Significance
Infectious diseases are responsible for one-third of all mortality worldwide. Innate immunity is critical for mounting host defenses that eliminate pathogens. Salmonella is a global food-borne pathogen that infects and replicates within macrophages. How inflammasomes—multimeric protein complexes that provide innate immune protection—function to restrict bacterial burden in macrophages remains unknown. We show that actin polymerization is critical for NLRC4 inflammasome activation in response to Salmonella infection. NLRC4 activation in Salmonella-infected cells prevents further uptake of bacteria by inducing cellular stiffness and antimicrobial responses, which prevent bacterial dissemination in the host. These results demonstrate a critical link between innate immunity and the actin cytoskeleton in the cellular defense against Salmonella infection.
Keywords: innate immunity, ASC, caspase-1, cytoskeleton, ROS
Abstract
Salmonellosis is one of the leading causes of food poisoning worldwide. Controlling bacterial burden is essential to surviving infection. Nucleotide-binding oligomerization domain-like receptors (NLRs), such as NLRC4, induce inflammasome effector functions and play a crucial role in controlling Salmonella infection. Inflammasome-dependent production of IL-1β recruits additional immune cells to the site of infection, whereas inflammasome-mediated pyroptosis of macrophages releases bacteria for uptake by neutrophils. Neither of these functions is known to directly kill intracellular salmonellae within macrophages. The mechanism, therefore, governing how inflammasomes mediate intracellular bacterial-killing and clearance in host macrophages remains unknown. Here, we show that actin polymerization is required for NLRC4-dependent regulation of intracellular bacterial burden, inflammasome assembly, pyroptosis, and IL-1β production. NLRC4-induced changes in actin polymerization are physically manifested as increased cellular stiffness, and leads to reduced bacterial uptake, production of antimicrobial molecules, and arrested cellular migration. These processes act in concert to limit bacterial replication in the cell and dissemination in tissues. We show, therefore, a functional link between innate immunity and actin turnover in macrophages that underpins a key host defense mechanism for the control of salmonellosis.
A critical step in disease pathogenesis for many clinically important bacteria is their ability to infect and survive within host cells such as macrophages. Salmonella enterica, a pathogen that resides and replicates within macrophages, causes a range of life-threatening diseases in humans and animals, and accounts for 28 million cases of enteric fever worldwide each year (1). S. enterica infects phagocytes by a process that requires cytoskeletal reorganization (2). This bacterium resides in a Salmonella-containing vacuole (SCV) within host macrophages, and this intracellular lifestyle enables them to avoid extracellular antimicrobial killing, evade adaptive immune responses, and potentially to spread to new sites to seed new infectious foci within host tissue, which eventually develop into granulomas (3). Survival and growth of S. enterica within phagocytes is critical for virulence (4) and host restriction of the intracellular bacterial load is, therefore, paramount in surviving salmonellosis. Salmonella delivers microbial effector proteins into the host cell via the type III secretion systems (T3SS), mediated by the Salmonella pathogenicity island-1 and -2 (SPI-1 and SPI-2), to subvert cellular functions and facilitate intracellular survival (5).
Microbes are recognized by macrophages through pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain-like receptors (NLRs), which initiate innate immune responses, including cytokine production and pathogen killing (6). NLRs drive the formation of inflammasomes—macromolecular protein complexes—comprising one or more NLRs, usually an adaptor protein (ASC) and the effector protein caspase-1, which then cleaves prointerleukin-1β (IL-1β) and pro–IL-18 into biologically active cytokines, and initiates macrophage cell death by pyroptosis (7). NLRC4, in concert with NAIPs 1, 2, 5, and 6, is a key PRR that forms an inflammasome complex upon sensing flagellin and/or the inner rod or needle proteins (PrgJ and PrgI, respectively) of the SPI-1 T3SS of S. enterica serovar Typhimurium (S. Typhimurium) (8–11). Activation of the NLRC4 inflammasome by Salmonella infection results in IL-1β and IL-18 production driven by an ASC-dependent pathway and macrophage pyroptosis driven by an ASC-independent pathway (12, 13). A second, noncanonical, NLR signaling pathway has been described, which requires caspase-11 to initiate delayed cell death and NLRP3 inflammasome activation (14–16). Effective clearance of Salmonella infection in host cells may therefore require a coordinated effort between different inflammasome signaling pathways.
We, and others, have shown that NLRC4 is important in regulating bacterial burden of S. Typhimurium in vivo (17–19). A recent study revealed that Salmonella-infected epithelial cells are extruded from the intestinal epithelium in a process that requires NLRC4 (20). The molecular mechanism behind how NLRC4 restricts bacterial burden in macrophages infected with Salmonella is still unknown. Here, we identify an actin-dependent mechanism that controls NLRC4-mediated regulation of bacterial replication in macrophages infected with S. Typhimurium. Activation of NLRC4 in infected macrophages mediates the production of reactive oxygen species (ROS) to inhibit bacterial replication and limits additional bacterial uptake by inducing mechanical stiffening the cell via actin polymerization. Overall, we describe a previously unidentified effector mechanism, governed by actin and the NLRC4 inflammasome, to control Salmonella infection in macrophages.
Results
The NLRC4 Inflammasome Controls Intracellular Bacterial Numbers in Macrophages.
