstFRET, AcpA, and cpA. High R means high stress and vice versa. (D) Histogram plot of R for HEK and MDCK cells. P < 0.05 by Student t test. (E) 3T3 and BAEC cell expressing force-free cpA or AcpA and HEK, MDCK, 3T3, and BAEC expressing Actin–GFP. Fluor, YFP fluorescence channel acquired from acceptor Venus R ratio image indicates force levels. R images are presented in ImageJ 16-pseudocolor map with a range of 1.2–2.0. (Scale bar, 20 μm.)
To measure the forces in actin and actinin in living cells, we introduced the construct’s DNA into MDCK and HEK cells and used the simple anisotropy FRET ratio R (the ratio of the two orthogonal dual-view polarized images) to measure changes in FRET (Fig. 2B and Fig. 1A equation). R is monotonic with r and force; high R equals high stress and low R equals low stress. We compared the properties of purified cpstFRET in the microscope and chimeric actinin in cells (Fig. 2B). The donor (CFP) emission (CFPex, CFPem) and acceptor (YFP) emission (YFPex, YFPem) in solution were similar to that in cells, with R = 2.7 and R = 2.5, consistent with results from the spectrofluorimeter. In solution, at basal/zero force, the FRET channel (CFPex, YFPem) yielded R ∼ 1.4. In MDCK cells, R of the actinin chimera increased to ∼1.7, showing constitutive stress. Actinin and actin both exhibited resting stress in MDCK and HEK cells (Fig. 2 C and D). The force-sensitive actinin–M–cpstFRET and AcpA cells showed high R, 1.63–1.72, meaning high stress, whereas the control cells containing the force-free probes cpA, cpstFRET, or Actinin–C–cpstFRET showed low stress, R ∼ 1.4. Both cell lines had spatial heterogeneity of the resting forces in actin and actinin. This highlights the requirement that mathematical models of mechanics not assume uniform stress. The fact that R was lower in cells than the probes in free solution means that there was resting tension on the probes, as we have shown for multiple chimeras in previous publications. We have found no floppy filamentous proteins in cells, and this may be particularly significant for communication purposes. Wang and coworkers (23) showed that the cytoskeleton can transmit information much faster than chemical messengers, and like the tin can telephone, this communication requires tension in the linker.
To show the proper targeting of actin probes, we expressed Actin–GFP in HEK, MDCK, 3T3, and BAEC cells. The histological distribution of Actin–GFP was similar to the actin force probes (AcpA and cpA) in all these cells (Fig. 2E). HEK and MDCK cells expressing Actin–GFP, AcpA, and cpA did not show clear stress fibers. Actin networks in these cells tend to be diffuse, and the cells usually do not show stress fibers. In contrast, BAEC and 3T3 cells expressing Actin–GFP and the actin probes showed clear stress fibers (Fig. 2E), demonstrating the correct integration of the probes into f-actin. The R images also showed resting force in actin in BAEC and 3T3 cells (Fig. 2E).
Force Modulation in Actin in Response to External Stimuli.
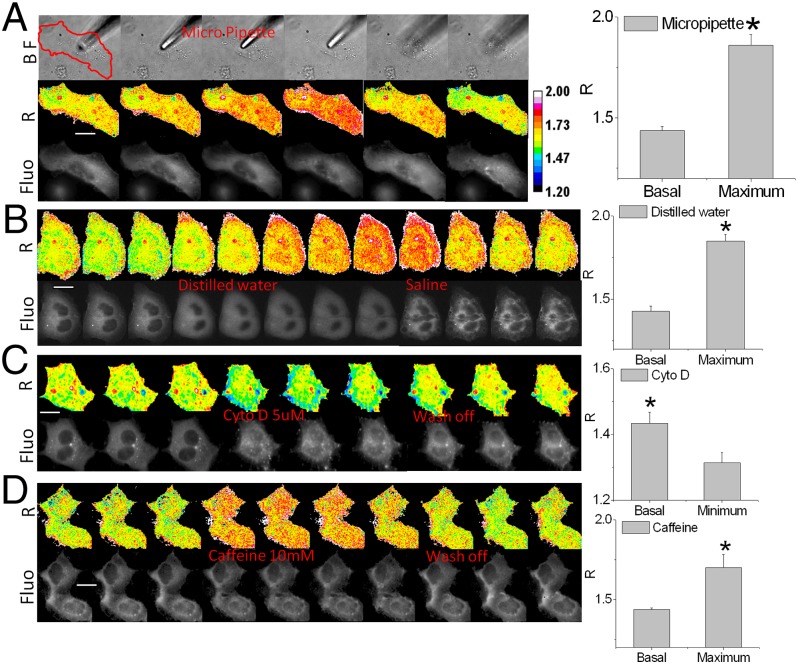
Actin is a major part of the cell cortex and helps define cell shape. External stimuli will modify structural protein stresses, and to measure these effects, we challenged transfected MDCK and HEK cells with a variety of mechanical and pharmaceutical stimuli (Fig. 3) (more details of the stimuli are presented in Materials and Methods). For direct mechanical stimuli, we pressed AcpA and cpA MDCK cells with a fire-polished micropipette (Fig. 3A and Fig. S1). MDCK cells showed an immediate increase in local stress, and the stress increased across the cell and intensified over the next minute and a half. This may represent the viscoelastic properties of the cytoskeleton in vivo or an effect of mechanosensitive ion channels (MSCs) letting in calcium. The increases in force were nonuniform in space and propagated through the cell. The stress returned to basal levels within 2 min after lifting the pipette, and some regions fell below resting levels (green areas). MDCK cells expressing the force-free cassette cpA did not show any change of R (Fig. S1) so that any possible biochemical alterations on the probe were ruled out.
