Significance
Many functions in the cell are performed by Brownian machines, macromolecular assemblies that use energy from the thermal environment for many of the conformational changes involved in their work cycles. Here we present a new approach capable of mapping the continuous motions of such nanomachines along their trajectories in the free-energy landscape and demonstrate this capability in the context of experimental cryogenic electron microscope snapshots of the ribosome, the nanomachine responsible for protein synthesis in all living organisms. We believe our approach constitutes a universal platform for the analysis of free-energy landscapes and conformational motions of molecular nanomachines and their dependencies on temperature, buffer conditions, and regulatory factors.
Keywords: cryo-electron microscopy, elongation cycle, manifold embedding, nanomachines, translation
Abstract
A Brownian machine, a tiny device buffeted by the random motions of molecules in the environment, is capable of exploiting these thermal motions for many of the conformational changes in its work cycle. Such machines are now thought to be ubiquitous, with the ribosome, a molecular machine responsible for protein synthesis, increasingly regarded as prototypical. Here we present a new analytical approach capable of determining the free-energy landscape and the continuous trajectories of molecular machines from a large number of snapshots obtained by cryogenic electron microscopy. We demonstrate this approach in the context of experimental cryogenic electron microscope images of a large ensemble of nontranslating ribosomes purified from yeast cells. The free-energy landscape is seen to contain a closed path of low energy, along which the ribosome exhibits conformational changes known to be associated with the elongation cycle. Our approach allows model-free quantitative analysis of the degrees of freedom and the energy landscape underlying continuous conformational changes in nanomachines, including those important for biological function.
Ideally, one would like to “see” the conformational changes of a Brownian machine as it traverses its work cycle trajectory over the energy landscape. This information is particularly relevant for a biologically important molecular machine such as the ribosome, which is responsible for protein synthesis in all living cells. During the so-called elongation process, the ribosome repeatedly links an amino acid carried in by transfer RNA (tRNA) to the nascent polypeptide chain, with the choice of amino acid in each cycle dictated by the genetic message on the mRNA. In the eukaryotic ribosome, this process is facilitated by elongation factors eEF1A and eEF2, both GTPases.
It is believed that many intermediate conformational states must be involved in the elongation cycle of the ribosome (1), but the evidence is inferred, albeit from an impressive array of experimental techniques. Both cryogenic electron microscopy (cryo-EM) (2) and X-ray crystallographic approaches (3) have been used to determine the structures of several biochemically “trapped” states along the conformational trajectory. However, it has been pointed out that these likely represent only a fraction of the relevant conformational states, that each biochemically trapped state may correspond to more than one conformational state, and that the observed intermediate structures may be affected by the trapping process itself (1). Powerful algorithms (4, 5) have been used to sort cryo-EM snapshots into a small number of discrete classes, each presumed to represent an intermediate state (6). In some cases, however, snapshots of major ribosomal regions with large conformational flexibility have defied classification into discrete states altogether, even by the most advanced analytical methods (7). Single-molecule FRET experiments have yielded evidence for discrete conformational changes in single, freely equilibrating pretranslocational ribosomes, and provided ensemble averages for such changes, but have been unable to provide data for short-lived intermediates (8, 9).
In a groundbreaking study, Fischer and coworkers (10) used cryo-EM to determine the structure and occupancy of different ribosomal conformational states as a function of time, obtaining the free-energy landscape through the Boltzmann factor (11). The specific process studied was that of back-translocation, a slow process (on the order of 30 min), in which the elongation cycle is partially reversed through interaction with another GTPase: LepA. Cryo-EM snapshots were classified in a hierarchical series of supervised (reference-based) steps to yield multiple conformations, differing mainly in specific preselected features: degree of intersubunit rotation, tRNA positions, and degree of head swivel of the small subunit—all changes known to be associated with the elongation work cycle of the ribosome.
There is general agreement, however, that the value of any approach capable of revealing conformational changes along specific trajectories is determined by the extent to which it provides unbiased quantitative insight into the conformational degrees of freedom, the ensemble kinetics, and the free-energy landscape traversed by the system under examination (1, 12, 13).