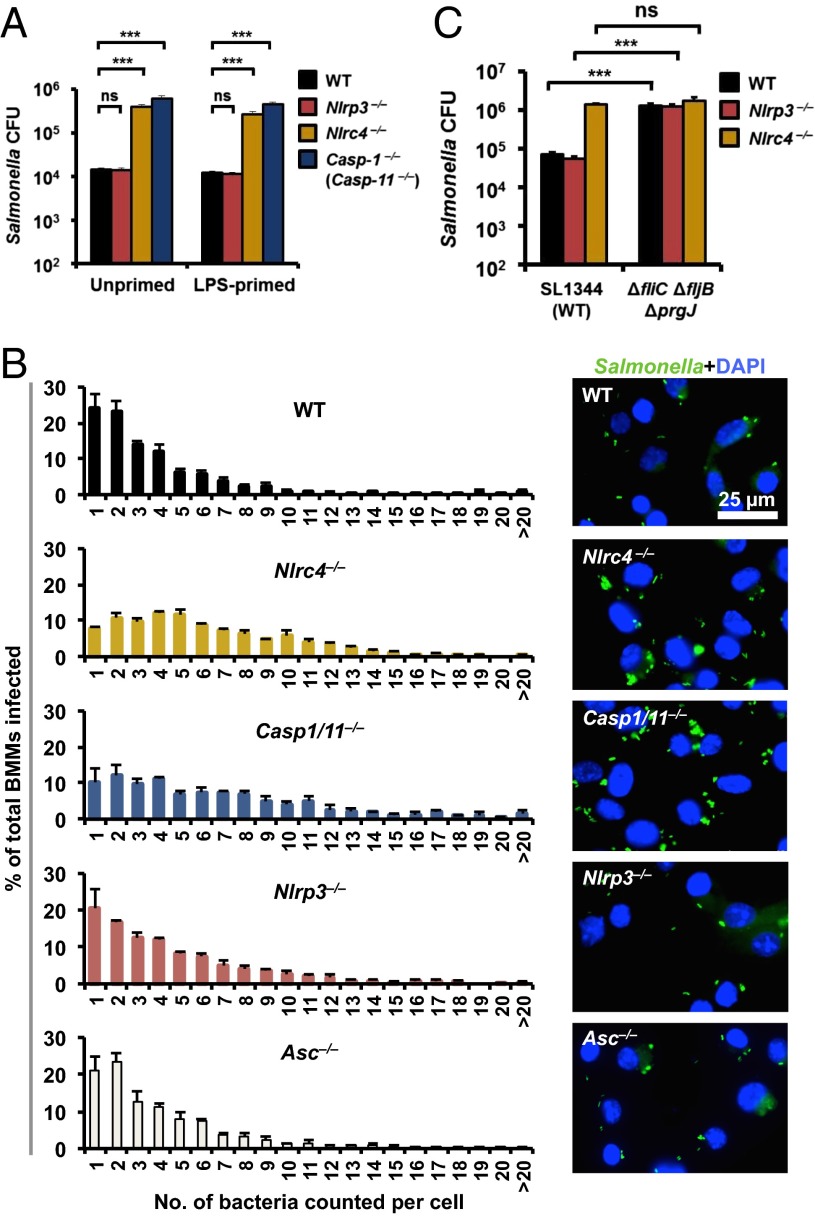
NLRC4 and NLRP3 inflammasomes are critical in controlling Salmonella infection in vivo (17–19); however, whether inflammasomes regulate intracellular bacterial load in macrophages is unknown. To investigate whether NLRC4 or NLRP3 regulate the burden of salmonellae in macrophages, we infected primary bone marrow-derived macrophages (BMMs) from WT, Nlrc4−/−, Nlrp3−/−, Asc−/−, Casp1−/−, or Casp11−/− mice with a SPI-1–competent S. Typhimurium [multiplicity of infection (MOI) 1] and quantified the number of bacteria using gentamicin protection assays. After 2 h of infection, we found a significantly higher Salmonella burden in Nlrc4−/− or Casp1−/− BMMs compared with WT BMMs (Fig. 1A). The Casp1−/− mouse strain was recently shown to be deficient in caspase-11 (herein referred to as Casp1/11−/−) (14). Casp11−/− BMMs infected with S. Typhimurium, however, contained similar number of bacteria compared with WT BMMs (Fig. S1), indicating that caspase-1, rather than caspase-11, controls bacterial numbers in macrophages at this time point. Confocal microscopy was performed on individual macrophages infected with a GFP-expressing strain of S. Typhimurium and the bacterial load per cell was enumerated. A higher number of bacteria per cell was found in Nlrc4−/− and Casp1/11−/− BMMs (most had 4–6 bacteria per cell) compared with WT cells (most had 1–2 bacteria per cell; Fig. 1B). These results indicate that the NLRC4–caspase-1 axis restricts bacterial numbers in macrophages.
Fig. 1.
The NLRC4–caspase-1 axis restricts high levels of intracellular S. Typhimurium numbers in macrophages. (A) Unprimed primary BMMs were infected with S. Typhimurium (MOI 1) for 1 h. Following 1-h infection, BMMs were treated with gentamicin (50 μg/mL) for 1 h to kill extracellular bacteria. Host cell lysates were plated onto LB agar and the number of viable intracellular bacteria was enumerated. (B) Unprimed BMMs were infected with S. Typhimurium expressing GFP (MOI 10) for 1 h, followed by gentamicin treatment (50 μg/mL) for 1 h to kill extracellular bacteria. The percentages of BMMs harboring different number of bacteria were determined by microscopy (WT, n = 1,134; Nlrc4−/−, n = 765; Casp1/11−/−, n = 708; Nlrp3−/−, n = 663; Asc−/−, n = 731). (C) Unprimed BMMs were infected with ΔfliCΔfljBΔprgJ S. Typhimurium (MOI 1) for 1 h and the number of viable intracellular bacteria was enumerated. Data are the mean of three independent experiments and error bars represent SEM. (A) One-way ANOVA with a Dunnett’s multiple comparisons test. (C) Two-way ANOVA with a Tukey’s multiple comparisons test. ***P < 0.001; ns, no statistical significance.
