Significance
Dynamic palmitoylation at the cell surface by the acyl transferase DHHC5 regulates a plethora of physiological processes, from endocytosis to synaptic plasticity. Here we report that DHHC5 is abundantly expressed in cell surface caveolar microdomains in cardiac muscle, and that the Na pump regulator phospholemman is a substrate for DHHC5. Palmitoylation of phospholemman C40 by DHHC5 reduces Na pump activity. DHHC5 interacts with phospholemman independent of its PSD-95/Discs-large/ZO-1 homology (PDZ) binding motif, via a region of its intracellular carboxyl tail that has not hitherto been implicated in substrate recognition. This suggests that it may be possible to selectively manipulate DHHC5 activity by blocking the recruitment of specific substrates to the active site by the carboxyl tail.
Keywords: phospholemman, sodium pump, palmitoylation, DHHC, ion transport
Abstract
The cardiac phosphoprotein phospholemman (PLM) regulates the cardiac sodium pump, activating the pump when phosphorylated and inhibiting it when palmitoylated. Protein palmitoylation, the reversible attachment of a 16 carbon fatty acid to a cysteine thiol, is catalyzed by the Asp-His-His-Cys (DHHC) motif-containing palmitoyl acyltransferases. The cell surface palmitoyl acyltransferase DHHC5 regulates a growing number of cellular processes, but relatively few DHHC5 substrates have been identified to date. We examined the expression of DHHC isoforms in ventricular muscle and report that DHHC5 is among the most abundantly expressed DHHCs in the heart and localizes to caveolin-enriched cell surface microdomains. DHHC5 coimmunoprecipitates with PLM in ventricular myocytes and transiently transfected cells. Overexpression and silencing experiments indicate that DHHC5 palmitoylates PLM at two juxtamembrane cysteines, C40 and C42, although C40 is the principal palmitoylation site. PLM interaction with and palmitoylation by DHHC5 is independent of the DHHC5 PSD-95/Discs-large/ZO-1 homology (PDZ) binding motif, but requires a ∼120 amino acid region of the DHHC5 intracellular C-tail immediately after the fourth transmembrane domain. PLM C42A but not PLM C40A inhibits the Na pump, indicating PLM palmitoylation at C40 but not C42 is required for PLM-mediated inhibition of pump activity. In conclusion, we demonstrate an enzyme–substrate relationship for DHHC5 and PLM and describe a means of substrate recruitment not hitherto described for this acyltransferase. We propose that PLM palmitoylation by DHHC5 promotes phospholipid interactions that inhibit the Na pump.
Protein palmitoylation, the reversible attachment of a 16 carbon fatty acid to a cysteine thiol via a thioester bond, is catalyzed by Asp-His-His-Cys motif-containing palmitoyl acyltransferases (DHHC–PATs); there are 23 human isoforms (1). These zinc-finger–containing enzymes typically have four transmembrane (TM) domains, with a conserved ∼50 amino acid cysteine-rich cytosolic core located between TM2 and -3, which contains a conserved DHHC motif, the active site. In contrast, the intracellular amino and carboxyl termini are poorly conserved, and likely contribute to DHHC isoform substrate selectivity (1). DHHC–PATs are expressed throughout the secretory pathway, but DHHC5 is widely recognized as one of very few cell-surface–localized PATs (2, 3). The final four amino acids of DHHC5 form a canonical class II PSD-95/Discs-large/ZO-1 homology (PDZ) binding motif, which interacts with postsynaptic density protein 95 (PSD-95) (2), although PSD-95 is not itself a DHHC5 substrate.
