Significance
We provide to our knowledge the first in vivo and in vitro evidence for H2O2-triggered heme transfer between proteins. Specifically, H2O2 binds to and labilizes cytochrome c peroxidase (Ccp1)’s heme by oxidizing the proximal Fe ligand (His175), which activates Ccp1 to transfer its heme to apoCta1, and apoCcp1 subsequently escapes from mitochondria. This sequence of H2O2-activated heme labilization, heme transfer between proteins, and protein relocalization defines a previously undefined mechanism of H2O2 signaling in cells. In contrast, established H2O2 signaling mechanisms are dominated by thiol-based redox changes.
Keywords: mitochondrial H2O2, heme transfer, cytochrome c peroxidase, catalase A, reverse translocation
Abstract
In exponentially growing yeast, the heme enzyme, cytochrome c peroxidase (Ccp1) is targeted to the mitochondrial intermembrane space. When the fermentable source (glucose) is depleted, cells switch to respiration and mitochondrial H2O2 levels rise. It has long been assumed that CCP activity detoxifies mitochondrial H2O2 because of the efficiency of this activity in vitro. However, we find that a large pool of Ccp1 exits the mitochondria of respiring cells. We detect no extramitochondrial CCP activity because Ccp1 crosses the outer mitochondrial membrane as the heme-free protein. In parallel with apoCcp1 export, cells exhibit increased activity of catalase A (Cta1), the mitochondrial and peroxisomal catalase isoform in yeast. This identifies Cta1 as a likely recipient of Ccp1 heme, which is supported by low Cta1 activity in ccp1Δ cells and the accumulation of holoCcp1 in cta1Δ mitochondria. We hypothesized that Ccp1’s heme is labilized by hyperoxidation of the protein during the burst in H2O2 production as cells begin to respire. To test this hypothesis, recombinant Ccp1 was hyperoxidized with excess H2O2 in vitro, which accelerated heme transfer to apomyoglobin added as a surrogate heme acceptor. Furthermore, the proximal heme Fe ligand, His175, was found to be ∼85% oxidized to oxo-histidine in extramitochondrial Ccp1 isolated from 7-d cells, indicating that heme labilization results from oxidation of this ligand. We conclude that Ccp1 responds to respiration-derived H2O2 via a previously unidentified mechanism involving H2O2-activated heme transfer to apoCta1. Subsequently, the catalase activity of Cta1, not CCP activity, contributes to mitochondrial H2O2 detoxification.
Cytochrome c peroxidase (Ccp1) is a monomeric nuclear encoded protein with a 68-residue N-terminal mitochondrial targeting sequence (1). This presequence crosses the inner mitochondrial membrane and is cleaved by matrix proteases (2, 3). Mature heme-loaded Ccp1 is found in the mitochondrial intermembrane space (IMS) in exponentially growing yeast (2, 3) but the point of insertion of its single b-type heme is unknown. Under strict anaerobic conditions, Ccp1 is present in mitochondria as the heme-free form or apoform (4). Once cells are exposed to O2 and heme biosynthesis is turned on, apoCcp1 converts rapidly to the mature holoenzyme by noncovalently binding heme (5).
It is well established that mature Ccp1 functions as an efficient H2O2 scavenger in vitro (6). Its catalytic cycle involves the reaction of ferric Ccp1 with H2O2 (Eq. 1) to form compound I (CpdI) with a ferryl (FeIV) heme and a cationic indole radical localized on Trp191 (W191+•). CpdI is one-electron reduced by the ferrous heme of cytochrome c (Cyc1) to compound II (CpdII) with ferryl heme (Eq. 2), and electron donation by a second ferrous Cyc1 returns CpdII to the resting Ccp1III form (Eq. 3):
| [1] |
| [2] |
| [3] |
Because Ccp1 production is not under O2/heme control (4, 5), CCP activity is assumed to be the frontline defense in the mitochondria, a major source of reactive oxygen species (ROS) in respiring cells (7). Contrary to the time-honored assumption that Ccp1 catalytically consumes the H2O2 produced during aerobic respiration (8), recent studies in our group reveal that the peroxidase behaves more like a mitochondrial H2O2 sensor than a catalytic H2O2 detoxifier (9–11). Notably, Ccp1 competes with complex IV for reducing equivalents from Cyc1, which shuttles electrons from complex III (ubiquinol cytochrome c reductase) to complex IV (cytochrome c oxidase) in the electron transport chain (12).
Because CCP activity in the IMS siphons electrons from energy production, an H2O2 sensor role for Ccp1 should be energetically more favorable for the cell. Key evidence for a noncatalytic role for Ccp1 in H2O2 removal is that the isogenic strain producing the catalytically inactive Ccp1W191F protein accumulates less H2O2 than wild-type cells (10). In fact, this mutant strain exhibits approximately threefold higher catalase A (Cta1) activity than wild-type cells (10) whereas CCP1 deletion results in a strain (ccp1Δ) with negligible Cta1 activity and high H2O2 levels (5). Unlike Cta1, which is the peroxisomal and mitochondrial catalase isoform in yeast (13), the cytosolic catalase Ctt1 (14) exhibits comparable activity in the wild-type, Ccp1W191F, and ccp1Δ strains (10). Given that both Ccp1 and Cta1 are targeted to mitochondria, we hypothesized that Ccp1 may transfer its heme to apoCta1 in respiring cells.
Cta1 is nuclear encoded with embedded mitochondrial and peroxisomal targeting sequences (15). Like Ccp1, each monomer noncovalently binds a b-type heme and mature Cta1 is active as a homotetramer. Synthesis of the Cta1 monomer is under O2/heme control such that the apoenzyme begins to accumulate only during the logarithmic phase of aerobic growth (16). Hence, its O2/heme independent production (4, 5) allows apoCcp1 to acquire heme while cells are synthesizing apoCta1. This, combined with our observation that Cta1 activity increases in respiring cells producing Ccp1 or Ccp1W191F but not in ccp1Δ cells (10), led us to speculate that respiration-derived H2O2 triggers heme donation from Ccp1 to apoCta1 within mitochondria.
What experimental evidence would support heme donation by Ccp1? It has been demonstrated that mutation of the proximal heme Fe ligand, His175, to a residue with weak or no Fe-coordinating ability produces Ccp1 variants (H175P, H175L, H175R, and H175M) that undergo mitochondrial processing but do not accumulate in isolated yeast mitochondria (17). Presumably, reduced heme affinity allows the Ccp1 variants to unfold and cross the outer mitochondrial membrane (17). Hence, we argued that if wild-type Ccp1 donated its heme, the apoprotein would likewise exit mitochondria. Consequently, we examine here age-dependent Ccp1–green fluorescent protein (Ccp1-GFP) localization in live cells chromosomally expressing Ccp1 C-terminally fused to GFP as well as the distribution of wild-type Ccp1 between subcellular fractions. Because weakening or removal of the proximal Fe ligand on His175 mutation reduces heme affinity (17), His175 oxidation in wild-type Ccp1 should have a similar effect, which we investigate here. We further speculated that in the absence of apoCta1 as an acceptor for its heme, more Ccp1 would remain trapped in the IMS so we compare mitochondrial Ccp1 levels in wild-type and cta1∆ cells. Our combined results support triggering of heme donation from Ccp1 to apoCta1 by respiration-derived H2O2. Such H2O2-activated heme transfer between proteins has not been reported to date and its implications in H2O2 signaling are discussed.