Here, we present a new analytical approach capable of mapping continuous conformational changes of nanomachines along any trajectory in the energy landscape, without timing information, supervision, or templates. Our unbiased approach determines the number of degrees of freedom exercised during the observations, the energy landscape explored by the nanomachine, the energy trajectory traversed during the work cycle, and the continuous conformational changes associated with the work cycle. These novel capabilities constitute a powerful platform for quantitative study of the conformational and energy trajectories of nanomachines, including those engaged in a wide range of important biological processes. As reported below, the first application of this approach to an experimental cryo-EM dataset of ribosomes purified from yeast not engaged in translation reveals a closed trajectory of minimum energy along which the ribosomes appear to idle with conformational changes reminiscent of those observed during the protein elongation cycle.
Conceptual Outline
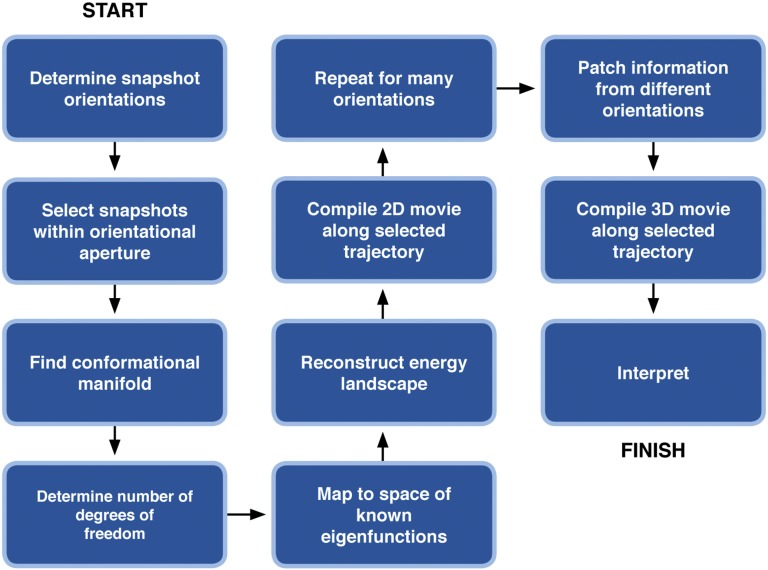
A flow diagram is shown in Fig. 1, with a more technical description following the conceptual outline. Details are presented in the SI Text. As used, for example, in early approaches to image classification in electron microscopy (14), a snapshot can be represented as a vector in high-dimensional space by regarding the intensity value at each pixel of the snapshot as a component of the vector. Distance is a measure of similarity between the snapshots in this space. A collection of snapshots produces one or more data clouds, with discrete conformations producing separate clouds. We have previously shown that this approach is able to determine 3D structure from a collection of ultra-low signal 2D snapshots of unknown orientation (15–18), and, in the presence of orientational heterogeneity and defocus variations, distinguish between discrete conformations with best-in-class performance (17).
Fig. 1.
Flowchart representation of the approach used to determine the free-energy landscape, work-cycle trajectory, and associated continuous conformational changes from experimental snapshots of nanomachines in unknown orientational and conformational states.
Here, we are concerned with continuous conformational changes, which produce correlations among the points representing the snapshots. These correlations define a hypersurface—a manifold—a concept found useful also in a previous approach to heterogeneity in cryo-EM (19). Using advanced machine-learning methods, we have demonstrated the ability to identify such manifolds in the presence of overwhelming noise and defocus variations (17).
Our approach begins with determining the orientations of the snapshots by any of a number of approaches (17), in this instance by a standard method used in electron microscopy (20). As shown previously (17, 19), this can be achieved without taking conformational heterogeneity into account, because the effects of orientational change dominate. Snapshots lying within a tight orientational aperture are then selected, and the manifold spanned by them is determined. This step in the analysis yields the conformational manifold (spectrum) in the selected viewing (projection) direction. Of course, the conformational changes may be governed by more than one parameter. In general, therefore, the conformational manifold is best described by a set of orthogonal coordinates. Such a description can be achieved by one of many well-established machine-learning techniques, which also reveal the intrinsic dimensionality of the manifold, and hence the number of degrees of freedom exercised by the system under observation.