An appreciation is now growing that protein palmitoylation turns over rapidly (in minutes) for certain proteins (4–8). For example, dynamic surface membrane protein palmitoylation by DHHC5 underlies a novel form of endocytosis, massive endocytosis (MEND), in which up to 70% of the cell surface membrane is internalized (7, 8). Calcium overload leading to mitochondrial stress causes transient openings of the mitochondrial permeability transition pore (MPTP) and release of CoA into the cytoplasm, where it is acylated to form a substrate for DHHC5 to palmitoylate surface membrane proteins (7). The clustering of acylated proteins in lipid-ordered domains leads to MEND by as-yet unidentified mechanisms in multiple cells types. Importantly, MEND occurs during reperfusion of anoxic cardiac muscle and is inhibited by interventions classically reported to reduce MPTP opening and protect against reperfusion injury, such as adenosine and cyclosporin A (8). DHHC5 knockout hearts in which MEND is essentially absent show significantly enhanced functional recovery following anoxia–reperfusion (8), strongly implicating DHHC5 and the MEND pathway in reperfusion injury.
Interestingly, MEND is accelerated in the presence of the Na pump regulatory subunit phospholemman (PLM) (7), which inhibits the Na pump when palmitoylated (9, 10), activates the pump when phosphorylated (11, 12), and relieves the pump from oxidative inhibition by becoming glutathionylated (13). The biochemical interplay of multiple posttranslational modifications on this small protein is complex. Palmitoylation occurs at both cytosolic cysteines (C) 40 and 42 (9), but glutathionylation only at C42 (13), where it presumably competes with palmitoylation. Phosphorylation of PLM by PKA promotes its palmitoylation (9). A number of alternative cellular functions have been proposed for PLM—including regulation of the cardiac Na/Ca exchanger (NCX1) (14), voltage-gated Ca channels (15), and cell volume through formation of a homooligomeric ion channel (16). Although palmitoylated PLM oligomers undoubtedly exist in cardiac muscle (17), their cellular function remains unclear.
In this study, we defined the molecular control of PLM palmitoylation and investigated the functional roles of the two PLM palmitoylation sites in the regulation of the Na pump. We report that PLM is a substrate for DHHC5, which is abundantly expressed in caveolar microdomains in cardiac muscle. Truncation analysis indicates interaction of PLM with and palmitoylation of PLM by DHHC5 requires a region of the DHHC5 carboxyl terminus close to the transmembrane domain, but is independent of the DHHC5 PDZ binding motif.
Results
DHHC Association with PLM.
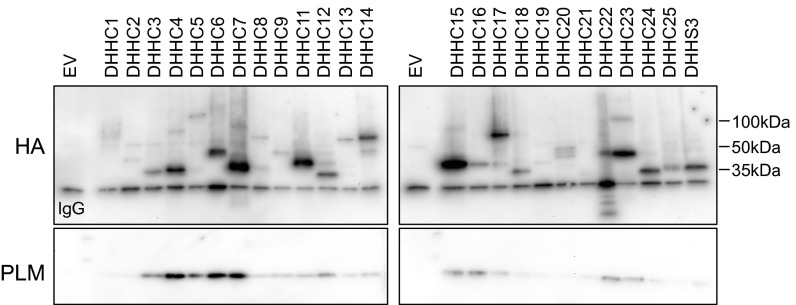
We hypothesized that palmitoylation of PLM may involve the formation of a stable complex with the palmitoylating DHHC enzyme(s). We therefore investigated the association of PLM and DHHC isoforms in coimmunoprecipitation experiments from FT-293 cells, constitutively and stably expressing human PLM, that were transfected with HA-tagged murine DHHCs (Fig. 1). DHHCs 4, 5, 6, and 7 consistently copurify PLM to a much greater extent than any other DHHC isoforms. DHHCs 3, 15, 16, 22, and 23 also consistently coimmunoprecipitate with PLM, but always to a lesser extent. Expression and solubilization of DHHC isoforms showed significant interisoform variability, and we were consistently unable to solubilize DHHC 8 (Fig. 1), so were unable to determine its ability to associate with PLM. Normalizing the quantity of purified PLM to the amount of each DHHC isoform immunoprecipitated identifies DHHC5 as a quantitatively significant binding partner of PLM.