We further confirmed these results by infecting WT BMMs with an isogenic mutant of S. Typhimurium lacking NLRC4 activators (ΔfliCΔfljB (deletion of flagellin proteins) and ΔprgJ). We found that WT BMMs failed to restrict bacterial numbers in the absence of NLRC4 activation (Fig. 1C). Gentamicin protection assays and single-cell analysis of Salmonella-infected Asc−/− or Nlrp3−/− BMMs revealed similar total numbers and distribution of bacteria per cell compared with WT BMMs (Fig. 1 A and B). Together, these results suggest that the NLRC4 inflammasome regulates Salmonella burden in macrophages via a mechanism that is independent of ASC.
NLRC4 Restricts Bacterial Replication in the Salmonella-Containing Vacuole.
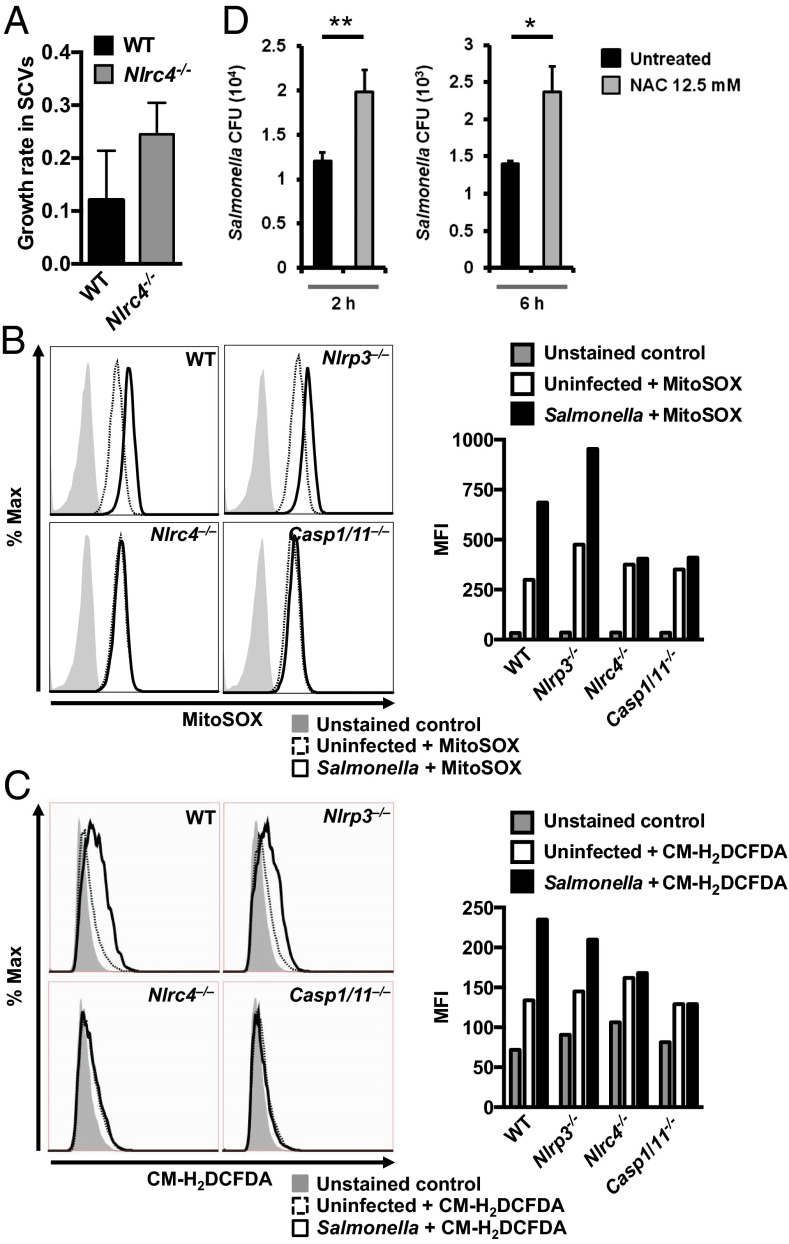
An elevated intracellular bacterial burden could occur by one or more of the following mechanisms: increased infection rate of macrophages, increased intracellular bacterial growth rate, or suppression of intracellular bacterial killing (21). When S. Typhimurium enters a macrophage, the cell must be able to limit bacterial replication to prevent overwhelming infection. The increased number of bacteria per cell in Nlrc4−/− BMMs suggests a role for NLRC4 in the inhibition of intracellular bacterial survival (Fig. 1). S. Typhimurium survive and replicate intracellularly in macrophages within SCVs of a diameter of 3–4 µm (Fig. S2). We used live confocal imaging to record the number of bacteria in each SCV over time and calculated the bacterial growth rate within each SCV from WT (n = 11) and Nlrc4−/− (n = 30) BMMs over 17 h. In WT BMMs that were not killed by pyroptosis, the bacterial growth rate in the SCV was 0.12 divisions per hour. Bacteria in the SCVs of Nlrc4−/− BMMs, in contrast, grew twice as fast—at a rate of 0.24 divisions per hour (Fig. 2A). These results indicate that Nlrc4−/− BMMs fail to effectively restrict bacterial replication in the SCV. Effective control of intracellular Salmonella by macrophages has been shown to be mediated by mitochondrial ROS (mROS) activity and hydrogen peroxide produced within the cell (22). We measured mROS activity and cellular hydrogen peroxide production in BMMs infected with S. Typhimurium for 30 min and found that WT BMMs generated mROS activity and hydrogen peroxide after infection with S. Typhimurium, whereas Nlrc4−/− or Casp1/11−/− BMMs failed to induce mROS or hydrogen peroxide (Fig. 2 B and C). Nlrp3−/− BMMs produced mROS and hydrogen peroxide at a level comparable to WT BMMs (Fig. 2 B and C), indicating that NLRC4, but not NLRP3, induces ROS in macrophages in response to Salmonella infection. Furthermore, inhibition of ROS in BMMs with N-acetyl-l-cysteine (NAC) resulted in a significantly increased number of intracellular bacteria compared with untreated controls following 2 and 6 h of infection (P = 0.007 for 2 h and P = 0.013 for 6 h; Fig. 2D). These results collectively demonstrate that NLRC4 contributes to the restriction of bacterial replication in SCVs.