Results
Ccp1-GFP and Ccp1 Are Selectively Exported from Mitochondria of Respiring Cells.
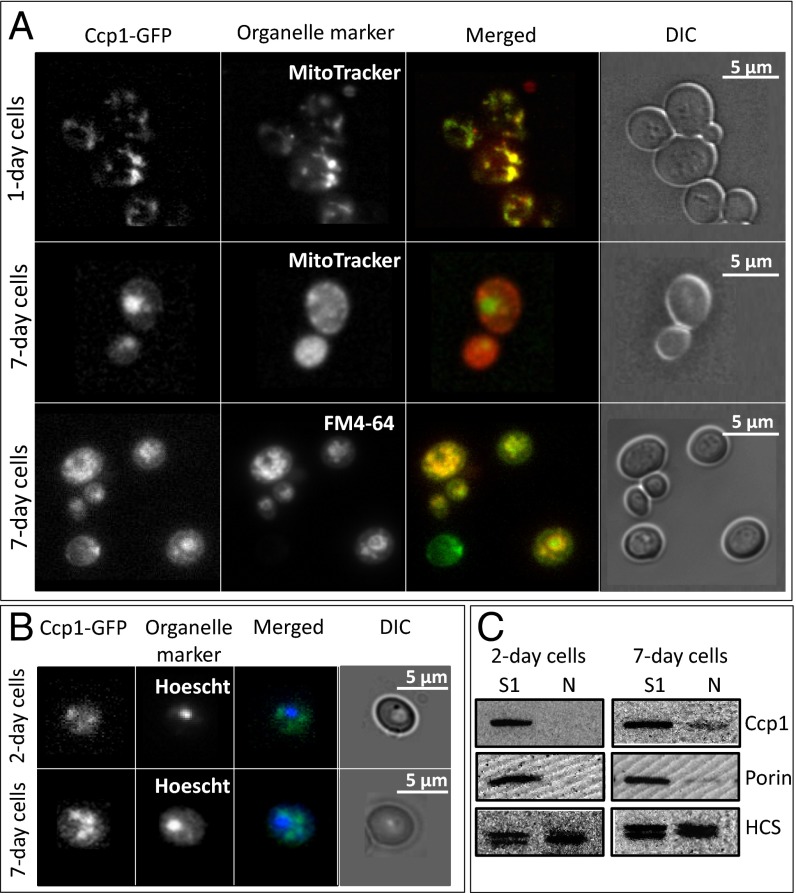
The cellular location of Ccp1-GFP was tracked in live cells by epifluorescence microscopy. GFP fluorescence and that of MitoTracker, a mitochondrial probe, overlap in 1-d cultures (Fig. 1A), consistent with previous reports that Ccp1-GFP is localized in the IMS of exponentially growing yeast (2). In contrast, cells from 7-d cultures exhibit distinct regions of GFP and MitoTracker fluorescence, indicating extramitochondrial localization of Ccp1-GFP. GFP fluorescence appears as large circular structures while the mitochondrial morphology changes from a reticular network in 1-d cells to diffuse spherical organelles in stationary-phase cells as previously reported (18). Notably, GFP fluorescence colocalizes with the vacuolar membrane marker FM4-64 in 7-d cells (Fig. 1A), suggesting that extramitochondrial Ccp1-GFP may be targeted to the vacuole.
Fig. 1.
Ccp1-GFP is mitochondrial in 1-d and vacuolar in 7-d yeast cells; Ccp1 is present in nuclei from 7-d cells. Live Ccp1-GFP–expressing cells were visualized for (A) GFP (480 nm, 535/25 nm), stained with MitoTracker (555 nm, 630/75 nm) and FM4-64 (480 nm, 550 nm long-pass filter) to visualize mitochondria and vacuole, respectively, and with (B) Hoescht (405 nm, 488/75 nm) to visualize nuclei. Phase-contrast microscopy (DIC) was used to monitor cell integrity. (C) Immunoblot analysis with anti-Ccp1 of total (S1) and nuclear (N) fractions isolated from 2-d and 7-d yeast. Porin and homocitrate synthase (HCS), which exists as 47-kDa and 49-kDa isoforms in yeast (51), were used as nuclear and mitochondrial outer membrane markers, respectively.
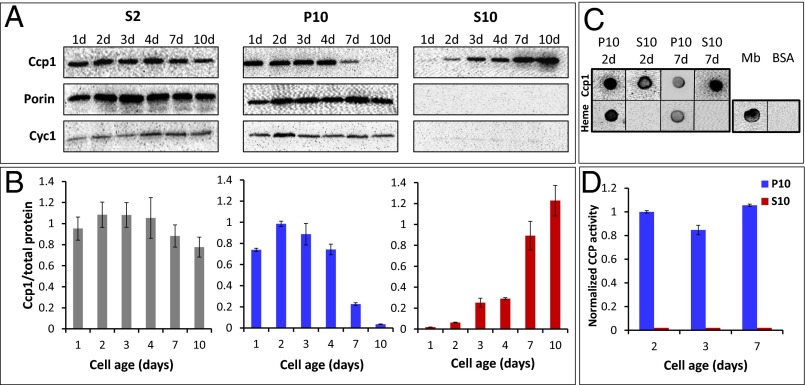
Probing subcellular fractions obtained by differential centrifugation with a polyclonal anti-Ccp1 antibody yields results in agreement with live-cell imaging. [We note here that anti-Ccp1 detects all of the chemical forms of Ccp1 of interest in our study (apoCcp1, holoCcp1, CpdI, and hyperoxidized Ccp1) with the same sensitivity (SI Appendix, Fig. S7 A and B)]. Western blotting reveals a relatively constant amount of Ccp1 or Ccp1-GFP in the denucleated (S2) fractions over 1–10 d, but the levels in mitochondria-free (S10) fractions increase dramatically at the expense of those in mitochondria-enriched (P10) fractions (Fig. 2 A and B and SI Appendix, Fig. S1 A and B). Notably, Ccp1 is barely detectable in P10 after 10 d but highly abundant in S10, indicating that during this period most of the protein escapes from mitochondria. Furthermore, processing of both Ccp1 and its GFP fusion by the matrix proteases (2) appears to be complete because the immature proteins with their additional 7-kDa mitochondrial targeting sequence would be detectable as anti-Ccp1 and anti-GFP bands above those of the mature proteins (Fig. 2A and SI Appendix, Fig. S1A). To establish whether Ccp1 export is selective or the result of nonspecific mitochondrial membrane damage, the fractions were probed with anti-porin and anti-Cyc1 as mitochondrial outer membrane and IMS markers, respectively. As shown in Fig. 2A and SI Appendix, Fig. S1A, the S10 fractions are not immunoreactive with these antibodies, so mitochondria remain intact. Thus, we conclude that apoCcp1 and apoCcp1-GFP selectively exit the mitochondria of respiring cells.