Fig. 1.
DHHC isoform association with PLM. PLM principally coimmunoprecipitates with DHHCs 4, 5, 6, and 7. Transiently expressed HA-tagged DHHC isoforms were immunoprecipitated from FT-293 cells stably expressing PLM. EV, empty-vector transfected cells. Blots are representative of three independent experiments. Light chain of the immunoprecipitating antibody is labeled in HA blots.
DHHC Expression Profile in Cardiac Muscle.
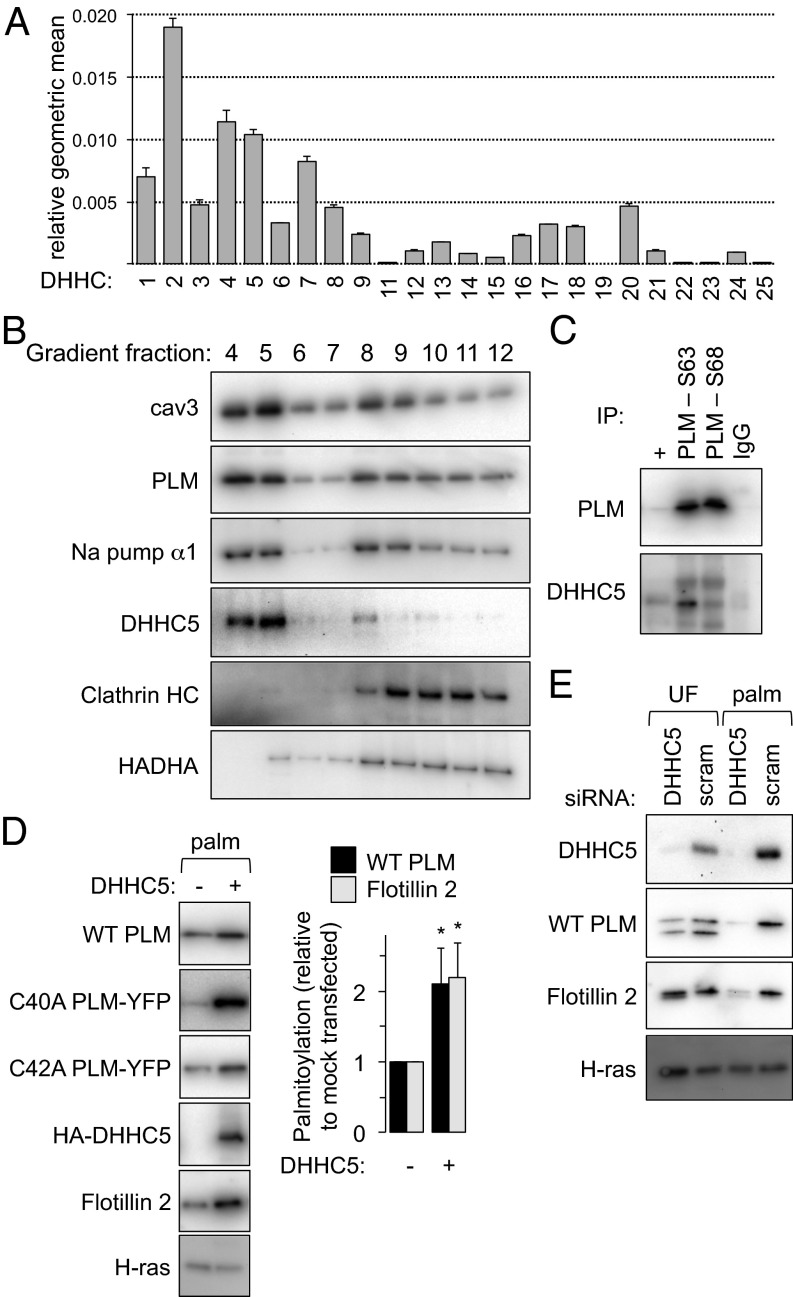
To identify those DHHC isoforms that are expressed in rat cardiac myocytes, we used a quantitative PCR approach similar to one that successfully determined the expression profile of DHHC isoforms in HEK cells (18). mRNA was prepared from four independent rat hearts. The expression profile of DHHCs in cardiac muscle is expressed as the geometric mean relative to β-actin and GAPDH (each heart was assessed in duplicate) in Fig. 2A. DHHCs 2, 4, and 5 are the most abundantly expressed in the heart at the level of mRNA, with DHHCs 19 and 22 below the level of detection for this assay.