Unfortunately, it is not possible to determine the conformational changes from such a description, because the local rates of change in multidimensional manifolds obtained from machine-learning techniques are, in general, unknown (21, 22) and cannot be easily related to the underlying changes in the system under observation (15, 23). To overcome this well-known difficulty, we introduce an additional step, in which the cloud of points is mapped to another coordinate system, where the local rates of change can be determined exactly, and related to the underlying conformational changes. This step leads to a representation of conformational change in terms of a universal parameter (metric). The density of points in this space can now be related to the energy landscape sampled by the system through the Boltzmann factor (10), with denoting the change in the Gibbs free energy, the Boltzmann constant, and the temperature. The locus of minimum energy in this landscape represents the trajectory traversed by the machine during its work cycle. In each viewing direction, 2D movies can be compiled to reveal the conformational changes along the path of minimum energy or indeed any chosen trajectory. 3D movies can be compiled by stepping along such a trajectory and, in each step, performing a 3D reconstruction by integrating the information from many different viewing directions.
Analytical Procedure
The analysis is illustrated below in more technical terms with reference to a set of 849,914 experimental cryo-EM snapshots of 80S ribosomes from yeast, obtained in the course of a study of translational initiation by a plant virus (SI Text). The procedure of purification rendered the ribosome free of mRNA and most of the tRNAs.
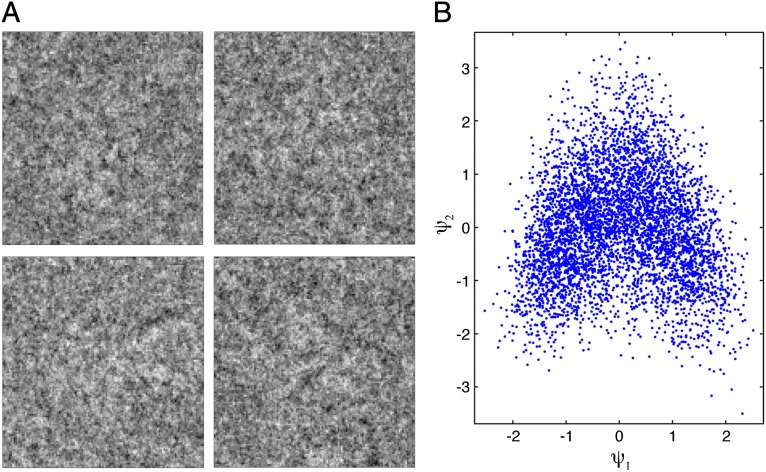
Manifolds defined by data clouds are, in general, nonlinear: the data lie on intrinsically curved rather than flat hypersurfaces. Such manifolds can be identified and described (embedded) by graph-theoretic machine-learning techniques often used for dimensionality reduction. The so-called diffusion map embedding algorithm used in our work yields a description of a curved manifold in terms of the orthogonal eigenfunctions (more precisely eigenvectors) of known operators, specifically the Laplace–Beltrami operator (21, 22, 24) (SI Text). We use a specially developed kernel to deal with the substantial defocus variations encountered in cryo-EM data (17) (SI Text). In Fig. 2, we show representative cryo-EM snapshots (Fig. 2A) and a typical data manifold (Fig. 2B) described in terms of the first two eigenvectors of the Laplace–Beltrami operator. [In general, there is no simple correspondence between the eigenvectors obtained from this nonlinear analysis, and those obtained with linear approaches, such as principal component analysis (PCA) and singular value decomposition (SVD).] The manifold produced by the experimental snapshots is, in fact, nonlinear, with an intrinsic dimensionality of five (SI Text).
Fig. 2.
(A) Representative cryo-EM snapshots, and (B) 2D view of a typical conformational manifold. The manifold is derived from a measure of similarity among ∼1,500 cryo-EM snapshots of ribosome particles viewed within a tight orientational aperture. The axes represent the first two eigenvectors obtained by the diffusion map algorithm with a kernel able to deal with defocus variations.
Fig. 2 describes the data in terms of the eigenfunctions of the Laplace–Beltrami operator with respect to an unknown metric (22). The absence of information on the metric precludes a consistent description of the conformational changes in terms of a known universal parameter. We solve this problem by mapping the manifold to another space, in which the eigenfunctions are known exactly. For this, we use an approach used in nonlinear Laplacian spectral analysis (NLSA) (25), a technique capable of performing SVD on nonlinear manifolds (nonlinear SVD for short) (SI Text). Briefly, one considers a collection of supervectors formed by concatenating snapshots falling within a window moving over the data vectors. The snapshots within each supervector are ordered according to the projections of the points representing them on a line through the origin, making an angle with, say, the horizontal axis (Fig. 2 and SI Text). By virtue of this projection, the arrangement picks out the conformational evolution along the selected line characterized by , with the random ordering of conformational changes along other directions assuming the character of noise. Nonlinear SVD (25) is then used to extract characteristic images (topos) and their evolutions (chronos) from these supervectors (SI Text). Each topo/chrono pair constitutes an element of a biorthogonal decomposition of the conformational changes along the selected line. Noise-reduced snapshots can be reconstructed from the topo/chrono pairs with significant (above-noise) singular values and embedded to obtain the manifold characteristic of the conformational changes along the selected line (SI Text). By construction, this manifold is one-dimensional, described in terms of known eigenfunctions, viz. , and governed by the single parameter (SI Text).