Fig. 2.
Ccp1 exits mitochondria as yeast begin respiring and extramitochondrial Ccp1 does not possess CCP activity. (A) Immunoblot analysis of equal volumes of denucleated (S2), mitochondrial (P10), and cytosolic (S10) fractions vs. cell age. Porin and Cyc1 are mitochondrial outer membrane and IMS markers, respectively. (B) The Ccp1 signals in A were quantified and normalized to the sum of the Coomassie bands in the same lane. (C) Dot blot analysis with anti-Ccp1 (Top row) and the ECL reagent (luminol/H2O2) to detect heme (Bottom row) from 2-d and 7-d P10 and S10 fractions. Myoglobin and BSA were used as positive and negative heme controls, respectively. (D) Normalized CCP activity in mitochondrial (P10) and cytosolic (S10) fractions. Specific activity was ratioed by the Ccp1 protein levels in B and normalized to the level for 2-d cells (SI Appendix, Table S2). Results in A and C are representative of three independent cultures (n = 3) and averages ± SD are plotted in B and D.
Ccp1 Is Targeted to the Nucleus and Possibly the Vacuole.
No overlap between the Hoechst nuclear dye and GFP fluorescence is observed at any cell age (Fig. 1B), indicating that Ccp1-GFP does not accumulate in the nucleus. Because the fusion protein (62 kDa) is above the size cutoff (60 kDa) for diffusion through the nuclear pore (19), nuclear enriched fractions (N) were probed for native Ccp1 (34 kDa). No Ccp1 was detected in nuclei isolated from 2-d cells but ∼10% was reproducibly detected in 7-d nuclei (Fig. 1C) and may function as a retrograde messenger (20) (Discussion). Ccp1, like Ccp1-GFP (Fig. 1A), also may be targeted to the vacuole in 7-d cells. However, this could not be confirmed because vacuoles, which are present in the S10 fraction of young cells, fragment during spheroplasting of 7-d cells.
Extramitochondrial Ccp1 Is Catalytically Inactive.
Staining for the intrinsic peroxidase activity of heme (21) confirmed that Ccp1 purified from 2-d and 7-d S10 subcellular fractions is in the apoform (Fig. 2C). Consistent with this absence of Ccp1 heme, no CCP activity is detected in the S10 fractions at any cell age (Fig. 2D and SI Appendix, Table S2) but adding exogenous hemin does not restore activity. Further experiments discussed below reveal that extramitochondrial apoCcp1 is oxidized, which prevents reformation of catalytically active Ccp1 on hemin addition. In contrast, the purified P10 mitochondrial-enriched fractions stain for heme (Fig. 2C) and are catalytically active (Fig. 2D). Ratioing CCP activity against Ccp1 protein level in Fig. 2B reveals that the activity of mitochondrial Ccp1 is relatively constant with cell age (Fig. 2D). Interestingly, the fractions containing Ccp1-GFP and Ccp1 exhibit very similar CCP activity (Fig. 2D and SI Appendix, Fig. S1C and Table S2), suggesting that fusion of Ccp1's C terminus to GFP does not interfere with CpdI reduction by Cyc1II (Eqs. 2 and 3).
Cta1 Activity Increases as Ccp1 Exits Mitochondria.
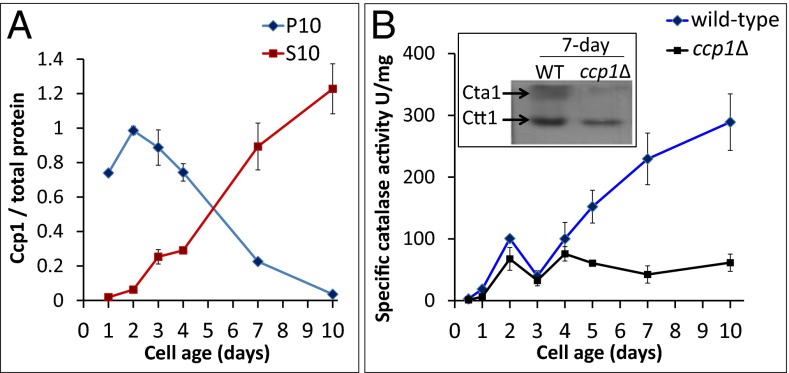
As mitochondrial Ccp1 levels drop in wild-type cells, their total catalase activity increases proportionally (Fig. 3). This led us to postulate that catalase may be an acceptor of Ccp1’s heme, which is supported by the depressed catalase activity in ccp1∆ cells (Fig. 3B). We separately monitored the activity of each catalase isoform, using an in-gel assay (10), and found comparable Ctt1 activity in the two strains, but approximately fivefold higher Cta1 activity in 7-d wild-type vs. ccp1∆ cells (Fig. 3B, Inset). This identifies apoCta1 as a mitochondrial recipient of Ccp1’s heme, which we further examined in catalase-null strains.
Fig. 3.
Catalase activity in wild-type yeast increases at the expense of mitochondrial Ccp1. (A) Ccp1 protein level vs. cell age in the S10 and P10 fractions from Fig. 2B. (B) Specific catalase activity in soluble protein extracts from wild-type and ccp1∆ cells vs. cell age. Results are averages of three independent cultures (n = 3) ± SD. (Inset) In-gel assay of Cta1 and Ctt1 catalase activities in 7-d soluble protein extracts (2.5 μg protein per lane).
Ccp1 Accumulates in Mitochondria of cta1Δ Cells.