Fig. 2.
DHHC expression profile and palmitoylation of PLM. (A) The expression profile of DHHCs in cardiac muscle is expressed as the geometric mean relative to β-actin and GAPDH (n = 4). An essentially identical expression profile was obtained normalizing to β-actin only or GAPDH only. (B) DHHC5 localizes to buoyant caveolin-enriched microdomains with PLM and the Na pump α-subunit in ventricular myocytes. Sonicated ventricular homogenates were separated using a standard discontinuous sucrose gradient. Fractions were collected from the top of the gradient. Caveolin-enriched membranes are found in fractions 4 and 5, whereas bulk sarcolemmal [marker protein, clathrin heavy chain (HC)] and mitochondrial membranes [marker protein, HADHA (hydroxyacyl CoA dehydrogenase)] are exclusively found in dense membranes. (C) PLM and DHHC5 are physically associated in ventricular muscle. PLM phosphorylation states (S63 and S68) were immunoprecipitated from ventricular lysates and immunoblotted as shown. +, unfractionated ventricular lysate. (D) DHHC5 overexpression increases PLM palmitoylation in FT-293 cells stably expressing wild-type PLM, C40A PLM–YFP and C42A PLM–YFP. Flotillin 2 palmitoylation is also elevated in the same cells (palm, palmitoylated proteins purified by Acyl-RAC, *P < 0.05). (E) DHHC5 was silenced in FT-293 cells. Palmitoylation of both PLM and Flotillin 2 was significantly reduced in the absence of DHHC5. Inhibiting palmitoylation of PLM increases its degradation rate (9), so the steady-state expression of PLM was reduced when DHHC5 was silenced. UF, unfractionated cell lysate.
DHHC5 Localization in Cardiac Muscle.
DHHC5 is one of very few cell-surface localized PATs (2, 3), is abundantly expressed in cardiac muscle, and associates with PLM, so we focused on the relationship between PLM and DHHC5. Like PLM and the Na pump, we found that DHHC5 was predominantly localized to buoyant caveolin-enriched microdomains in rat ventricular myocytes (Fig. 2B). We therefore investigated the association of PLM and DHHC5 by coimmunoprecipitation from rat ventricular myocyte lysates (Fig. 2C). PLM phosphorylated at S63 or S68 both coimmunoprecipitate with DHHC5. We were unable to assess copurification of DHHC5 with unphosphorylated PLM as our antibody specific for unphosphorylated PLM is raised in rabbit and showed significant interference when blotting for DHHC5. The pool of PLM phosphorylated at S63 is not associated with the Na pump in cardiac muscle (17), so our data suggest a direct interaction between DHHC5 and PLM, rather than one dependent upon another pump subunit.
Effect of DHHC5 Overexpression and Silencing on PLM Palmitoylation.
Given the physical interaction between PLM and DHHC5, we investigated whether PLM is a DHHC5 substrate by overexpressing DHHC5 in FT-293 cells constitutively and stably expressing PLM. Overexpression of HA-tagged DHHC5 increased palmitoylation of PLM in these cells, as well as increasing the palmitoylation of the established DHHC5 substrate flotillin 2 (Fig. 2D). When we overexpressed DHHC5 in cells expressing PLM–YFP C40A or PLM–YFP C42A, both variants showed increased palmitoylation, suggesting that both sites are DHHC5 substrates.