Fig. 3.
cpstFRET reports changes in actin force in response to mechanical and chemical stimuli. (A) MDCK cells expressing AcpA were indented by a micropipette; the red line outlines the cell. Fluo, FRET image of the cell; R, anisotropy ratio images showing force in actin. (The experiments were repeated n ≥ 5.) Histograms show maximum changes in the ratio R (P < 0.05 by Student t test). (B) MDCK cells challenged by anisotonic osmotic pressure. Replacing Hepes buffer with distilled water swelled the cells and increased force in actin, and returning them to saline shrank the swollen cells and lowered stress below resting levels in places (green). (C) HEK AcpA cells treated with 5 μM of cytochalasin D. (D) HEK AcpA cells treated with 10 mM of caffeine to elevate calcium Ca+2 and induce contraction. Each experiment was repeated n ≥ 5. Histograms on the right show the maximum ratio R changes under each condition. P < 0.05 by Student's t test, calibration bar with 16-color map. (Scale bar, 20 μm.)
As another form of mechanical stimulation, we challenged the cells with hypo-osmotic stress by switching the bath from saline to distilled water (it may be surprising to realize that many nucleated cells do not lyse under these conditions). The cells swelled as expected, and the tension in actin reached the maximum within 5 min. Returning the bath to saline returned the stress to basal levels, and the cell shrank (Fig. 3B). Control probes showed no change in R (Movie S1). These data confirm the tight coupling of cell volume to cytoskeletal stress in three dimensions throughout the cell volume rather than being confined to the cell cortex (24) and back up the predictions from atomic force microscopy studies (24).
Disrupting f-actin in HEK AcpA cells with cytochalasin D (5 μM) released the basal tension (Fig. 3C). Actin tension in AcpA HEK cells fell within 10 min of CytoD treatment and recovered within 10 min of washout. Again, force-free cpA controls did not show any changes (Movie S2), showing that the changes in FRET are not biochemically mediated but driven by forces. We also treated the cells with 10 mM of caffeine (Fig. 3D) to elevate calcium Ca+2. Within 1 min, R increased dramatically, and washout rapidly returned the forces to basal levels. The R of control cells was unaffected (Movie S3). These simple stimuli have in common changes in mechanical force within the actin network, and we presume those changes will lead to activation of downstream messenger cascades.
Increased Cytoskeletal Forces in Reprogrammed Cells.
Mechanical and physical factors affect stem cell renewal and differentiation (3). Varying the mechanical properties of the extracellular matrix can redirect stem cell linage commitment (19). Other mechanical cues correlated with fate commitment of mesenchymal stem cells are cell shape and the mean stress in the cytoskeleton (15). Because actin transfers physical cues between the cell exterior and interior, it likely plays a major role in stem cell linage commitment (14, 25). However, there have been no measurements available on the forces in specific structural proteins in adult cell reprogramming and dedifferentiation. We have now made those measurements for actin and actinin.
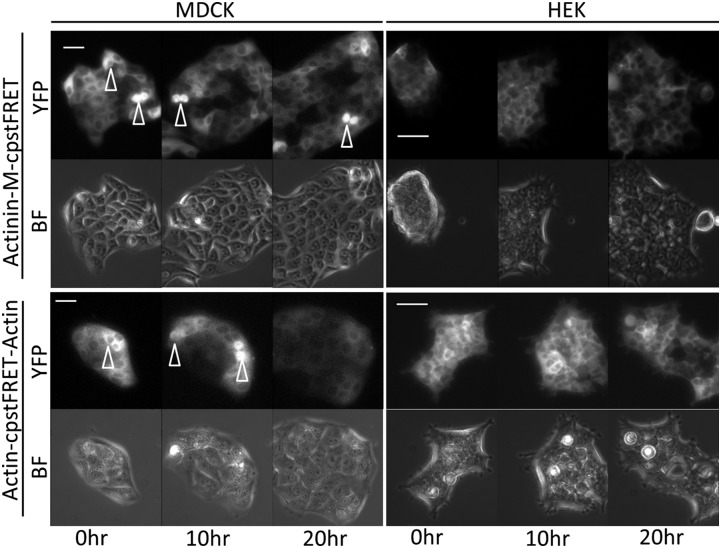
Transient expression of the probes proved inconvenient because the cells gradually lost the plasmids over time and the fluorescence faded. We therefore created stable cell lines expressing probe chimeras (Fig. 4, Fig. S2, and Table S1). These “prelabeled” HEK and MDCK cell lines underwent regular cell cycles and the proliferation, showing that the probes were not toxic (Movies S4 and S5). We also observed no morphological differences relative to the wild-type cells.
Fig. 4.
Stable cell lines expressing actin-sensor and actinin-sensor chimeras show normal cell physiology. We created 13 cell lines (Table S1). The MDCK and HEK stable cell lines were cultured in media in a 5% CO2 chamber on a heated stage. A 20-h time lapse sequence from each cell line was used to monitor cell proliferation (BF, bright field; YFP, YFP channel signal from cpVenus). Using the Zeiss Definite Focus, we monitored at least five cell colonies simultaneously. Arrowheads indicate the dividing cells. All cells went through mitosis and proliferation. (Scale bar, 50 μm.)