Given a sufficiently dense collection of radial lines, each making an angle with the horizontal axis, the conformational changes can be described in any direction in the multidimensional space of conformations by the parameter . The mapping from a space of unknown metric and hence eigenfunctions to one characterized by known eigenfunctions governed by a single parameter allows a consistent description of the conformational changes (SI Text and Fig. S1). This description of conformational changes in a given projection direction can be related to descriptions in other projection directions by assuming that the same conformational spectrum is viewed in all projection directions. In other words, the histograms of occupancy vs. conformational parameter for different projection directions all represent the same spectrum of conformations and can thus be equalized. This equalization allows a universal description of the conformational spectrum across all projection directions. For any selected projected direction, the energy landscape along any line can be determined from the density of points along that line via the Boltzmann factor. Energy landscapes of higher dimension (with the dimensionality given by the number of eigenvectors) can be reconstructed by a tomographic extension of this approach (Fig. 3 and SI Text). Here, we concentrate on the first two eigenvectors, which contain some of the most interesting biological information (see below).
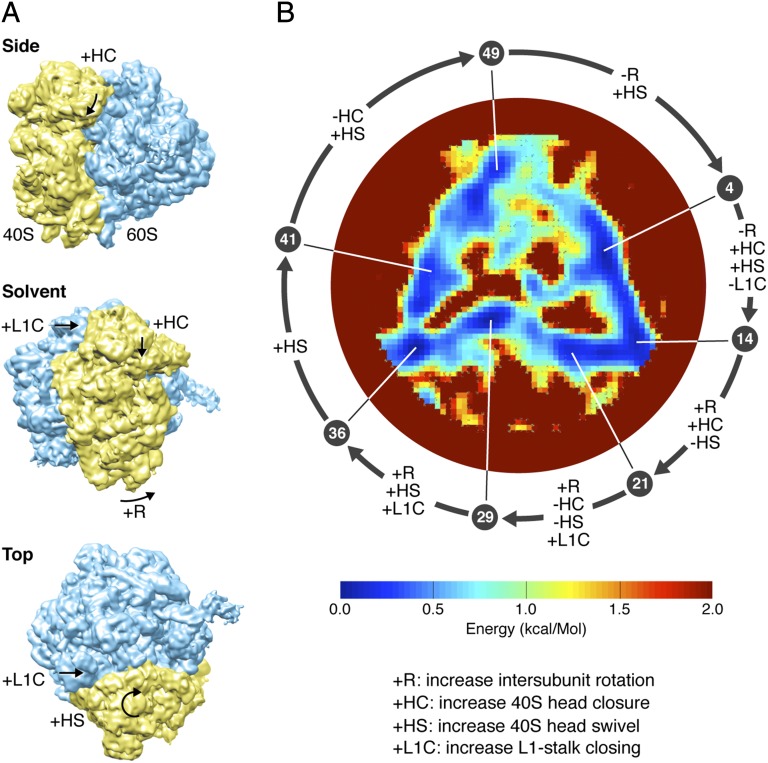
Fig. 3.
(A) Three views of a cryo-EM map of the 80S ribosome from yeast (32), with arrows indicating four key conformational changes associated with the elongation work cycle of the ribosome. (B) The energy landscape traversed by the ribosome. The color bar shows the energy scale. The energy range has been truncated at 2 kcal/mol to show details of the triangular trough. The error in energy determination along the closed triangle is 0.05 kcal/mol. The roughly triangular minimum free-energy trajectory is divided into 50 states. The arrows indicate the structural changes between 7 selected states, each identified by its place in the sequence of 50 states.