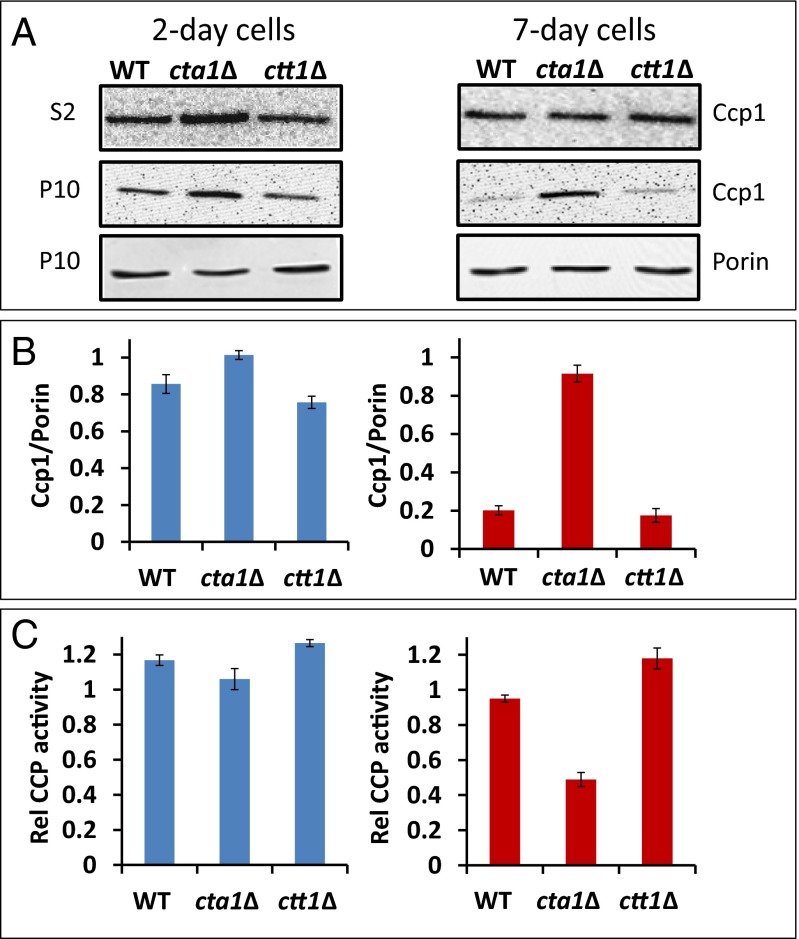
If apoCta1 is indeed an acceptor of Ccp1 heme, then holoCcp1 should accumulate in the mitochondria of cells deleted for Cta1. On probing denucleated S2 and mitochondrial-enriched P10 fractions, we detect similar Ccp1 protein levels in 2-d wild-type, ctt1Δ, and cta1Δ strains but 4.5-fold more Ccp1 in 7-d cta1Δ mitochondria (Fig. 4 A and B). Hence, more heme-loaded Ccp1 is trapped in mitochondria when apoCta1 is not present as a heme acceptor. Although the total CCP activity of cta1Δ cells is double that of wild-type or ctt1Δ cells (SI Appendix, Table S3), ratioing CCP activity against 7-d mitochondrial Ccp1 protein levels (Fig. 4B) reveals that Ccp1 in cta1Δ mitochondria is <50% active (Fig. 4C). We attribute this to Ccp1 hyperoxidation by the elevated H2O2 levels found in the cta1Δ strain (22), a conclusion supported by the results presented in the next section.
Fig. 4.
Ccp1 accumulates in the mitochondria of cta1∆ yeast. (A) Immunoblot analysis of denucleated (S2) and mitochondrial (P10) fractions isolated from wild-type (WT), cta1∆, and ctt1∆ cells. (B) The Ccp1 signals for the P10 fractions in A were quantified and normalized to the porin signal (mitochondrial outer membrane marker) for 2-d (blue bars) and 7-d (red bars) cells. (C) Specific CCP activity (µmol⋅min−1⋅mg−1 total protein) of the P10 fractions in B ratioed by their Ccp1 protein levels to give the relative amount of active CCP remaining in mitochondria. Results in A are representative of three independent cultures (n = 3) and averages ± SD are plotted in B and C.
Heme-Mediated Ccp1 Hyperoxidation by H2O2 Results in Heme Labilization.
Intracellular H2O2 levels rise ∼10-fold at the diauxic shift around day 2 (Fig. 5A) as cells switch to respiratory metabolism (23). Low levels of Cyc1 are likely present in the IMS during these early phases of oxygen adaptation because CYC1 transcription is induced by oxygen (24) and repressed by glucose (25). Thus, we hypothesize that the burst in respiration-derived H2O2 overwhelms the available reducing capacity of Cyc1II, hyperoxidizes Ccp1, and labilizes its heme, which is donated to apoCta1 as the latter accumulates in respiring mitochondria. To test this hypothesis, we compared heme transfer in vitro to apomyoglobin (apoMb) from untreated Ccp1 and Ccp1 hyperoxidized by 10 M eq of H2O2 because the peroxidase is known to consume this quantity of H2O2 in vitro before heme modification (26). ApoMb, the prototypical heme acceptor (27), was selected because of its high heme affinity (Kd = 3 × 10−15 M) (28) and high stability (29). Following incubation with apoMb, ∼6 µM and ∼12 µM heme are lost from Ccp1 and hyperoxidized Ccp1, respectively (SI Appendix, Table S4), based on the decrease in their Soret absorbance (SI Appendix, Fig. S2B). Thus, hyperoxidized Ccp1 donates twice as much heme as the untreated peroxidase. In the absence of an acceptor, the Soret band of hyperoxidized Ccp1 does not decrease (SI Appendix, Table S4 and Fig. S2B) so no heme escapes to the solvent. In fact, heme transfer between the proteins is stoichiometric because heme-loaded Mb and apoCcp1 increase by the same concentration (SI Appendix, Table S4 and Fig. S2 B and D).
Fig. 5.
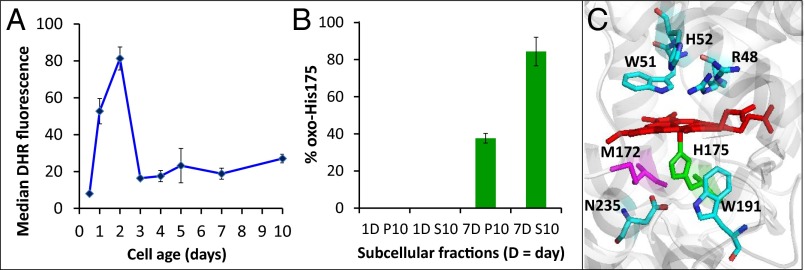
Ccp1 is oxidized at His175. (A) FACS measurements of H2O2 in wild-type cells stained with dihydrorhodamine 123 (DHR) (10). Data points are the median fluorescence per cell (in relative fluorescence units, RFU) measured for 10,000 cells per sample. (B) Percentage of oxidized His175 in mitochondrial (P10) and extramitochondrial (S10) Ccp1 from 1-d and 7-d yeast cells based on tryptic peptide peak areas in the LC-MS spectra (note that no His175 oxidation is detected in 1-d cells). See SI Appendix, Fig. S6 and SI Materials and Methods for further experimental details. (C) Ribbon diagram of the heme-binding site of Ccp1 showing the proximal Fe ligand His175 (green). Residues important in the activation of H2O2 following its binding to the vacant sixth coordination site of FeIII are also shown. This diagram was generated using PyMOL software with the coordinates from Protein Data Bank 1ZBY.