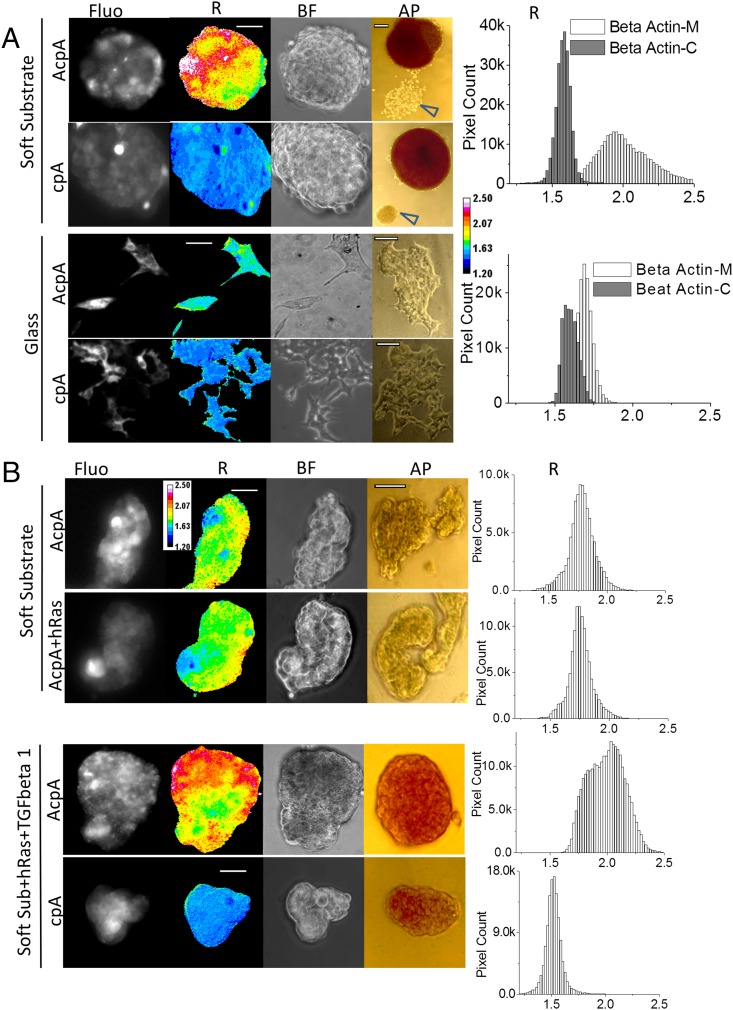
Specialized somatic cells can be reprogrammed to pluripotent stem cells (PSCs), induced pluripotent stem cells (IPS) with a combination of four transcription factors (TFs) (26, 27), and there has been much effort to reduce the number of potentially carcinogenic TFs by using chemical compounds (28, 29). Remarkably, HEK cell reprogramming requires no TFs but simply culturing on low-adhesion soft substrates such as agarose (30), emphasizing again the critical role of physical cues in reprogramming. To create labeled cells exhibiting stemness, we cultured stable HEK cell lines on soft polydimethylsiloxane (PDMS) (Fig. 5A). Within 3 d, the majority of cells formed tightly packed multicellular spherical embryonic bodies (EBs). These cells were alkaline phosphatase (AP) positive, as expected for EB stem cells. Some cells remained attached on PDMS and did not show AP activity (Fig. 5A, arrowheads). There was no AP activity in HEKs cultured on glass. These data support the critical role of mechanics in dedifferentiation. The majority of cells in EBs showed much higher actin tension than cells cultured on glass, suggesting increased force may be essential to HEK cell reprogramming. As a repeated control for biochemical effects on the chromophores, we found that cells expressing stress-free cpA had low basal R values and showed no changes between the cells in EBs and cells attached to glass. Also surprising was that EBs did not show a significant force increase in actinin (Fig. S3A). This visibly demonstrates that cytoskeletal stress is not uniform, even for actin and its cross-linkers, and one cannot reliably generalize about the forces in one protein from forces in another.
Fig. 5.
Actin force elevation in stem-like cells reprogrammed from HEK and MDCK stable cell lines. (A) HEK actin stable cell lines cultured on soft substrate (PDMS) and glass. AcpA, HEK stable line expressing actin–cpstFRET–actin; AP, alkaline phosphatase staining of EBs and cells; BF, bright-field images of EBs and cells; cpA, HEK stable line expressing cpstFRET–actin; Fluo, FRET signal; R, anisotropy ratio representing tension in actin. Arrowheads indicate HEK cells attached to the substrate showing no AP activity, and these cells were not reprogrammed. Pixel count distribution plots on the right were generated from all pixels from >10 R images. R images are presented with a 16-color map of 1.20–2.50. (Scale bar, 50 μm.) (AP scale bar, 100 μm.) (B) MDCK actin stable cell lines cultured on soft substrate (PDMS). AcpA, MDCK stable line expressing actin–cpstFRET–actin cassette; cpA, MDCK stable line expressing cpstFRET–actin; +hRas, MDCK stable cell lines expressing hRas gene; +TGFbeta 1, cell cultures were supplied with 5 μg/mL TGF-β1. hRas MDCK stable lines were derived from MDCK AcpA or cpA stable cell lines. Pixel count distribution plots on the right were created using all pixel values from at least 10 R images. R images are presented with a 16-color map of Image J with a range of 1.20–2.50. (Scale bar, 50 μm.) (AP scale bar, 100 μm.)
As another test for stemness and actin force, we examined the behavior of a different cell line, MDCK. MDCK cells cultured on PDMS also aggregated into clumps (Fig. 5B), but these cells did not form spheroidal EBs. In contrast to HEK lines, these clumps of AcpA-labeled cells did not show increased force in actin compared with cells cultured on glass (Fig. 5B and Fig. S3B). AP staining was also negative, indicating that the lumps did not contain stem cells. TGF-β1 and Harvey rat sarcoma viral oncogene homolog are reported to potentiate the epithelial–mesenchymal transition (EMT) of MDCK cells (31, 32). Human mammary epithelia cells undergoing EMT show stem cell properties and the expression of stem cell markers (33). Therefore, we introduced hRas into MDCK AcpA and cpA cell lines and created more cell lines with stable expression of hRas (Table S1). The hRas–MDCK–AcpA cells cultured on soft PDMS aggregated to clumps, but they exhibited no increase in actin stress or AP activity (Fig. 5B). However, when we added TGF-β1 to hRas MDCK cells, and cultured them on a soft substrate, both MDCK AcpA and cpA cells transformed to stem-like cells, and AcpA cells displayed elevated force in actin and had AP activity (Fig. 5B). Control cells with force-free cpA showed no FRET differences relative to the parents cultured on glass (Fig. S3B). On glass, the TGF-β1–treated hRas MDCK cells showed fibroblast-like morphology, probably as a result of EMT. None of the cells cultured on glass showed AP activity (Fig. S3). Both HEK and MDCK, with different mechanisms of reprogramming, showed elevated force in actin when they exhibited stemness. These cell lines may reside in intermediate states between highly specialized cells and PSCs, and a high force in actin may shift the balance toward pluripotent states, whereas low force may reverse the balance.