Fig. 2.
NLRC4 regulates mitochondrial ROS and H2O2 production to restrict bacterial replication by Salmonella in the SCV. (A) The bacterial growth rate in each SCV was determined in WT and Nlrc4−/− BMMs using live confocal imaging of SCVs (WT, n = 11; Nlrc4−/−, n = 30) over 17 h following inoculation. The growth rate of S. Typhimurium was calculated using the formula ∆n/t, where ∆n is the number of bacteria in a SCV immediately before the death of a macrophage minus the number of bacteria following the formation of a SCV, and t is the length of time that the macrophage had survived over the course of the infection. (B and C) Unprimed BMMs were infected with S. Typhimurium (MOI 10) for 30 min and stained with MitoSOX stain or CM-H2DCFDA for 30 min. (B) Levels of mROS and (C) H2O2 were measured using flow cytometry. (D) WT BMMs were treated with NAC for 1.5 h and then infected with S. Typhimurium (MOI 1) for 1 h, followed by gentamicin treatment for a total of 2 or 6 h. Lysates from BMMs were plated on LB agar and the number of viable intracellular bacteria was enumerated. Data are representative of three (C and D) or four (B) independent experiments. Error bars indicate SEM. Two-tailed t test, *P < 0.05; **P < 0.01.
NLRC4 Modifies Actin Polymerization to Activate Inflammasome Responses to Salmonella Infection.
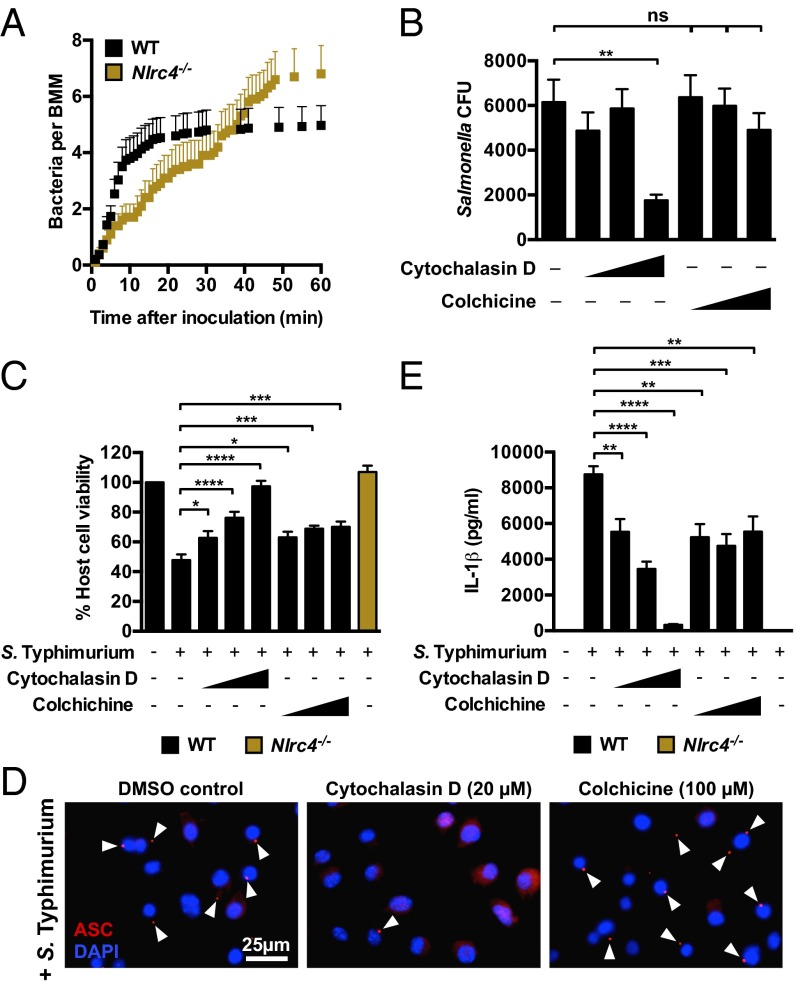
Our live-cell imaging experiments using confocal microscopy revealed important differences in the growth rate of bacteria in WT and Nlrc4−/− macrophages. To further investigate the dynamics of the infection process, we used live-cell imaging to track and quantify infection events of individual WT and Nlrc4−/− macrophages infected with S. Typhimurium-GFP (MOI of 10) every 60 s for 1 h. WT BMMs were susceptible to infection for the first 10 min, but susceptibility to infection plateaued after 10 min, whereas Nlrc4−/− BMMs were initially less susceptible to infection compared with WT BMMs (0–30 min), but remained readily susceptible to infection over time (Fig. 3A). The failure to limit bacterial uptake in the absence of NLRC4 ultimately resulted in a greater number of total infection events per macrophage after 1 h of infection (6.8 ± 1.0 bacteria per Nlrc4−/− BMMs vs. 5.0 ± 0.7 bacteria per WT BMMs).
Fig. 3.
NLRC4 inflammasome functions are linked to the actin cytoskeleton. (A) Unprimed BMMs were infected with S. Typhimurium expressing GFP (MOI 10) and tracked by confocal live imaging for 1 h. The number of bacteria internalized into each macrophage was counted every 60 s. (B) The number of viable intracellular bacteria recovered from WT BMMs infected with S. Typhimurium in the presence of the vehicle control DMSO, cytochalasin D (0.2, 2, and 20 µM) or colchicine (1, 10, and 100 µM) was counted. (C) Host cell viability and (D) levels of IL-1β secreted from LPS-primed WT BMMs infected with S. Typhimurium for 1 h in the presence of the vehicle control DMSO, cytochalasin D, or colchicine. (E) Unprimed WT BMMs were infected with S. Typhimurium for 1 h and stained for ASC (red) and DNA (blue). Data are the mean of three independent experiments and error bars represent SEM. One-way ANOVA with a Dunnett’s multiple comparisons test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not statistically significant.