Ferric heme (hemin) released from reconstituted Mb has an exact mass that differs by <1 ppm from that of authentic hemin (SI Appendix, Fig. S3). Also, the absorption spectrum of reconstituted Mb is indistinguishable from that of the native protein (SI Appendix, Fig. S2C) so we conclude that unmodified heme is transferred from hyperoxidized Ccp1 to apoMb. In contrast, ∼16 oxygen adducts are detected in the mass spectrum of hyperoxidized Ccp1 (SI Appendix, Fig. S4B), which confirms our previous reports (30, 31) that Ccp1’s residues are extensively oxidized by excess H2O2 in the absence of its reducing substrate, Cyc1II (Eqs. 2 and 3). Liquid chromatography (LC)-MS/MS analysis of tryptic peptides from hyperoxidized Ccp1 reveals that 42% of His175 is converted to oxo-histidine (SI Appendix, Fig. S5 and Table S5). Weakening of the axial ligand on His175 oxidation will labilize Ccp1’s heme and we observe a doubling of heme transfer from hyperoxidized Ccp1 to apoMb (SI Appendix, Table S4).
To establish whether Ccp1 is hyperoxidized in vivo, we isolated the protein from P10 and S10 subcellular fractions. No oxo-histidine is detected in Ccp1 isolated from 1-d cells but ∼85% of His175 is found to be oxidized in the extramitochondrial protein from 7-d respiring cells compared with ∼35% His175 oxidation in the mitochondrial protein (Fig. 5B). The extensive oxidation of His175 in extramitochondrial Ccp1 indicates that the peroxidase is indeed overwhelmed by H2O2 in vivo. This serves to labilize the heme and we anticipate that heme transfer is more efficient in mitochondria than in vitro (SI Appendix, Fig. S2) because apoMb is not a biological partner of Ccp1. The lack of CCP activity on hemin addition to extramitochondrial Ccp1 can also be attributed to decreased heme affinity on His175 oxidation.
Importantly, we note that apoCcp1 is not oxidized by H2O2 (SI Appendix, Fig. S4D). Activation of H2O2 by heme peroxidases involves binding to the heme FeIII (Eq. 1) and Ccp1 can sequentially bind and reduce multiple molecules of H2O2, using its polypeptide as an electron source in the absence of Cyc1II (30, 31). Thus, on activating H2O2, Ccp1’s heme promotes its own transfer by catalyzing the oxidation of its polypeptide host, including its Fe ligand, His175 (Fig. 5 B and C). We conclude that Ccp1 hyperoxidation by respiration-derived H2O2 triggers heme transfer to apoCta1 in mitochondria and that His175 oxidation is critical in this process.
Discussion
Evidence for Ccp1 as a Heme Donor.
Immature preCcp1 is targeted to yeast mitochondria where it is processed to the heme-loaded mature holoprotein (2, 3, 17). In this state it is trapped in the IMS, and hence it is classified as an IMS protein in exponentially growing fermenting yeast cells (2, 3, 17). However, using a combination of live-cell imaging, immunoanalysis of subcellular fractions, and CCP activity assays, we demonstrate that respiration triggers apoCcp1 exit from mitochondria. Reverse translocation of proteins processed by mitochondria depends on the stability of their folded forms (32). Previously, we investigated Ccp1 denaturation and uncovered a conformational stability at the low end of the range reported for globular proteins (33) so it is not surprising that heme binding is required for Ccp1’s retention in the IMS (17). Also, mature holoCcp1 added to isolated mitochondria does not enter these organelles (17), demonstrating that holoCcp1 does not cross the outer mitochondrial membrane. Therefore, escape of Ccp1 from mitochondria is tantamount to its acting as a mitochondrial heme donor. Incidentally, Ccp1’s substrate Cyc1 also requires heme for IMS retention (34). Mature Cyc1 possesses a covalently bound heme and deletion of its heme lyase (Cyc3) or mutation of residues that serve as sites of heme attachment (Cys19 and/or Cys22) results in cytoplasmic accumulation of apoCyc1, which adopts an unfolded structure in vitro (35).
Cta1 as a Mitochondrial Acceptor of Ccp1 Heme.
The synthesis in yeast of both catalase isoforms is heme regulated. However, catalase T protein (Ctt1) activity is detected during the early phases of heme synthesis (16, 36, 37) whereas Cta1 accumulates as the apoprotein in exponentially growing yeast (16, 36, 37). The heme donor to Ctt1 is unknown but Cta1 activity increases in parallel with extramitochondrial apoCcp1 buildup (Fig. 3 A and B), which links holoCta1 formation to H2O2-induced labilization of Ccp1’s heme. The negligible Cta1 activity found in ccp1∆ cells (Fig. 3B) and the high Ccp1 protein levels in cta1∆ mitochondria (Fig. 4B) further identify apoCta1 as an acceptor of Ccp1 heme.
HoloCcp1 accumulates during heme synthesis (4, 5) so heme-mediated hyperoxidation of the protein occurs when H2O2 sharply increases in respiring yeast (Fig. 5A). Hyperoxidized Ccp1 donates its heme to apoCta1 and Cta1 activity consumes mitochondrial H2O2. Hence, H2O2 levels are elevated in cta1∆ cells (22) despite their twofold higher CCP activity than that in wild-type cells (SI Appendix, Table S3). Trapping more Ccp1 in cta1∆ mitochondria does not compensate for the absence of Cta1’s H2O2 scavenging activity. On the contrary, Ccp1 from cta1∆ mitochondria is only 50% active (Fig. 4C), which we attribute to its hyperoxidation by H2O2 due to limited availability of reducing equivalents from Cyc1II in the IMS.
Heme Labilization by His175 Oxidation.
Irreversible histidine oxidation has been associated with the toxic effects of peroxides in aging and neurodegeneration (38). However, in peroxide resistance protein (PerR), H2O2 binds to the FeII center of iron-replete PerR, which results in oxidation of the metal’s His37 and His91 ligands to 2-oxo-histidine (39, 40). This promotes iron release and apoPerR dissociation from DNA with the induction of target genes including kat A (catalase), ahpCF (alkyl hydroperoxide reductase), and hemAXCDBL (heme biosynthesis operon) (40). Analogous to the nonheme FeII of PerR, the heme FeIII of Ccp1 binds H2O2, which under stress conditions (high H2O2, low Cyc1II) oxidizes the proximal His175 ligand to oxo-histidine (Fig. 5 and SI Appendix, Fig. S5) and activates Cta1 catalysis (Fig. 3). Thus, PerR and mitochondrial Ccp1 sense and mediate a H2O2 stress signal by similar iron-dependent mechanisms to regulate key defensive responses at the transcriptional level for PerR and at the posttranslational level for Ccp1. To the best of our knowledge, we provide the first report of heme labilization in a heme protein by ligand oxidation. Binding of nitric oxide to the sixth coordinate position cleaves or weakens the proximal FeII-His bond in some heme proteins, including soluble guanylate cyclase (41) and nitrophorin isoform 7 (42), but this process has not been associated with heme transfer.