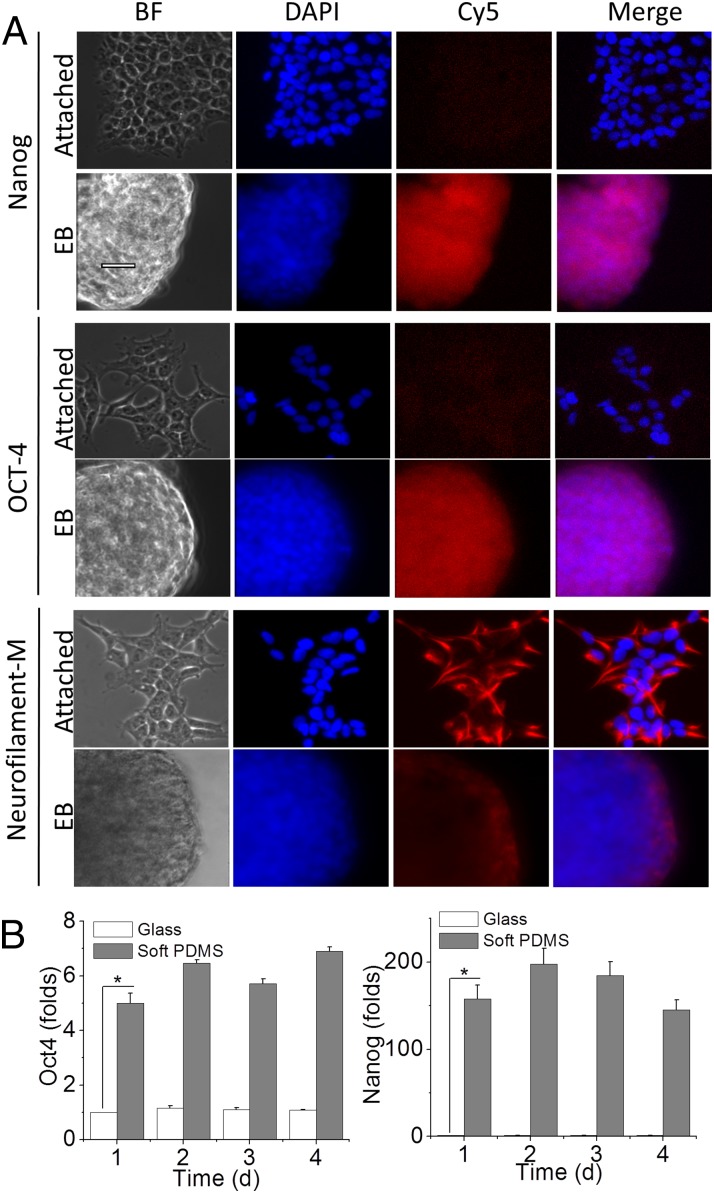
To further show the stemness of cells on soft substrates, we examined the up-regulation of two other key marker proteins of stem cells, transcription factor OCT4 and Nanog29,30 (Fig. 6A). HEK cells cultured on glass coverslips showed little or no OCT4 and Nanog expression, whereas the EBs generated by 3 d culturing on soft PDMS showed strong OCT4 and Nanog immunostaining. We confirmed the immunostaining data with real-time RT-PCR (Fig. 6B). Both OCT4 and Nanog expression were up-regulated in HEKs cultured on soft PDMS. The OCT4 expression on PDMS increased 5–7-fold over HEKs cultured on coverslips, whereas the Nanog expression increased more than 150-fold. We also checked the neuron progenitor marker neurofilament-M. HEK cells showed high expression of the protein, which is consistent with them being closely related to neurons (34), and we found robust staining of neurofilament-M in glass-attached HEK cells (Fig. 6A). When cultured on soft PDMS and forming EBs, the cells had much lower immunostaining of neurofilament-M, indicating the soft, low-adhesion substrate induced morphological as well as genomic transitions.
Fig. 6.
Stem cell markers in HEK-derived EBs. (A) HEK cells cultured on a coverslip (attached) and PDMS (EB, embryonic body) were immunostained with anti-Nanog, OCT4, and Neurofilament-M antibodies. Then, the cells were treated with secondary antibodies conjugated with Cy5. The nuclei were stained with DAPI. BF, bright field. (Scale bar, 50 μm.) (B) Real-time RT-PCR of HEK cultured on coverslip glass and soft PDMS for 1, 2, 3, and 4 d. The histogram shows the average fold of increase of OCT4 and Nanog from three sets of independently prepared samples at each time point (*P < 0.05 by Student t test).
Forces Promote Cell Reprogramming and Retaining Stemness.