Cytoskeletal rearrangement, particularly changes in actin conformation, is an important process for entry of Salmonella into epithelial cells (23); however, it is unknown whether NLRC4 regulates cytoskeletal function to control infection in macrophages. We hypothesized that NLRC4 might alter cytoskeletal function to reduce bacterial uptake in macrophages. Inhibition of host cell actin polymerization and reduced actin availability, using cytochalasin D (24, 25), significantly impaired Salmonella uptake into macrophages, whereas colchicine (a microtubule polymerization inhibitor) failed to significantly inhibit bacterial uptake after 2 h of infection (Fig. 3B). Cytochalasin D also dose-dependently inhibited Salmonella-induced NLRC4-dependent pyroptosis (Fig. 3C).
NLRC4 is unusual among the inflammasome-forming NLRs in that it can activate caspase-1 directly by CARD–CARD domain interactions as well as through association with the adaptor protein ASC (26, 27). Our data suggest that changes in actin polymerization are an important effector mechanism for NLRC4-dependent ASC-independent antimicrobial effects induced by the cell, and we wondered whether this would also be true for ASC-dependent NLRC4 inflammasome activation by S. Typhimurium. Inflammasome activation drives the formation of a large macromolecular complex, or “speck,” within the cell whereby ASC forms the platform for recruitment of effector proteins in response to Salmonella infection (17, 28). We infected BMMs with S. Typhimurium and immunolocalized ASC at different time-points and observed the formation of an ASC speck within 5 min postinfection (Fig. S3). The formation of macromolecular protein complexes can be facilitated by cytoskeletal reorganization. We found that cytochalasin D, but not colchicine, inhibited the formation of these rapidly induced ASC inflammasome specks in Salmonella-infected BMMs (Fig. 3D). ASC speck formation is critical for the production of IL-1β, and in our experiments cytochalasin D indeed dose-dependently blocked early NLRC4-dependent Salmonella-induced IL-1β production, whereas colchicine did not dose-dependently inhibit Salmonella-induced IL-1β production (Fig. 3E). We further confirmed these results and found that cytochalasin D, but not colchicine, inhibited flagellin-induced ASC speck formation and IL-1β secretion in BMMs (Fig. S4). These results suggest that actin polymerization provides a mechanism by which NLRC4 activates innate immunity in response to S. Typhimurium.
Activation of NLRC4 Modulates Macrophage Deformability and Movement.
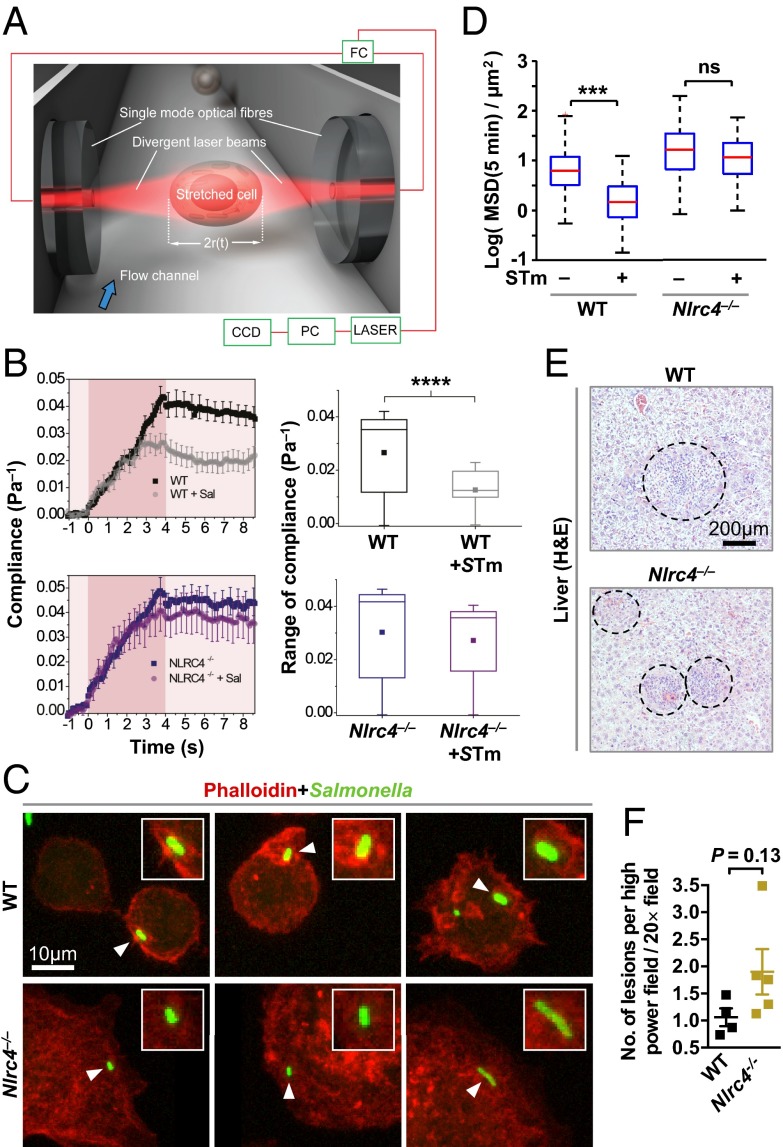
We hypothesized that NLRC4-dependent changes in actin polymerization should impact cytoskeletal functions within the cell; to investigate this, we used an optical stretcher, an established biophysical instrument (29), to determine whether there is a dynamic change in the cytoskeletal function of WT and Nlrc4−/− BMMs in response to Salmonella infection. The optical stretcher employs two counterpropagating laser beams to trap and deform individual macrophages and measure changes in their deformability or compliance (the inverse of stiffness), which is dependent on the functionality of the cytoskeleton (Fig. 4A) (30). When coupled to a fluorescence detector, it is possible to trap and specifically stretch cells that had been infected with GFP-expressing S. Typhimurium. We analyzed uninfected WT or Nlrc4−/− BMMs or BMMs infected with S. Typhimurium-GFP at an MOI of 10 for 10 min using the optical stretcher. The BMMs were trapped and stretched for 4 s, followed by quantification of their compliance. We found that uninfected WT or Nlrc4−/− BMMs exhibited identical viscoelastic properties. Infected WT BMMs, however, exhibited a change in their viscoelastic properties, such that the macrophages became less deformable or stiffer compared with uninfected WT cells (Fig. 4B). Remarkably, Nlrc4−/− BMMs infected with S. Typhimurium showed little change in cell compliance, indicating that infection-induced cell stiffening did not occur (Fig. 4B).