Why Cells Produce Ccp1 as a Sensor.
In the absence of a known cellular mechanism for oxo-histidine reduction, it has been speculated that oxidized apoPerR may be degraded (39, 40). Hyperoxidized Ccp1 also may be degraded because Ccp1-GFP is associated with the vacuole in 7-d cells (Fig. 1A). Why would yeast use Ccp1 as a sacrificial H2O2 sensor instead of producing active Cta1 before cells begin to respire? The H2O2 spike around the diauxic shift triggers a beneficial stress response known as mitohormesis (43) that results in increased superoxide dismutase (Sod2) activity and depressed O2•− levels (10). We reported that the Ccp1W191F strain, which possesses high catalase activity during early exponential growth due to extensive Ccp1W191F hyperoxidation, mounts a weak mitohormesis response and has a short lifespan (10, 11). Thus, it appears that H2O2 dismutation by Cta1 activity during the early phases of oxygen adaption abrogates or attenuates the beneficial H2O2 stress signal. In contrast, because CCP activity also depends on Cyc1II levels (Eqs. 2 and 3), wild-type Ccp1 temporally controls the signal from respiration-derived H2O2 to trigger mitohormesis and modulate lifespan (10, 11).
The Role of Extramitochondrial ApoCcp1.
A large fraction of apoCcp1-GFP and possibly untagged apoCcp1 translocate to the vacuole (Fig. 1A). This may be the cell’s mechanism for removing or recycling hyperoxidized apoCcp1 and/or vacuolar apoCcp1 may be involved in signaling analogous to yeast enolase (44). Around 10% of apoCcp1 translocates to the nucleus (Fig. 1B). There it likely conveys an oxidative stress signal to the nuclear transcription factor Skn7 when yeast are challenged with exogenous H2O2 (45). We found ccp1∆ cells to be considerably more H2O2 sensitive than wild-type or ccp1W191F cells because of their inability to up-regulate catalase and peroxiredoxin activities on H2O2 challenge (10). Ccp1 is not present in the deletion mutant to transmit a stress signal to Skn7, which regulates the expression of many antioxidant enzymes, including cytosolic Ctt1 (46, 47). Hence, reverse translocation of apoCcp1 to the nucleus provides a retrograde message (20) vital in the cell’s response to exogenous H2O2, a process that merits further investigation.
Conclusions
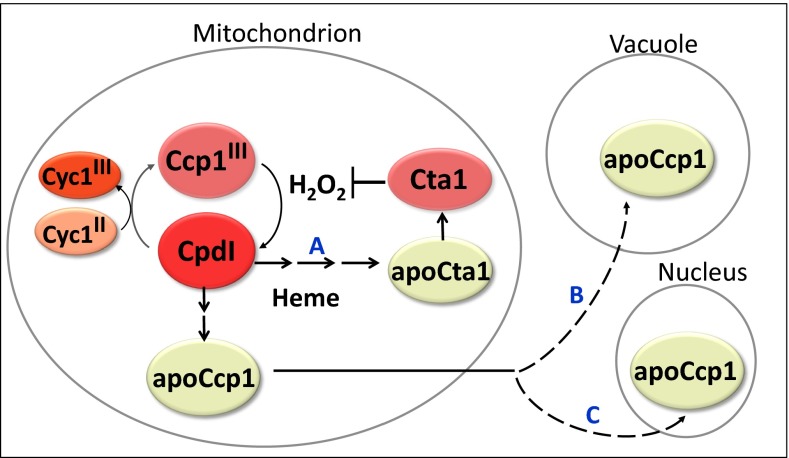
Fig. 6 summarizes our model of respiration-triggered heme transfer from Ccp1 to Cta1 in mitochondria. A key step in this model is the heme-mediated oxidation by H2O2 of the proximal Fe ligand, His175. Before we identified Ccp1 as a heme-based H2O2 sensor (10), Bacillus subtilis PerR was the only documented nonthiol H2O2 sensor (40), whereas well-characterized H2O2 sensor proteins such as Yap1 in yeast and OxyR in bacteria undergo reversible thiol oxidation upon exposure to H2O2 (48, 49). Over 70 y of research portrays Ccp1 as an antioxidant enzyme that functions to protect yeast mitochondria by catalytically consuming H2O2 (6, 50). Based on our current and previous investigations of its physiological functions (9–11), Ccp1 may serve in future as a paradigm of heme-based H2O2 sensing and heme transfer.
Fig. 6.
Respiration-triggered heme transfer in yeast mitochondria. H2O2 generated during mitochondrial respiration oxidizes Ccp1 to compound I (CpdI). The oxidizing equivalents from H2O2 can be reduced by Cyc1II or transferred to the peroxidase’s residues, including the proximal Fe ligand His175 to yield oxo-histidine. Formation of the latter labilizes the heme group, which is transferred either directly or via unidentified intermediate(s) to apoCta1 (path A), and the nascent Cta1 activity detoxifies H2O2. ApoCcp1 has low conformational stability and undergoes reverse translocation to the vacuole (path B) and the nucleus (path C).
Materials and Methods
Yeast strains used in this study (SI Appendix, Table S1) are in the BY4741 genetic background. The wild-type strain was purchased from the European Saccharomyces cerevisiae Archive for Functional Analysis (EUROSCARF) and the Ccp1-GFP–expressing strain was purchased from Invitrogen Life Sciences. The cta1∆ and ctt1∆ strains were kind gifts from Christopher Brett (Concordia University), and the DNA template for recombinant His6-Ccp1 expression was generously provided by Yu Li (University of Illinois at Urbana–Champaign, Urbana, IL). SI Appendix, SI Materials and Methods outlines the live-cell imaging of Ccp1-GFP by fluorescence microscopy, the isolation and immunoblotting of yeast nuclei and subcellular fractions, the isolation of Ccp1 from yeast by anion-exchange chromatography, heme blotting, assays of CCP and catalase activity, expression and purification of recombinant Ccp1 from Escherichia coli, investigation of heme transfer in vitro, and mass spectrometric analysis of hyperoxidized Ccp1.