Overexpression of DHHC isoforms may result in nonspecific substrate palmitoylation, so we also silenced DHHC5 and measured the impact on PLM palmitoylation (Fig. 2E). Palmitoylation of both PLM and flotillin 2 were significantly reduced when DHHC5 was silenced. In both overexpression and silencing experiments we observed no change in palmitoylation of H-ras, which is palmitoylated by DHHC9 in the Golgi apparatus (19).
DHHC5 Truncations.
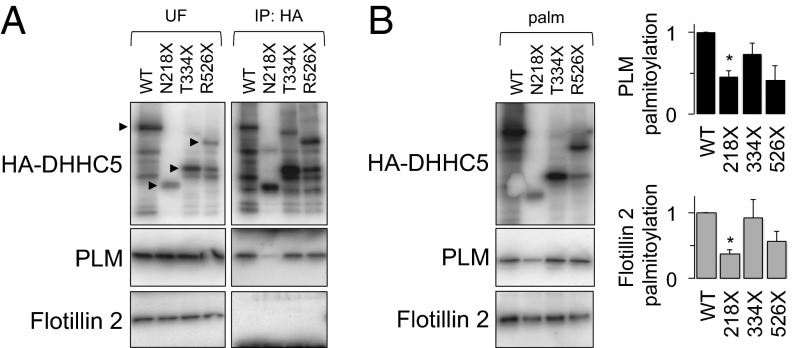
We investigated the region(s) of DHHC5 that interacts with PLM, and the functional importance of interaction(s) for PLM palmitoylation by making truncations based on a DHHC5 disorder prediction (Fig. S1, the sites of truncation are marked in the disorder prediction). DHHC5 was truncated in its disordered intracellular carboxyl terminus (C-tail) by introducing stop codons to produce mutants N218X, T334X, and R526X. Wild-type and truncated DHHC5 were transiently expressed with constitutively expressed PLM in FT-293 cells, and their association was assessed by immunoprecipitating the HA-tagged DHHC5. Fig. 3A indicates that although wild type, R526X, and T334X DHHC5 copurify with PLM to the same extent, association of N218X DHHC5 with PLM is essentially abolished. Interestingly, we were unable to detect the association of the DHHC5 substrate flotillin 2 with wild-type or truncated DHHC5 in the same experiment, suggesting that DHHC5 can palmitoylate certain substrates without forming stable complexes (Fig. 3A).
Fig. 3.
The DHHC5 C-tail is required for palmitoylation of PLM. (A) Effect of DHHC5 truncations on its association with PLM and flotillin 2. Note the sensitivity to proteolysis characteristic of disordered proteins. PLM associates with DHHC5 until a region between N218 and T334 is removed. Flotillin 2 is not found associated with DHHC5 in these experiments. UF, unfractionated cell lysate. (B) Influence of DHHC5 truncations on substrate palmitoylation. Palmitoylation of PLM is significantly reduced when the region of DHHC5 that interacts with PLM is removed. Flotillin 2 palmitoylation is also reduced in the presence of the N218X truncation compared with wild-type DHHC5. (palm, palmitoylated proteins, *P < 0.05).
We next investigated whether palmitoylation of PLM by DHHC5 was influenced by DHHC5 truncations. DHHC catalytic activity relies on autopalmitoylation within the DHHC motif, followed by transfer of the palmitoyl group to the substrate (20), which is unlikely to be influenced by mutations outside the DHHC-containing cysteine-rich domain. Fig. 3B indicates that, although all DHHC5 truncation mutants are palmitoylated, and therefore likely retain catalytic activity, mutant N218X is no longer able to palmitoylate PLM. Hence the interaction with and palmitoylation of this DHHC5 substrate requires a region of DHHC5 between N218 and T334. Interestingly, despite its failure to associate with any DHHC5 constructs expressed, palmitoylation of flotillin 2 was also reduced in cells expressing N218X DHHC5 compared with cells expressing WT DHHC5, suggesting that the C-tail may also play a role in enabling flotillin 2 palmitoylation.