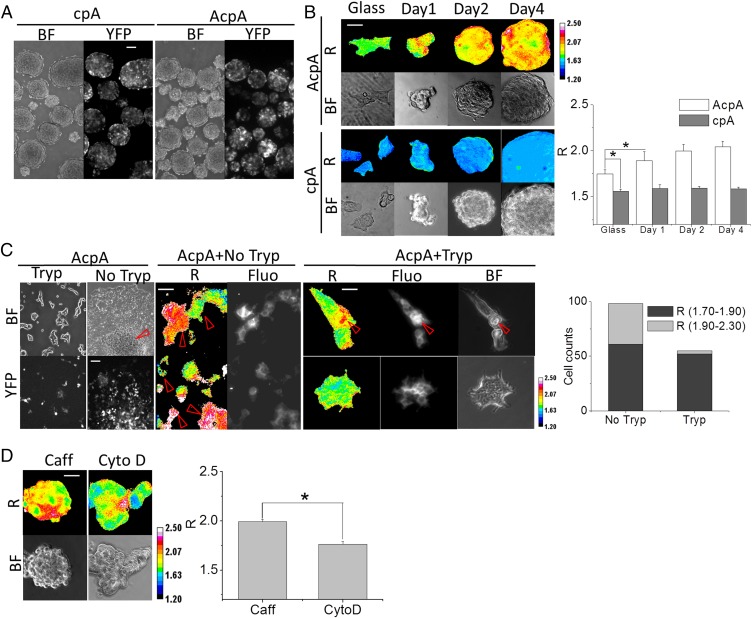
After 4 d of culture on soft PDMS, HEK AcpA and cpA stable cell lines dedifferentiated into stem-like cells with nearly 100% efficiency (Fig. 7 A and B). The cells proliferated and attached to each other to form EBs within 24 h. Actin tension in EBs increased at the same time. Force-free cpA probe in control cells under the same culture conditions did not show any change in R. Hydrophobic surfaces with low adhesion, such as soft PDMS, can trigger HEK cell reprogramming, and reprogramming is associated with increased tension in actin. Transferring the EBs to glass coverslips shifted the cells toward differentiated states (Fig. 7C). After 2 d of culturing, cells from trypsinized HEK EBs reattached to glass and displayed typical HEK morphology. Intact EBs cultured on coverslips (with no trypsin treatment) started spreading on the surface at the periphery of the EBs. Trypsin treatment to loosen the cells had its own effects, which may apply whenever cells are trypsinized. After 2 d in culture, 30 out of 96 cells obtained from EBs without trypsin treatment retained high stress in actin, whereas only one out of 52 cells from trypsinized EBs retained high stress.
Fig. 7.
Actin stress in dedifferentiated HEK cells and stemness retention. (A) HEK force-free cpA and AcpA cultured on PDMS for 3 d (BF, bright field; YFP, YFP channel fluorescence). (Scale bar, 100 μm.) (B) Time course of R in HEK cpA and AcpA cells. R, the anisotropy ratio is positively correlated to force. Histograms show the average R of five images at each time point. (Scale bar, 50 μm.) P < 0.05 by Student t test. (C) HEK AcpA cells cultured on a coverslip. AcpA HEK EBs were transferred to coverslips with (Tryp) or without trypsin (No Tryp) treatment. BF and YFP channels show the attached cells and fluorescence. AcpA+NoTryp displays R in attached cells derived from EBs without trypsin digestion; AcpA+Tryp displays R of two clusters of HEK cells differentiated from cells from trypsinized EBs. (Scale bar, 50 μm.) The histogram gives the proportion of high-force versus low-force cells from the two groups of HEK cells derived from trypsinized and nontrypsinized EBs. (D) HEK AcpA cells cultured on PDMS and supplied with Caff (caffeine) and CytoD (cytochalasin D). The histogram shows the average R of five images from each condition. (Scale bar, 50 μm.) P < 0.05 by Student t test. All R images have a 16-color map of 1.20–2.50.
We treated the reprogrammed cells with drugs that might alter actin tension. HEK AcpA cells on PDMS were treated with 5 mM of caffeine to elevate calcium Ca+2 (to likely enhance forces in actin; Fig. 3D) or 5 μM of cytochalasin D (to reduce forces in actin; Fig. 3C). After 2 d, cells grown in caffeine media formed EBs and showed elevated actin tension characteristic of stemness. CytoD-treated cells did not form EBs but accumulated in clusters of cells with lower force as differentiated cells. These data imply that high forces in actin may be required for cells to retain stemness.
Discussion
Mechanical forces affect all life forms, but until recently there had been no reliable method to track mechanical forces in specific proteins in living cells. This changed with the development of genetically coded FRET-based force probes (2, 10). To calibrate the intrinsic force sensitivity of these probes, we used DNA springs to stretch the probes in solution and showed that the probes responded clearly to forces in the range of 10 pN (7, 10, 11). We have calibrated the strain sensitivity of the earlier linear linked probes and showed FRET to be linear as predicted (2). (In the strain experiment, we attached the probes at each end to a rubber sheet and biaxially stretched the sheet while performing FRET.) We have explored a variety of fluorescence imaging techniques to improve FRET imaging and reduce cross-talk, and we have found, as previously suggested (16, 17), that fluorescence anisotropy provided the most sensitive measurement (Figs. 1, 2, and 3) and is simple to implement.
Our FRET images do not explicitly report the stress in individual proteins, as the microscope resolution is the order of a cubic micrometer, much larger than a single molecule. Thus, our images are averages across the voxel and can include force gradients. The probes are most useful in reporting stress gradients in time and space. Although the DNA spring calibration showed that the probe was readily modulated with stress in the range of 10 pN, the most significant measure of sensitivity comes from the fact that we could observe changes in FRET with reversible perturbations that are physiologically relevant.
The cpstFRET angular force probe (7) used here has a wider dynamic range and smaller molecular weight than the earlier linear linker probes. The angular probe uses the dipole orientation dependence of FRET, allowing FRET efficiencies, in principle, from 0% to 100% (2, 8, 11). The dimeric structure of cpstFRET mimics the dimer subunit structure of many multimeric cytoskeleton proteins such as actin and tubulin (Figs. 1 and 2). As we have shown for a variety of stimulation modalities and stem cell reprogramming, the probes appear to be applicable to all physiological variables (35).