Supplementary Material
Acknowledgments
Profs. Christopher Brett and Yu Li are thanked for providing the cta1∆ and ctt1∆ knockout strains and for the DNA template for recombinant His6-Ccp1, respectively. We also are grateful to Marc Ouellet and Profs. Christopher Brett and Alisa Piekny (Concordia University) for helpful discussions. This work was funded by grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada and Concordia University to A.M.E., who also holds a Senior Concordia University Research Chair. M.K. and D.M. acknowledge doctoral scholarships from NSERC and FRQNT (Fonds de recherche du Québec – Nature et technologies), respectively, and additional awards from Concordia University and the Quebec Network for Research on Protein Function, Structure, and Engineering (PROTEO).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409692111/-/DCSupplemental.
References
- 1.Kaput J, Goltz S, Blobel G. Nucleotide sequence of the yeast nuclear gene for cytochrome c peroxidase precursor. Functional implications of the pre sequence for protein transport into mitochondria. J Biol Chem. 1982;257(24):15054–15058. [PubMed] [Google Scholar]
- 2.Suppanz IE, Wurm CA, Wenzel D, Jakobs S. The m-AAA protease processes cytochrome c peroxidase preferentially at the inner boundary membrane of mitochondria. Mol Biol Cell. 2009;20(2):572–580. doi: 10.1091/mbc.E07-11-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michaelis G, et al. Mitochondrial signal peptidases of yeast: The rhomboid peptidase Pcp1 and its substrate cytochrome C peroxidase. Gene. 2005;354:58–63. doi: 10.1016/j.gene.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Sels AA, Cocriamont C. Induced conversion of a protein precursor into cytochrome C peroxidase during adaptation of yeast to oxygen. Biochem Biophys Res Commun. 1968;32(2):192–198. doi: 10.1016/0006-291x(68)90368-9. [DOI] [PubMed] [Google Scholar]
- 5.Djavadi-Ohaniance L, Rudin Y, Schatz G. Identification of enzymically inactive apocytochrome c peroxidase in anaerobically grown Saccharomyces cerevisiae. J Biol Chem. 1978;253(12):4402–4407. [PubMed] [Google Scholar]
- 6.Volkov AN, Nicholls P, Worrall JAR. The complex of cytochrome c and cytochrome c peroxidase: The end of the road? Biochim Biophys Acta. 2011;1807(11):1482–1503. doi: 10.1016/j.bbabio.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Perrone GG, Tan S-X, Dawes IW. Reactive oxygen species and yeast apoptosis. Biochim Biophys Acta. 2008;1783(7):1354–1368. doi: 10.1016/j.bbamcr.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 8.Kwon M, Chong S, Han S, Kim K. Oxidative stresses elevate the expression of cytochrome c peroxidase in Saccharomyces cerevisiae. Biochim Biophys Acta. 2003;1623(1):1–5. doi: 10.1016/s0304-4165(03)00151-x. [DOI] [PubMed] [Google Scholar]
- 9.Jiang H, English AM. Phenotypic analysis of the ccp1Delta and ccp1Delta-ccp1W191F mutant strains of Saccharomyces cerevisiae indicates that cytochrome c peroxidase functions in oxidative-stress signaling. J Inorg Biochem. 2006;100(12):1996–2008. doi: 10.1016/j.jinorgbio.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 10.Martins D, Kathiresan M, English AM. Cytochrome c peroxidase is a mitochondrial heme-based H2O2 sensor that modulates antioxidant defense. Free Radic Biol Med. 2013;65:541–551. doi: 10.1016/j.freeradbiomed.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 11.Martins D, Titorenko VI, English AM. Cells with impaired mitochondrial H2O2 sensing generate less •OH radicals and live longer. Antioxid Redox Signal. 2014;21(10):1490–1503. doi: 10.1089/ars.2013.5575. [DOI] [PubMed] [Google Scholar]
- 12.Skulachev VP. Cytochrome c in the apoptotic and antioxidant cascades. FEBS Lett. 1998;423(3):275–280. doi: 10.1016/s0014-5793(98)00061-1. [DOI] [PubMed] [Google Scholar]
- 13.Cohen G, Rapatz W, Ruis H. Sequence of the Saccharomyces cerevisiae CTA1 gene and amino acid sequence of catalase A derived from it. Eur J Biochem. 1988;176(1):159–163. doi: 10.1111/j.1432-1033.1988.tb14263.x. [DOI] [PubMed] [Google Scholar]
- 14.Hartig A, Ruis H. Nucleotide sequence of the Saccharomyces cerevisiae CTT1 gene and deduced amino-acid sequence of yeast catalase T. Eur J Biochem. 1986;160(3):487–490. doi: 10.1111/j.1432-1033.1986.tb10065.x. [DOI] [PubMed] [Google Scholar]
- 15.Petrova VY, Drescher D, Kujumdzieva AV, Schmitt MJ. Dual targeting of yeast catalase A to peroxisomes and mitochondria. Biochem J. 2004;380(Pt 2):393–400. doi: 10.1042/BJ20040042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimniak P, Hartter E, Woloszczuk W, Ruis H. Catalase biosynthesis in yeast: Formation of catalase A and catalase T during oxygen adaptation of Saccharomyces cerevisiae. Eur J Biochem. 1976;71(2):393–398. doi: 10.1111/j.1432-1033.1976.tb11126.x. [DOI] [PubMed] [Google Scholar]
- 17.Kaput J, Brandriss MC, Prussak-Wieckowska T. In vitro import of cytochrome c peroxidase into the intermembrane space: Release of the processed form by intact mitochondria. J Cell Biol. 1989;109(1):101–112. doi: 10.1083/jcb.109.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yaffe MP. The cutting edge of mitochondrial fusion. Nat Cell Biol. 2003;5(6):497–499. doi: 10.1038/ncb0603-497b. [DOI] [PubMed] [Google Scholar]
- 19.Dingwall C, Laskey RA. Protein import into the cell nucleus. Annu Rev Cell Biol. 1986;2:367–390. doi: 10.1146/annurev.cb.02.110186.002055. [DOI] [PubMed] [Google Scholar]
- 20.Whelan SP, Zuckerbraun BS. Mitochondrial signaling: Forwards, backwards, and in between. Oxid Med Cell Longev. 2013;2013:351613. doi: 10.1155/2013/351613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feissner R, Xiang Y, Kranz RG. Chemiluminescent-based methods to detect subpicomole levels of c-type cytochromes. Anal Biochem. 2003;315(1):90–94. doi: 10.1016/s0003-2697(02)00658-9. [DOI] [PubMed] [Google Scholar]
- 22.Mesquita A, et al. Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc Natl Acad Sci USA. 2010;107(34):15123–15128. doi: 10.1073/pnas.1004432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg AA, et al. Effect of calorie restriction on the metabolic history of chronologically aging yeast. Exp Gerontol. 2009;44(9):555–571. doi: 10.1016/j.exger.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Burke PV, Raitt DC, Allen LA, Kellogg EA, Poyton RO. Effects of oxygen concentration on the expression of cytochrome c and cytochrome c oxidase genes in yeast. J Biol Chem. 1997;272(23):14705–14712. doi: 10.1074/jbc.272.23.14705. [DOI] [PubMed] [Google Scholar]