With this information in hand, the structural evolution of the system, including 2D movies of the conformational changes and the system’s thermodynamic properties, can be quantitatively investigated in any viewing direction (SI Text and Movie S1). For any point on the energy landscape, a 3D structure map can be compiled by integrating the 2D information from many viewing directions into a 3D representation (Fig. 4) (SI Text and Movies S2–S4). Thus, 3D movies can be compiled by stepping along any selected trajectory in the energy landscape and reconstructing a 3D structure map in each step. The conformational changes along the closed minimum-energy trajectory of Fig. 3B are summarized in Fig. 3 and exemplified in Fig. 4. The associated movies (Movies S2–S4) present the ribosome as it evolves, with each movie corresponding to a different viewing direction selected for optimal visibility of the biologically relevant domain motions (Fig. 3A). We note that, by construction, these movies are based on similarity rather than time. Accordingly, identical trajectories in opposite directions cannot be distinguished.
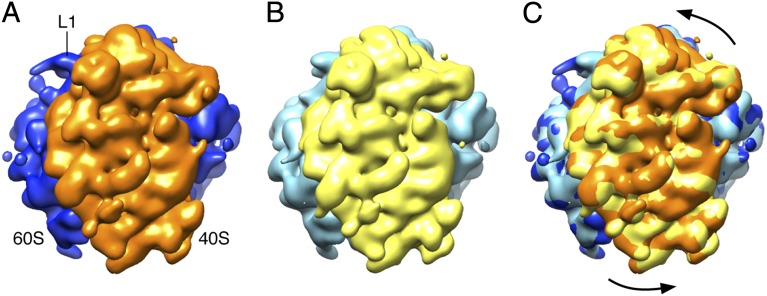
Fig. 4.
Example of conformational changes along the trajectory: ratchet-like motion. (A) Unrotated ribosome, map 14 in Fig. 3B. (B) Maximally rotated ribosome, map 36. (C) Superposition of the two maps. The full set of frames showing continuous conformational changes is shown in three common viewing directions in Movies S2–S4.
Results
The trough of minimum free energy is seen to form a closed, roughly triangular path (Fig. 3B), with variations of <0.9 kcal/mol (1.5kBT at room temperature) in the energy positions of the deepest points. The energy difference between the lowest and highest points of the entire landscape, corresponding to states with the highest and lowest nonzero occupancy in the experiment, amount to 3.8 ± 0.65 kcal/mol (6.5 ± 1.0kBT at room temperature), with the uncertainty stemming chiefly from the low occupancy of high-energy states. The energy range covered nonetheless indicates that so-called transition states with energies several times the thermal energy have been probed in our experiment.
We examine the 3D movies showing the conformational changes along the closed triangular trajectory, analyzing in detail seven structure maps along the way (SI Text and Movie S5). Using rigid-body fitting of domains while making use of a published cryo-EM map of the yeast ribosome (26) for reference, we observe combinations of four motions previously described in the literature and associated with the elongation cycle, as follows: (i) ratchet-like intersubunit rotation, a counterclockwise rotation of the small subunit about an axis normal to the subunit interface (Fig. 3A, solvent view) (27, 28); (ii) rotation (closing movement) of the L1 stalk toward the intersubunit space (Fig. 3A, solvent and top views) (29); (iii) small subunit head swivel, a rotation of the subunit head about its long axis (Fig. 3A, top view) (30); and (iv) small subunit head closure, a “nodding” motion of the head (Fig. 3A, side and solvent views) (31). The first three motions are correlated and known to be associated with the process of mRNA–tRNA translocation (30, 32), whereas the fourth is known to accompany the selection of cognate aminoacyl tRNA during the process of decoding (31).
Proceeding clockwise along the minimum free-energy pathway, these motions occur in the sequence encountered during the elongation cycle: starting at the apex of the triangle and moving to the right side of the base (Fig. 3B), we see the small subunit head closure as a prominent motion, corresponding to the tRNA selection step. Next, proceeding along the base of the triangle, three coordinated movements of small subunit body, head swivel, and L1 stalk occur—all expected during translocation—while the small subunit head remains in the closed position. Finally, moving up from the left side of the base to the apex, we observe the reversal of head closing (the conformational change associated with the disengagement of the mRNA from the decoding center), as well as the reversal of the intersubunit motion.