We reprogrammed HEK and MDCK cell lines into stem-like cells and examined how actin forces correlate with stemness (Fig. 5). The PDMS substrate was sufficient for HEK and MDCK cell reprogramming. Even though HEK cells are called human embryonic kidney cells, they did not exhibit AP activity, a marker of stemness, or other marker genes such as OCT4 and Nanog. PDMS is hydrophobic as well as soft, and we do not yet know which of these properties promoted cell reprogramming. Engler et al. performed the first evaluation of the role of substrate stiffness in the fate commitment of human mesenchymal stem cells, and they kept the surface chemistry of substrate the same while varying the stiffness. After several weeks in culture, the cells committed to lineages specified by substrate stiffness, hinting that the softness of the substrate in our experiments, rather than hydrophobicity, might be the key factor in dedifferentiation (19). We found that the mechanical effects on reprogramming were reversible by placing EBs on glass or placing individual cells isolated from EBs on glass (Fig. 7).
Induced PSCs (iPSCs) reprogrammed by viral introduction of OSKM TFs have potential for regenerative medicine. However, two issues have limited the utility of patient-orientated human iPSCs: (i) The integration of viral transgenes and the oncogenes c-MYC and KLF4 TFs into the genome poses a high risk of tumorigenesis, and (ii) the reprogramming efficiency is low with the four TFs (26, 36). Substantial effort has been devoted to reduce the number of required TFs using additional pharmaceuticals (28, 29, 37). Tested drugs include those that manipulate levels of histone methylation, acetylation, DNA methylation, and MAPK/ERK pathways (29), but these studies all omitted the influence of mechanical factors. We have now demonstrated dramatic changes in actin forces associated with reprogramming and dedifferentiation, so the probes promise to be useful as assay tools to screen for mechanical modifying drugs.
The probes permit researchers to study the role of mechanics under normal and pathologic conditions in cells, tissues, organs, and animals. The probes are nontoxic based on the stability of labeled cell lines and transgenic animals (38) so that transgenic animals can be readily created, providing prelabeled tissues of all kinds (Fig. 4, Fig. S2, and Table S1). Although we have emphasized physiologically relevant perturbations, the results are transferrable directly to pathology. There is probably no disease that does not involve a change in the shape of some cells, and that can only occur through changes in force. These induced changes can have direct effects, as in athletic injuries, traumatic brain injury, or muscular dystrophy, or indirectly, as in the mechanical changes noted for metastatic cancer cells (39) or in anemia (40, 41).
Materials and Methods
cDNA Cloning and Plasmid Construction.
We created the angular-dependent FRET force sensor cpstFRET by tandemly linking circularly permutated YFP/Venus and at the same position circularly permutated CFP/Cerulean at position 174 (7). The cpstFRET gene was incorporated into pEGFP-C1 vector (Clontech) with the EGFP gene preremoved from the plasmid. Then, we cloned human β-actin from an HEK cDNA library and incorporated the gene at the C-terminal of cpstFRET, creating pEG–cpstFRET–β-actin (cpA). We inserted another actin gene at the N-terminal of cpstFRET and created pEG–β-Actin–cpstFRET– β-actin (AcpA). For human alpha-actinin constructs, we purchased pEGFP-N1 alpha-actinin plasmid from Addgene (Plasmid 11908). We created pEG–actinin–sstFRET with sstFRET inserted between two spectrin repeats in actinin and pEG–Actinin–C–sstFRET with sstFRET tagged to the C-terminal of actinin (11). We replaced sstFRET with cpstFRET and created pEG–Actinin–cpstFRET and pEG–Actinin–C–cpstFRET. We also subcloned cpstFRET, cpCerulean, and cpVenus into pET-52b (+) vector (Novagen) for prokaryotic protein expression. A detailed list of primers and restriction enzyme sites is provided in Table S2.
Production and Purification of Recombinant Proteins.
Recombinant proteins were expressed in the BL21(DE3) strain (Novagen) of Escherichia coli after induction with 1 mM isopropyl β-d-thiogalactopyranoside at 16 °C for 24 h. The proteins were purified using Ni-NTA agarose, and bacteria were lysed with BugBuster Protein Extraction Reagents (EMD Millipore) according to the manufacturer’s instructions. The purified protein solutions were dialyzed to remove imidazole and extra salts.
Spectrometry and Microscopy Measurements of Fluorescence Polarization.
We diluted the protein solution in 10 mM Tris⋅HCl to 10 μg/mL, pH 7.4, in a 50-μL quartz cuvette and recorded emission spectra with a PTI spectrofluorimeter (Photon Technology International). For cpstFRET, we fixed the excitation at 433 nm and recorded emission from 450 nm to 600 nm; for cpCerulean, we used 433-nm excitation and recorded emission from 450 nm to 600 nm, and for the cpVenus protein, we set excitation at 515 nm and recorded emission from 520 nm to 600 nm. We measured fluorescence polarization by exciting the protein solutions with vertically polarized light and scanned the parallel and perpendicular emission spectra. We calculated the anisotropy, r, or the simpler intensity ratio R using the equations shown in Fig. 1; both are negatively correlated to FRET efficiency but positively correlated with force.
We used a wide-field fluorescence microscope to image fluorescence polarization. Cells or protein solutions were imaged on an inverted Zeiss Axio Observer A1 equipped with an Andor Ixon DV897 back-illuminated cooled CCD camera. The light source was a LED light engine, model 6-LCR-XA (Lumencor), which uses electronic shutters to switch excitation wavelengths for CFP or YFP. We positioned a polarizing filter (Chroma) in front of the light engine to generate polarized excitation. For epifluorescence imaging, we used an ET-Dual CFP/YFP filter cube set (430/24x, 500/20x, 470/24m, 535/30m) (Chroma Technology). The excitation filter passes 433 nm and 515 nm of light, and the emission filter passes 475 nm of CFP emission (polarized) and 527 nm of acceptor emission/FRET (depolarized). For CFP channel recording, we used 433 nm of Ex and recorded 475 nm of Em; for YFP channel recording, we used 515 nm of Ex and recorded 525 nm of Em; for FRET channel recording, we used 433 nm of Ex and recorded 475 nm + 525 nm of Em. The total recorded FRET signals are the sum of polarized CFP and depolarized FRET signals. The ratio of the two changes with FRET efficiency. We used the Duel View (Photometrics) image splitter to separate the vertical and horizontal polarized images. The anisotropy value r or ratio R was calculated using the equations shown in Fig. 1. Because of the different N.A. of 63× objectives used for individual cells and the 20× objectives for imaging EBs and clusters of cells, the ratio R from 20× objectives is 0.2 higher than that obtained with 63× objectives.