Quantitative Analysis of PLM Palmitoylation Sites.
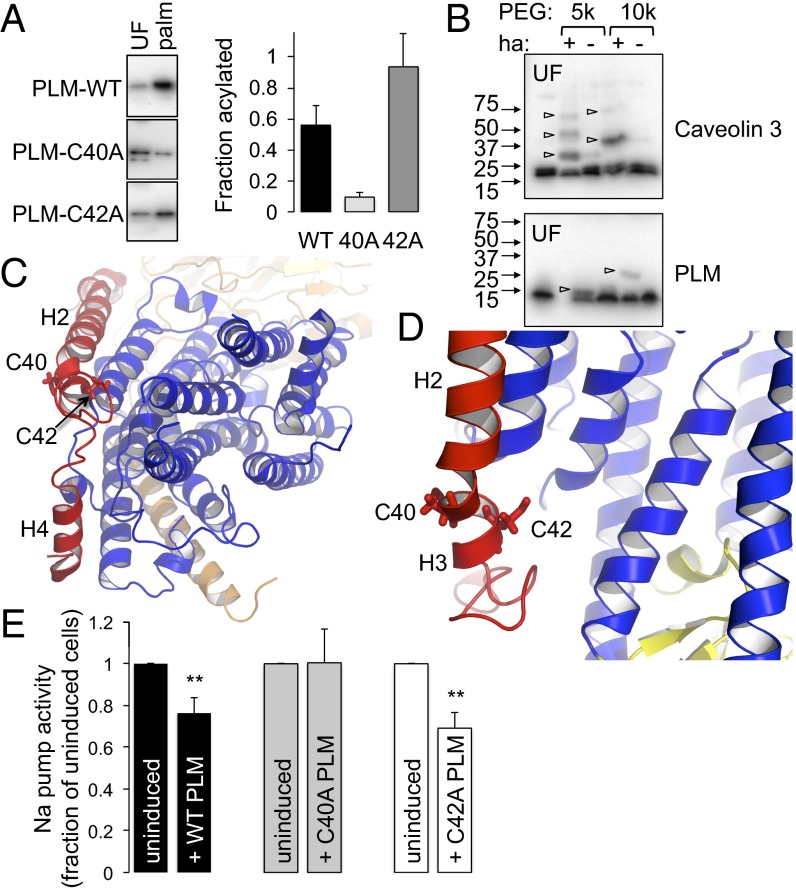
We used resin-assisted capture of acylated proteins (Acyl-RAC) to assess the palmitoylation of wild type, C40A and C42A PLM–YFP in stably transfected cell lines, which are characterized in Fig. S2. Fig. 4A indicates that although both PLM cysteines are clearly palmitoylated (because both mutants were purified by Acyl-RAC), C40 is the principal palmitoylation site in FT-293 cells (because PLM–YFP C42A is substantially enriched by the Acyl-RAC procedure), with only a modest amount of palmitoylation at position C42.
Fig. 4.
Analysis of PLM palmitoylation sites. (A) Palmitoylated proteins (palm) were purified by Acyl-RAC from FT-293 cells expressing wild-type, C40A, and C42A PLM. The relative enrichment compared with unfractionated cell lysates (UF) indicates PLM C40 is the principal palmitoylation site. (B) PEG switch assay from ventricular myocytes. Free cysteines were alkylated and lysates incubated in the presence of 5 kDa or 10 kDa PEG maleimide ± hydroxylamine (ha) to hydrolyze thioester bonds. PEGylation of previously palmitoylated cysteines retards mobility on SDS/PAGE. Caveolin 3 is triply palmitoylated and PLM singly palmitoylated in ventricular myocytes (arrowheads). (C) Position of PLM palmitoylation sites relative to the pump α-subunit. View is approximately perpendicular to the membrane from the cytoplasm. PLM is red, α-subunit transmembrane region is blue, and β-subunit is wheat. Helices H2, H3, and H4 of PLM are labeled. (D) C42 of PLM is orientated toward the α-subunit, and C40 away. The α-subunit P domain is yellow. (E) Na pump activity was measured as ouabain-sensitive 86Rb uptake by cells following induction of wild-type (WT), C40A, or C42A PLM–YFP. For simplicity, pump activity is expressed as a percentage of the activity in the same cells in which PLM expression was not induced. Wild-type and C42A PLM inhibit the Na pump, but C40A PLM does not (n = 7; **P < 0.01).