Discussion
The energy landscape constructed by the analysis of the ribosome ensemble shows multiple interconnected paths of relatively low energy. Of these, a path of roughly triangular shape clearly stands out, encompassing roughly one third of the molecules. Comparison of 3D reconstructions from data at selected points along this path reveals conformational changes previously observed in the elongation cycle of fully programmed, translating ribosomes. Because the ribosomes captured in our snapshots were not engaged in protein synthesis, lacking mRNA and aminoacylated tRNAs, we must assume that their idle motions in the thermal environment sample the conformational space permitted by the degrees of freedom, thus exhibiting the conformational changes that would be productive in the presence of the ligands of the translational machinery (mRNA, aa-tRNA, eEF2, and eEF1A). Indeed, idling of the pretranslocational ribosome in the absence of elongation factors along the direction of the most prominent conformational changes (intersubunit motion and opening/closing of L1 stalk) has been previously observed by single-molecule FRET (8, 9) and cryo-EM (33). Idling of the empty ribosome along the same path has also been inferred from a series of X-ray structures (34). It remains to be established by the approach outlined in this paper whether fully programmed ribosomes follow the same path identified here. As to the other paths traversing the free-energy landscape (Fig. 3B), it is tempting to speculate that they may represent alternative routes of the molecular machine under different buffer and temperature conditions.
We now discuss the salient features of our approach. First, the usefulness of movies of Brownian machines has been rightly questioned, because each trajectory of a single machine is strongly influenced by stochastic factors and thus is unique (1). The movies presented here, however, integrate information from a large ensemble of snapshots, each stemming from an object viewed only once. By using manifolds to capture the properties of the entire dataset, and nonlinear singular value analysis to suppress noise, our approach offers an efficient means for extracting the ensemble kinetics, i.e., the information common to a collection of objects, each viewed in an initially unknown orientational and conformational state.
Most successful experimental studies of the conformational spectra of biological machines have been hitherto restricted to sorting snapshots into a small number of classes (10, 11), using templates in some form, relying on timing information, or a combination of these tools (10). In contrast, our approach naturally yields detailed conformational trajectories and energy landscapes without a priori information or assumptions and at moderate computational expense (SI Text).
Of course, the maximum number of detectable conformational states is limited by various factors. These factors include the frequency (via the Shannon–Nyquist theorem) with which the conformational spectrum has been sampled, the information content of the individual snapshots, and, ultimately, the atomic nature of the object itself. In our analysis, the conformational signal stems primarily from the most heavily sampled projection directions, in each of which the conformational spectrum is randomly sampled by up to 5,000 snapshots. The number of meaningfully distinct conformational states is governed by the signal-to-noise ratio, which determines the statistical confidence with which neighboring states can be distinguished. In this study, the requirement for neighboring conformational states to be separated by 3 SDs () means that ∼50 conformational states can be distinguished (SI Text). This number is about an order of magnitude larger than previously achieved without timing information or templates. In combination with the recent availability of large cryo-EM datasets with near-atomic resolution (7, 35), our approach promises the possibility to extract conformational information with unprecedented detail.
In recent years, increasing computing power has fueled efforts to simulate the conformational trajectories of molecular machines, particularly the ribosome (36), by molecular dynamics. Experimental results obtained with the new approach presented here offer the promise to guide these efforts and provide the means for verifying important modeling assumptions.
Conclusions
Given our increasingly detailed knowledge of the structure of biological machines in general, and the ribosome in particular, it has been suggested that the “heroic age” of biostructure determination is drawing to a close and that further progress requires the study of ensemble kinetics and conformational energy landscapes (1). As shown by the results presented here, our approach offers a powerful platform for the quantitative study of continuous conformational motions over the energy landscapes traversed by biological machines in the course of their operation. This development may significantly accelerate the dawn of the predicted new era in biology. More generally, the way to the quantitative study of the structure, kinetics, and operational cycles of nanomachines is now open.
Supplementary Material
Acknowledgments
We thank D. Giannakis, K. Sanbonmatsu, and M. Schmidt for helpful comments on an early version of the manuscript and M. Thomas-Baum for assistance in the preparation of the illustrations. This research was supported by US Department of Energy, Office of Science, Basic Energy Sciences Award DE-FG02-09ER16114 (to A.O.); the Howard Hughes Medical Institute and National Institutes of Health Grants R01 GM29169 and GM55440 (to J.F.); and National Science Foundation MCB Grant 1157906 (to A.E.S.). The publication of this work was supported by US National Science Foundation Award STC 1231306.
Footnotes
The authors declare no conflict of interest.