Cell Culture and Gene Expression.
MDCK and HEK cells were cultured in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS and antibiotics. Cells were spread on 35-mm coverslips and allowed to grow for 24 h. Fugene 6 (Promega) was used to deliver 0.5–1 μg of plasmid DNA per coverslip to the cells. Gene expression was examined on the microscope 24–36 h following transfection. Cells on coverslips were mounted on a stage equipped with a Bioptechs temperature controller to keep the cells at 37 °C. Because of strong autofluorescence (433 nm of Ex, 527 nm of Em) from serum containing media, the culture media was substituted with 37 °C prewarmed saline Hepes buffer, pH 7.4, for all FRET measurements.
Mechanical and Biochemical Acute Stimuli.
A micromanipulator (SENSAPEX) was used to indent cells. The micropipette tips were fire-polished and sealed. To place the pipette on top of the MDCK AcpA and cpA cells, we focused the microscope to the top of the cells and lowered the micropipette until a shadow of the tip appeared. The micropipette was then lowered into direct contact with the cell, producing a visible deformation. FRET and bright-field (BF) images were taken at 30-s intervals. We treated HEK cells with 10 mM of caffeine in Hepes buffer and made images every 60 s. The caffeine was washed out with buffer after 5 min. We treated HEK cells with 5 μM of cytochalasin D and acquired images at 10-min intervals. We stimulated cells with cycles of hypotonic and isotonic solutions by simply exchanging the buffer with distilled water and returning to saline and taking images every 60 s. All FRET signals were acquired using 433-nm excitation and the dual-band emission (475 nm + 527 nm) filter cube and the Dual View outfitted with a polarizing splitter. The resulting vertical and horizontal polarized images were used to calculate the anisotropy ratio R and anisotropy r.
Stable Cell Line Development.
We developed multiple cell lines with various genes (Table S1). The founder cell lines, genes, and antibiotics used for screening are listed in Table S1. The founder cell lines were spread on 35-mm polystyrene tissue culture Petri dishes and then transfected after 24 h in culture. For each cell line, four nontransfected dishes were used to determine the proper concentration of antibiotics. We set a concentration and then added 1×, 2×, 3×, and 4× of that to the nontransfected dishes. The concentration causing 70–80% cell death within 2–3 d was used for screening. Two days after transfection, antibiotics were added to the dishes, and they were placed in an incubator until the cells reached confluence. Cells were then trypsinized and transferred to 50-mL cell culture flasks (BD, Falcon). The same concentration of antibiotics was applied, and cells were allowed to grow to confluence. The cells were subcultured using a 1:10 dilution for three rounds. We checked cell fluorescence, and if 80–90% of the cells were fluorescent, we used these lines with multiple cloning. If the fluorescent cells were less than 30% of the population, we did further screening using 96-well flat-bottom Corning Costar cell culture plates (Invitrogen). We counted and diluted the cells to 150/10 mL of media and then added 100 μL per well. The majority of the wells had 1–2 cells. The culture plates were incubated for 15 d and individual colonies developed from single-cell cultures. We screened the colonies under the microscope and picked ones with relatively bright fluorescence and grew up the cells. The resulting cell lines are single cloning stable lines with nearly 100% identical genotype and phenotype.
Stem Cell Reprogramming and Culture.
We reprogrammed HEK and MDCK to have stemness properties. For HEK, trypsinized cells were spread on PDMS-coated coverslips or 35-mm Petri dishes. PDMS was prepared from the SYLGARD 184 Silicone Elastomer Kit (DOW Corning) with an elastomer base to curing agent ratio of 25:1. The cells growing on PDMS formed EBs in 1–4 d. Strong autofluorescence from the serum containing media required us to transfer cells to saline for imaging. The EBs were collected and centrifuged at 200 g for 5 min. After removing the supernatant, the pellets were washed once with Hepes buffered saline. The pellets were resuspended and transferred to 35-mm tissue culture Petri dishes with a glass bottom for imaging. For MDCK cells, we first introduced the hRas gene to transform them using the pBabe–puro Ras V12 (Plasmid 1768, Addgene). After a cell line with stable expression of hRas was established, cells were trypsinized and plated on PDMS-coated 35-mm Petri dishes supplied with TGF-β1 (5 ng/mL). Cells were cultured for 7 d, and ∼10% of the cells turned into EBs.
Quantitative Real-Time PCR Analysis.
Total RNA was extracted from HEK293 cells using TRIzol reagent (Invitrogen). The cDNA was synthesized by reverse transcription of the RNA using a High Capacity cDNA Archive kit (Invitrogen, USA) with random hexamers. The expression levels of the target genes were quantified by quantitative RT-PCR with the SYBR Green PCR Master Mix and the 7300 Real-Time PCR system (Applied Biosystems). GAPDH was used as an internal expression control, and all data were analyzed using the GAPDH gene expression as a reference.
Statistical Analysis.
All data are expressed as mean ± SD or ± SEM as noted. Each experiment was repeated at least three times, and >5 cells were imaged and analyzed under each condition. We evaluated statistical significance with the Student's t test for comparison between two mean values. The R of EBs is presented as pixel count distribution from >10 EBs. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Suchyna for thoughtful discussion, Dr. Phil Gottlieb for advice on the chemistry, Mary Teeling for cell culture, and Joanne Pazik for plasmid DNA constructs. This work was supported by grants from the National Institutes of Health, US Department of Defense, and The Children’s Guild of Buffalo.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411683111/-/DCSupplemental.