- 25.Wright RM, Poyton RO. Release of two Saccharomyces cerevisiae cytochrome genes, COX6 and CYC1, from glucose repression requires the SNF1 and SSN6 gene products. Mol Cell Biol. 1990;10(3):1297–1300. doi: 10.1128/mcb.10.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erman JE, Yonetani T. A kinetic study of the endogenous reduction of the oxidized sites in the primary cytochrome c peroxidase-hydrogen peroxide compound. Biochim Biophys Acta. 1975;393(2):350–357. doi: 10.1016/0005-2795(75)90061-6. [DOI] [PubMed] [Google Scholar]
- 27.Paul J, Chen W, Ohlsson P-I, Smith M. Heme transfer reactions: An important prerequisite for synthetic oxygen carriers. Turkish J Chem. 1998;22(2):103–108. [Google Scholar]
- 28.Hargrove MS, et al. Stability of myoglobin: A model for the folding of heme proteins. Biochemistry. 1994;33(39):11767–11775. doi: 10.1021/bi00205a012. [DOI] [PubMed] [Google Scholar]
- 29.Adams PA. The kinetics of the recombination reaction between apomyoglobin and alkaline haematin. Biochem J. 1977;163(1):153–158. doi: 10.1042/bj1630153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fox T, Tsaprailis G, English AM. Fluorescence investigation of yeast cytochrome c peroxidase oxidation by H2O2 and enzyme activities of the oxidized enzyme. Biochemistry. 1994;33(1):186–191. doi: 10.1021/bi00167a024. [DOI] [PubMed] [Google Scholar]
- 31.Tsaprailis G, English AM. Different pathways of radical translocation in yeast cytochrome c peroxidase and its W191F mutant on reaction with H(2)O(2) suggest an antioxidant role. J Biol Inorg Chem. 2003;8(3):248–255. doi: 10.1007/s00775-002-0407-6. [DOI] [PubMed] [Google Scholar]
- 32.Yogev O, Naamati A, Pines O. Fumarase: A paradigm of dual targeting and dual localized functions. FEBS J. 2011;278(22):4230–4242. doi: 10.1111/j.1742-4658.2011.08359.x. [DOI] [PubMed] [Google Scholar]
- 33.Tsaprailis G, Chan DW, English AM. Conformational states in denaturants of cytochrome c and horseradish peroxidases examined by fluorescence and circular dichroism. Biochemistry. 1998;37(7):2004–2016. doi: 10.1021/bi971032a. [DOI] [PubMed] [Google Scholar]
- 34.Dumont ME, Cardillo TS, Hayes MK, Sherman F. Role of cytochrome c heme lyase in mitochondrial import and accumulation of cytochrome c in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11(11):5487–5496. doi: 10.1128/mcb.11.11.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher WR, Taniuchi H, Anfinsen CB. On the role of heme in the formation of the structure of cytochrome c. J Biol Chem. 1973;248(9):3188–3195. [PubMed] [Google Scholar]
- 36.Hörtner H, et al. Regulation of synthesis of catalases and iso-1-cytochrome c in Saccharomyces cerevisiae by glucose, oxygen and heme. Eur J Biochem. 1982;128(1):179–184. doi: 10.1111/j.1432-1033.1982.tb06949.x. [DOI] [PubMed] [Google Scholar]
- 37.Woloszczuk W, Sprinson DB, Ruis H. The relation of heme to catalase apoprotein synthesis in yeast. J Biol Chem. 1980;255(6):2624–2627. [PubMed] [Google Scholar]
- 38.Uchida K, Kawakishi S. Identification of oxidized histidine generated at the active site of Cu,Zn-superoxide dismutase exposed to H2O2. Selective generation of 2-oxo-histidine at the histidine 118. J Biol Chem. 1994;269(4):2405–2410. [PubMed] [Google Scholar]
- 39.Traoré DAK, et al. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat Chem Biol. 2009;5(1):53–59. doi: 10.1038/nchembio.133. [DOI] [PubMed] [Google Scholar]
- 40.Lee J-W, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440(7082):363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 41.Goodrich LE, Paulat F, Praneeth VKK, Lehnert N. Electronic structure of heme-nitrosyls and its significance for nitric oxide reactivity, sensing, transport, and toxicity in biological systems. Inorg Chem. 2010;49(14):6293–6316. doi: 10.1021/ic902304a. [DOI] [PubMed] [Google Scholar]
- 42.He C, Neya S, Knipp M. Breaking the proximal Fe(II)-N(His) bond in heme proteins through local structural tension: Lessons from the heme b proteins nitrophorin 4, nitrophorin 7, and related site-directed mutant proteins. Biochemistry. 2011;50(40):8559–8575. doi: 10.1021/bi201073t. [DOI] [PubMed] [Google Scholar]
- 43.Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis) Exp Gerontol. 2010;45(6):410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 44.Decker BL, Wickner WT. Enolase activates homotypic vacuole fusion and protein transport to the vacuole in yeast. J Biol Chem. 2006;281(20):14523–14528. doi: 10.1074/jbc.M600911200. [DOI] [PubMed] [Google Scholar]
- 45.Charizanis C, Juhnke H, Krems B, Entian KD. The mitochondrial cytochrome c peroxidase Ccp1 of Saccharomyces cerevisiae is involved in conveying an oxidative stress signal to the transcription factor Pos9 (Skn7) Mol Gen Genet. 1999;262(3):437–447. doi: 10.1007/s004380051103. [DOI] [PubMed] [Google Scholar]
- 46.Martins D, English AM. Catalase activity is stimulated by H(2)O(2) in rich culture medium and is required for H(2)O(2) resistance and adaptation in yeast. Redox Biol. 2014;2:308–313. doi: 10.1016/j.redox.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J, et al. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J Biol Chem. 1999;274(23):16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 48.Toledano MB, Delaunay A, Monceau L, Tacnet F. Microbial H2O2 sensors as archetypical redox signaling modules. Trends Biochem Sci. 2004;29(7):351–357. doi: 10.1016/j.tibs.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Kiley PJ, Storz G. Exploiting thiol modifications. PLoS Biol. 2004;2(11):e400. doi: 10.1371/journal.pbio.0020400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erman JE, Vitello LB. Yeast cytochrome c peroxidase: Mechanistic studies via protein engineering. Biochim Biophys Acta. 2002;1597(2):193–220. doi: 10.1016/s0167-4838(02)00317-5. [DOI] [PubMed] [Google Scholar]
- 51.Chen S, Brockenbrough JS, Dove JE, Aris JP. Homocitrate synthase is located in the nucleus in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;272(16):10839–10846. doi: 10.1074/jbc.272.16.10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.