Data deposition: The 50 3D maps generated along the free-energy trajectory (clockwise in Fig. 3B) have been deposited in the Electron Microscopy Data Bank (accession no. EMD-6044).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419276111/-/DCSupplemental.
References
- 1.Moore PB. 2012. How should we think about the ribosome? Annu Rev Biophys 41(1):1–19.
- 2.Villa E, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci USA. 2009;106(4):1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12(6):674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheres SHW. Maximum-likelihood methods in cryo-EM. Part II: Application to experimental data. Methods Enzymol. 2010;482:295–320. doi: 10.1016/S0076-6879(10)82012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheres SH. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012;415(2):406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank J. Intermediate states during mRNA-tRNA translocation. Curr Opin Struct Biol. 2012;22(6):778–785. doi: 10.1016/j.sbi.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343(6178):1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30(3):348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30(5):578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466(7304):329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 11.Agirrezabala X, et al. Structural characterization of mRNA-tRNA translocation intermediates. Proc Natl Acad Sci USA. 2012;109(16):6094–6099. doi: 10.1073/pnas.1201288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank J, Gonzalez RL., Jr Structure and dynamics of a processive Brownian motor: The translating ribosome. Annu Rev Biochem. 2010;79(9):381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munro JB, Sanbonmatsu KY, Spahn CMT, Blanchard SC. Navigating the ribosome’s metastable energy landscape. Trends Biochem Sci. 2009;34(8):390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Heel M, Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6(2):187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- 15.Giannakis D, Schwander P, Ourmazd A. The symmetries of image formation by scattering. I. Theoretical framework. Opt Express. 2012;20(12):12799–12826. doi: 10.1364/OE.20.012799. [DOI] [PubMed] [Google Scholar]
- 16.Schwander P, Giannakis D, Yoon CH, Ourmazd A. The symmetries of image formation by scattering. II. Applications. Opt Express. 2012;20(12):12827–12849. doi: 10.1364/OE.20.012827. [DOI] [PubMed] [Google Scholar]
- 17.Schwander P, Fung R, Ourmazd A. Conformations of macromolecules and their complexes from heterogeneous datasets. Philos Trans R Soc Lond B Biol Sci. 2014;369(1647):20130567. doi: 10.1098/rstb.2013.0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosseinizadeh A, et al. High-resolution structure of viruses from random diffraction snapshots. Philos Trans R Soc Lond B Biol Sci. 2014;369(1647):20130326. doi: 10.1098/rstb.2013.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu J, Gao H, Frank J. Unsupervised classification of single particles by cluster tracking in multi-dimensional space. J Struct Biol. 2007;157(1):226–239. doi: 10.1016/j.jsb.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: New techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53(3):251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 21.Belkin M, Niyogi P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput. 2003;15(6):1373–1396. [Google Scholar]
- 22.Coifman RR, et al. Geometric diffusions as a tool for harmonic analysis and structure definition of data: Diffusion maps. Proc Natl Acad Sci USA. 2005;102(21):7426–7431. doi: 10.1073/pnas.0500334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coifman RR, Shkolnisky Y, Sigworth FJ, Singer A. Reference free structure determination through eigenvectors of center of mass operators. Appl Comput Harmon Anal. 2010;28(3):296–312. doi: 10.1016/j.acha.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coifman RR, Kevrekidis IG, Lafon S, Maggioni M, Nadler B. Diffusion maps, reduction coordinates, and low dimensional representation of stochastic systems. Multiscale Model Simulation. 2008;7(2):842–864. [Google Scholar]
- 25.Giannakis D, Majda AJ. Nonlinear Laplacian spectral analysis for time series with intermittency and low-frequency variability. Proc Natl Acad Sci USA. 2012;109(7):2222–2227. doi: 10.1073/pnas.1118984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor DJ, et al. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17(12):1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406(6793):318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 28.Budkevich T, et al. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell. 2011;44(2):214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114(1):123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 30.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468(7324):713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci. 2003;28(5):259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 32.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci USA. 2007;104(50):19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32(2):190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang W, Dunkle JA, Cate JHD. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325(5943):1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504(7478):107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitford PC, Blanchard SC, Cate JHD, Sanbonmatsu KY. Connecting the kinetics and energy landscape of tRNA translocation on the ribosome. PLOS Comput Biol. 2013;9(3):e1003003. doi: 10.1371/journal.pcbi.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.