References
- 1.Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 2.Meng F, Suchyna TM, Sachs F. A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ. FEBS J. 2008;275(12):3072–3087. doi: 10.1111/j.1742-4658.2008.06461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li D, et al. Role of mechanical factors in fate decisions of stem cells. Regen Med. 2011;6(2):229–240. doi: 10.2217/rme.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 5.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317(5838):663–666. doi: 10.1126/science.1139857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kung C. A possible unifying principle for mechanosensation. Nature. 2005;436(7051):647–654. doi: 10.1038/nature03896. [DOI] [PubMed] [Google Scholar]
- 7.Meng F, Sachs F. Orientation-based FRET sensor for real-time imaging of cellular forces. J Cell Sci. 2012;125(Pt 3):743–750. doi: 10.1242/jcs.093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466(7303):263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo J, Sachs F, Meng F. Fluorescence-based force/tension sensors: A novel tool to visualize mechanical forces in structural proteins in live cells. Antioxid Redox Signal. 2014;20(6):986–999. doi: 10.1089/ars.2013.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng F, Suchyna TM, Lazakovitch E, Gronostajski RM, Sachs F. Real time FRET based detection of mechanical stress in cytoskeletal and extracellular matrix proteins. Cell Mol Bioeng. 2011;4(2):148–159. doi: 10.1007/s12195-010-0140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng F, Sachs F. Visualizing dynamic cytoplasmic forces with a compliance-matched FRET sensor. J Cell Sci. 2011;124(Pt 2):261–269. doi: 10.1242/jcs.071928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borghi N, et al. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci USA. 2012;109(31):12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heisenberg CP, Bellaïche Y. Forces in tissue morphogenesis and patterning. Cell. 2013;153(5):948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Connelly JT, et al. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat Cell Biol. 2010;12(7):711–718. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- 15.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 16.Rizzo MA, Piston DW. High-contrast imaging of fluorescent protein FRET by fluorescence polarization microscopy. Biophys J. 2005;88(2):L14–L16. doi: 10.1529/biophysj.104.055442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rizzo V. Lights, camera, actin! The cytoskeleton takes center stage in mechanotransduction. Focus on “Mapping the dynamics of shear stress-induced structural changes in endothelial cells.”. Am J Physiol Cell Physiol. 2007;293(6):C1771–C1772. doi: 10.1152/ajpcell.00453.2007. [DOI] [PubMed] [Google Scholar]
- 18.Choy JS, et al. DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc. 2010;132(6):1782–1783. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 20.Tseng CY, Wang A, Zocchi G, Rolih B, Levine AJ. Elastic energy of protein-DNA chimeras. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80(6 Pt 1):061912. doi: 10.1103/PhysRevE.80.061912. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Wang A, Qu H, Zocchi G. Protein-DNA chimeras: Synthesis of two-arm chimeras and non-mechanical effects of the DNA spring. J Phys Condens Matter. 2009;21(33):335103. doi: 10.1088/0953-8984/21/33/335103. [DOI] [PubMed] [Google Scholar]
- 22.Sjöblom B, Salmazo A, Djinović-Carugo K. Alpha-actinin structure and regulation. Cell Mol Life Sci. 2008;65(17):2688–2701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Na S, et al. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc Natl Acad Sci USA. 2008;105(18):6626–6631. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spagnoli C, Beyder A, Besch S, Sachs F. Atomic force microscopy analysis of cell volume regulation. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;78(3 Pt 1):031916. doi: 10.1103/PhysRevE.78.031916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Treiser MD, et al. Cytoskeleton-based forecasting of stem cell lineage fates. Proc Natl Acad Sci USA. 2010;107(2):610–615. doi: 10.1073/pnas.0909597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 28.Huangfu D, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26(11):1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y, et al. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2(6):525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Su G, et al. The effect of forced growth of cells into 3D spheres using low attachment surfaces on the acquisition of stemness properties. Biomaterials. 2013;34(13):3215–3222. doi: 10.1016/j.biomaterials.2013.01.044. [DOI] [PubMed] [Google Scholar]
- 31.Nicolás FJ, Lehmann K, Warne PH, Hill CS, Downward J. Epithelial to mesenchymal transition in Madin-Darby canine kidney cells is accompanied by down-regulation of Smad3 expression, leading to resistance to transforming growth factor-beta-induced growth arrest. J Biol Chem. 2003;278(5):3251–3256. doi: 10.1074/jbc.M209019200. [DOI] [PubMed] [Google Scholar]
- 32.Oft M, et al. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10(19):2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 33.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16(8):869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- 35.Bowman CL, Gottlieb PA, Suchyna TM, Murphy YK, Sachs F. Mechanosensitive ion channels and the peptide inhibitor GsMTx-4: History, properties, mechanisms and pharmacology. Toxicon. 2007;49(2):249–270. doi: 10.1016/j.toxicon.2006.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451(7175):141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 37.Zhu S, et al. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7(6):651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng F, Suchyna TM, Lazakovitch E, Gronostajski RM, Sachs F. Real time FRET based detection of mechanical stress in cytoskeletal and extracellular matrix proteins. Cell Mol Bioeng. 2011;4(2):148–159. doi: 10.1007/s12195-010-0140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bastatas L, et al. AFM nano-mechanics and calcium dynamics of prostate cancer cells with distinct metastatic potential. Biochim Biophys Acta. 2012;1820(7):1111–1120. doi: 10.1016/j.bbagen.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Bae C, Gnanasambandam R, Nicolai C, Sachs F, Gottlieb P. Xerocytosis is caused by mutations that alter the kinetics of the mechanosensitive channel PIEZO1. Proc Natl Acad Sci USA. 2013;110(12):E1162–E1168. doi: 10.1073/pnas.1219777110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart AK, et al. Loss-of-function and gain-of-function phenotypes of stomatocytosis mutant RhAG F65S. Am J Physiol Cell Physiol. 2011;301(6):C1325–C1343. doi: 10.1152/ajpcell.00054.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.