Fig. 4.
NLRC4 activation induces physical stiffening of macrophages by limiting actin availability for bacterial uptake and impairs bacterial dissemination. (A) The principle and setup of the optical stretcher. Two diverging and counterpropagating laser beams emanating from single-mode optical fibers are used to trap and deform single suspended cells. A 50:50 intensity-ratio fiber coupler (FC) is used to split the output of a fiber laser into the two optical fibers. A personal computer (PC) is used for laser control and data acquisition by video microscopy via a CCD camera. (B) Unprimed primary BMMs were infected with GFP-expressing S. Typhimurium (MOI 10) for 10 min. BMMs were then serially trapped and stretched outwardly along the laser beam axis. Shown are (Left) the average creep compliance responses of the various cells during the 4 seconds of stretching (indicated by darker red) and (Right) box plots of the average compliance during the stretch. (C) A 3D reconstruction of phalloidin-stained F-actin cytoskeleton (red) in unprimed immortalized BMMs infected with GFP-expressing S. Typhimurium (MOI of 10) for 5 min. (D) Unprimed primary BMMs were imaged for 3 h and then infected with S. Typhimurium (MOI 10) and imaged for 30 min. For each cell, the movies were divided in sections of 10 min and cell movement was analyzed. Data shown are from uninfected cells (over the 3-h time period) and from cells infected with S. Typhimurium for 20–30 min. The mean square displacement (MSD) of infected cells compared with uninfected cells (τ = 2–10 min) is fitted with a power law . From the resulting fit, the value of the MSD at 5 min is calculated. In the box plot, the red bars represent the median of the distribution, the blue edges of the box indicate the 25th and 75th percentile, and dashed bars indicate the extreme points. (E) Histopathology of livers from WT (n = 4) and Nlrc4−/− (n = 5) mice i.v. infected with S. Typhimurium for 7 d. Dashed circles indicate extent of lesions. (F) Livers were fixed and lesions were counted from four liver sections from each mouse. Two-tailed t test, ***P < 0.001; ****P < 0.0001; ns, not statistically significant.
We further confirmed the impact of NLRC4 on actin functionality with the use of phalloidin staining and 3D microscopy to reconstruct the F-actin cytoskeleton to visualize actin reorganization in response to Salmonella infection. In WT immortalized BMMs infected with a strain of S. Typhimurium expressing GFP at an MOI of 10, F-actin formed a dense network, indicated by the intense red staining around the bacterium at 5 min postinfection (Fig. 4C). Nlrc4−/− immortalized BMMs infected with S. Typhimurium showed diffused actin staining proximal to the bacteria, which failed to form a striking stranded pattern observed in WT cells (Fig. 4C). This process could result in a localized “stiff” region to limit the size of the SCV and may restrict bacterial growth.
A consequence of cytoskeletal rearrangements that lead to increased cell stiffness can be impaired cellular movement (31). To investigate whether Salmonella-induced cellular stiffness mediated by NLRC4 affects macrophage migration, we tracked the movement of uninfected primary WT and Nlrc4−/− BMMs for 3 h before infection and for the first 30 min postinfection. Both WT and Nlrc4−/− BMMs moved freely before infection (Fig. 4D, Fig. S5, and Movie S1). Following infection with S. Typhimurium, the movement of WT BMMs ceased rapidly, whereas the movement of Nlrc4−/− BMMs was unaffected, despite the increasing intracellular bacterial burden (Fig. 4D and Movie S1). Our data therefore suggest that Salmonella infection activates the NLRC4 inflammasome to cause major cytoskeletal reorganization in macrophages, reducing macrophage movement, and susceptibility to infection. To confirm the physiological relevance of cellular movement in the control of salmonellosis, we performed histological analysis of liver tissue sections to investigate the number of infectious foci (lesions or granulomas induced by Salmonella and macrophage recruitment (3)) from mice infected with S. Typhimurium. An increased in bacterial burden is associated with an increase in lesion number (3). We saw an increase in bacterial burden and the number of infectious foci in Nlrc4−/− mice compared with WT mice (Fig. 4 E and F and Fig. S6). Taken together, our results suggest that NLRC4 activation by Salmonella infection changes cytoskeletal dynamics and leads to important physiological functions, including reduced cellular movement, suppression of bacterial uptake, and production of antimicrobial molecules, which together leads to a reduced intracellular bacterial burden that can be effectively cleared by the cell.
Discussion
Inflammasome activation provides host protection against infectious agents. The NLRC4 inflammasome is activated in response to Salmonella infection, inducing caspase-1–dependent cleavage and release of bioactive IL-1β and IL-18 and pyroptosis. Pyroptosis of macrophages leads to release of bacteria for uptake by other cell types, such as neutrophils, but also permits bacterial dissemination (18). Neutrophils, for example, do not undergo NLRC4-mediated pyroptosis and provide a major source of IL-1β during Salmonella infection (32), which indicates that pyroptosis is not always required for the control of pathogen burden. Here, we have shown that macrophages that resist pyroptosis can directly control and restrict intracellular salmonellae. Salmonella-induced NLRC4 inflammasome activation is intimately linked to actin polymerization, where F-actin filaments are recruited to the bacteria within 5 min of infection. Efficient reorganization of the actin cytoskeleton is required for rapid ASC speck formation, a hallmark of inflammasome assembly (17, 28), which occurs within 5 min of infection.
Mechanical correlates of actin reorganization can be measured using an optical stretcher, which determines the dynamic change of the cytoskeleton and changes in its compliance. We found that Salmonella infection and activation of the NLRC4 inflammasome induces cellular stiffness in macrophages such that the cell has an impaired ability to take up more bacteria. This finding raises the question of whether limiting the uptake of bacteria in a cell is of benefit to the host.
Host macrophages infected by Salmonella may direct their resources to killing the residing bacteria before uptake of more bacteria. This strategy may avoid overwhelming infection, thereby allowing more effective clearance of a small number of intracellular bacteria at a time and to avoid pyroptosis. Importantly, we found that activation of the NLRC4 inflammasome is required for generating ROS and restrict intracellular bacterial replication in the SCV.
Bacterial uptake concomitantly induces cell stiffness and impairs cellular movement. The use of actin to target and surround the invading bacteria may also reduce the total amount of actin available for cellular movement, which reduces macrophage migration and dissemination of infected cells to other areas of the tissue. Indeed, mice lacking NLRC4 exhibit increased numbers of infectious foci in liver tissues and are more susceptible to salmonellosis. A previous study has identified that β-actin and γ-actin are targets for caspase-1 cleavage (33). It is possible that Salmonella-induced NLRC4 inflammasome activation drives cleavage and modification of actin to arrest cellular movement. Inflammasome-induced cellular stiffening and arrest may represent a general mechanism for the control of intracellular bacterial infection. Earlier work showed that CD4+ T lymphocytes infected with Shigella flexneri exhibit an impaired ability to migrate in vitro and in vivo (34, 35). Shigella can drive inflammasome activation through both NLRC4 and NLRP3 (36, 37), but it remains to be determined whether increased cell stiffness and impaired movement in immune cells are a consequence of inflammasome activity induced by pathogens other than Salmonella, such as Shigella or Listeria. In conclusion, our work has identified a key cellular mechanism driven by the NLRC4 inflammasome and actin polymerization to reduce intracellular Salmonella burden in macrophages. The fate of a macrophage and the bacterium, therefore, depends on the host–microbial engagement of the NLRC4 inflammasome, which is governed by the number of bacteria that infect and are taken up by the cell, and the ability of the bacteria to replicate or resist bacterial killing.
Materials and Methods
Bacterial Strains.
S. Typhimurium strain SL1344 and S. Typhimurium SL1344 ΔfliCΔfljBΔprgJ and the GFP-expressing strain JH3016 were used and described previously (17, 38).
Fluorescence and Confocal Microscopy and Analysis.
To investigate the number of bacteria per macrophage, primary BMMs were infected with S. Typhimurium JH3016 (38), a derivative of SL1344 that expresses GFP, for 1 h, followed by gentamicin treatment (50 μg/mL) for 1 h to kill extracellular bacteria. BMMs were washed twice with PBS and fixed in 4% (wt/vol) paraformaldehyde for 15 min. Cells were counterstained in DAPI mounting medium (Vector Labs). The number of bacteria per infected BMM was counted, and at least 200 infected BMMs of each genotype were counted in each of the three independent experiments.
To visualize the formation of the ASC speck in primary BMMs, cells were infected with S. Typhimurium strain SL1344 for 5, 10, and 20 min. BMMs were washed twice with PBS and fixed in 4% (wt/vol) paraformaldehyde for 15 min. Blocking was performed using 10% (vol/vol) normal goat serum (Dako) in 0.1% saponin (wt/vol; Sigma) for 1 h. Cells were stained with a rabbit anti-ASC antibody (1:500 dilution, AL177, AG-25B-0006-C100; Adipogen) for 40 min followed by Alexa Fluor 568 anti-rabbit (1:250 dilution; Life Technologies) for 50 min. Cells were counterstained in DAPI mounting medium (Vector Labs). Cells were visualized and imaged using a Leica DM6000 B fluorescence microscope.
For phalloidin staining and 3D reconstruction, immortalized BMMs were detached using Accutase solution (Sigma) and seeded onto glass-bottomed well chambers in serum limiting DMEM for 4 h. Supernatant was aspirated and S. Typhimurium JH3016 (MOI 10) was added. After 5 min, samples were aspirated and washed with PBS, fixed with 4% (wt/vol) paraformaldehyde for 10 min, blocked with 5% (wt/vol) BSA for 60 min, and stained with anti-Salmonella O antigen antibody (1:1,000 dilution, R30956901, Remel, Thermo Scientific) for 60 min. Samples were washed with PBS and stained with Alexa Fluor 488 anti-rabbit IgG (1:1,000 dilution; Life Technologies) and phalloidin-568 (1:50 dilution; Life Technologies) for 60 min. Samples were washed with PBS three times and mounted with DAPI mounting medium (Vector Labs). Samples were imaged on the Leica SP5 laser scanning confocal microscope (63×, 1.4 N.A.) and analyzed using LAS AF software (Leica).
Optical Stretcher.
We used a microfluidic optical stretcher—a specific dual-beam optical trap capable of inducing well-defined stresses on whole single cells in suspension—to measure the overall deformability and creep compliance of cells (29, 30, 39).
All work involving live animals complied with the University of Cambridge Ethics Committee regulations and was performed under the Home Office Project License number 80/2572. Detailed information is presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank K. A. Fitzgerald (University of Massachusetts Medical School) and E. Creagh (Trinity College) for supplying the knockout mouse strains. Financial support for this work was provided by a Cambridge International Scholarship (to S.M.M.), European Research Council Starting Investigator Grant “LightTouch” 282060 (to J.R.G.), EU-FP7 Marie-Curie Initial Training Network Grant Transpol 264399 (to E.C., G.N., P.C., and J.R.G.), Biotechnology and Biological Sciences Research Council (BBSRC) Grants BB/H003916/1 and BB/K006436/1, and BBSRC Research Development Fellowship BB/H021930/1 (to C.E.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419925111/-/DCSupplemental.
References
- 1.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82(5):346–353. [PMC free article] [PubMed] [Google Scholar]
- 2.Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 3.Mastroeni P, Grant A, Restif O, Maskell D. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat Rev Microbiol. 2009;7(1):73–80. doi: 10.1038/nrmicro2034. [DOI] [PubMed] [Google Scholar]
- 4.Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83(14):5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6(1):53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 6.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 7.Tschopp J, Martinon F, Burns K. NALPs: A novel protein family involved in inflammation. Nat Rev Mol Cell Biol. 2003;4(2):95–104. doi: 10.1038/nrm1019. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477(7366):596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 9.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477(7366):592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Zhao Y, Shi J, Shao F. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc Natl Acad Sci USA. 2013;110(35):14408–14413. doi: 10.1073/pnas.1306376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rayamajhi M, Zak DE, Chavarria-Smith J, Vance RE, Miao EA. Cutting edge: Mouse NAIP1 detects the type III secretion system needle protein. J Immunol. 2013;191(8):3986–3989. doi: 10.4049/jimmunol.1301549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430(6996):213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 13.Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 2010;8(6):471–483. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kayagaki N, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479(7371):117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 15.Broz P, et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490(7419):288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurung P, et al. Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-β (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. J Biol Chem. 2012;287(41):34474–34483. doi: 10.1074/jbc.M112.401406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Man SM, et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc Natl Acad Sci USA. 2014;111(20):7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miao EA, et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol. 2010;11(12):1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franchi L, et al. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13(5):449–456. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sellin ME, et al. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 2014;16(2):237–248. doi: 10.1016/j.chom.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Gog JR, et al. Dynamics of Salmonella infection of macrophages at the single cell level. J R Soc Interface. 2012;9(75):2696–2707. doi: 10.1098/rsif.2012.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West AP, et al. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472(7344):476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicks SW, Galán JE. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol. 2013;11(5):316–326. doi: 10.1038/nrmicro3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brenner SL, Korn ED. The effects of cytochalasins on actin polymerization and actin ATPase provide insights into the mechanism of polymerization. J Biol Chem. 1980;255(3):841–844. [PubMed] [Google Scholar]
- 25.Brenner SL, Korn ED. Substoichiometric concentrations of cytochalasin D inhibit actin polymerization. Additional evidence for an F-actin treadmill. J Biol Chem. 1979;254(20):9982–9985. [PubMed] [Google Scholar]
- 26.Poyet JL, et al. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276(30):28309–28313. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 27.Proell M, Gerlic M, Mace PD, Reed JC, Riedl SJ. The CARD plays a critical role in ASC foci formation and inflammasome signalling. Biochem J. 2013;449(3):613–621. doi: 10.1042/BJ20121198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Man SM, et al. Salmonella infection induces recruitment of Caspase-8 to the inflammasome to modulate IL-1β production. J Immunol. 2013;191(10):5239–5246. doi: 10.4049/jimmunol.1301581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guck J, et al. The optical stretcher: A novel laser tool to micromanipulate cells. Biophys J. 2001;81(2):767–784. doi: 10.1016/S0006-3495(01)75740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lautenschläger F, et al. The regulatory role of cell mechanics for migration of differentiating myeloid cells. Proc Natl Acad Sci USA. 2009;106(37):15696–15701. doi: 10.1073/pnas.0811261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guck J, Lautenschläger F, Paschke S, Beil M. Critical review: Cellular mechanobiology and amoeboid migration. Integr Biol (Camb) 2010;2(11-12):575–583. doi: 10.1039/c0ib00050g. [DOI] [PubMed] [Google Scholar]
- 32.Chen KW, et al. The neutrophil NLRC4 inflammasome selectively promotes IL-1β maturation without pyroptosis during acute Salmonella challenge. Cell Reports. 2014;8(2):570–582. doi: 10.1016/j.celrep.2014.06.028. [DOI] [PubMed] [Google Scholar]
- 33.Lamkanfi M, et al. Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol Cell Proteomics. 2008;7(12):2350–2363. doi: 10.1074/mcp.M800132-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konradt C, et al. The Shigella flexneri type three secretion system effector IpgD inhibits T cell migration by manipulating host phosphoinositide metabolism. Cell Host Microbe. 2011;9(4):263–272. doi: 10.1016/j.chom.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Salgado-Pabón W, et al. Shigella impairs T lymphocyte dynamics in vivo. Proc Natl Acad Sci USA. 2013;110(12):4458–4463. doi: 10.1073/pnas.1300981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willingham SB, et al. Microbial pathogen-induced necrotic cell death mediated by the inflammasome components CIAS1/cryopyrin/NLRP3 and ASC. Cell Host Microbe. 2007;2(3):147–159. doi: 10.1016/j.chom.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki T, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3(8):e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hautefort I, Proença MJ, Hinton JC. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl Environ Microbiol. 2003;69(12):7480–7491. doi: 10.1128/AEM.69.12.7480-7491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guck J, et al. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys J. 2005;88(5):3689–3698. doi: 10.1529/biophysj.104.045476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.