Acyl-RAC is incapable of distinguishing singly from multiply palmitoylated proteins, so to determine whether PLM is singly or doubly palmitoylated in cardiac myocytes, we developed a PEG switch assay to measure palmitoylation. Free cysteines in lysates from rat ventricular myocytes were blocked with maleimide in the presence of SDS, and after removal of the alkylating agent, lysates were treated with either 5 kDa or 10 kDa PEG maleimide in the presence or absence of neutral hydroxylamine (ha). Cleavage of the thioester bond by ha allows cysteine PEGylation, which is visualized as an increase in molecular mass using SDS/PAGE. We validated this technique using the constitutively palmitoylated protein caveolin 3 (Fig. 4B). Caveolins are palmitoylated at three cysteine residues (21), and three PEGylated forms of caveolin 3 were clearly visible when using 5 kDa PEG maleimide. We only detected a single PEGylated form of PLM in the same samples (Fig. 4B), suggesting that the majority of PLM in cardiac myocytes is singly palmitoylated.
PLM Palmitoylation at C40 Inhibits the Na Pump.
Molecular models of the PLM/Na pump complex (Fig. 4 C and D) indicate that the side chain of PLM C42 is orientated toward the pump α-subunit, and C40 is orientated away. Palmitoylation of C42 (with incorporation of the palmitate into the lipid bilayer) may reorientate PLM helix 3 (H3) toward the α-subunit to inhibit the pump, possibly through an effect on the recently described Na entry pathway (22) or an influence on the transition between E1 and E2 states (23). Conversely, palmitoylation of C40 on the opposite side of H3 would oppose such a movement by reorientating H3 in the opposite direction. This raises the possibility that although the overall effect of PLM palmitoylation on the pump is inhibitory, the individual palmitoylation sites may have opposing effects on pump activity through their reorientating effects on PLM H3. Palmitoylation-induced rearrangements of PLM H3 may also influence the interaction of other cytosolic elements of PLM with the pump α-subunit.
We measured Na pump activity as the ouabain-sensitive uptake of 86Rb by cells expressing inducible wild-type, C40A, and C42A PLM–YFP. For simplicity, pump activity following induction of PLM is expressed as a percentage of the activity in the absence of PLM in each experiment. As we have reported previously (9), induction of wild-type PLM–YFP inhibits the Na pump (Fig. 4E). In contrast, PLM–YFP C40A is without effect on pump activity, whereas PLM–YFP C42A inhibits the pump (Fig. 4E). Under these conditions, neither PLM itself (9) nor the palmitoylation status of PLM (Fig. S2) influences cell surface expression of the pump α-subunit. Thus, functional effects on the pump are the consequence of an influence of PLM palmitoylation on pump activity (rather than cell surface expression). Hence the inhibitory effect of PLM palmitoylation on the pump is via palmitoylation of PLM C40, which is consistent with our finding that this is the principal palmitoylation site.
Discussion
Here we establish an enzyme–substrate relationship between DHHC5 and PLM, the mechanism by which DHHC5 targets PLM and the palmitoylation site in PLM that regulates the Na pump.
Substrate Recruitment by DHHC5.
The importance of DHHC5 in a variety of biological processes, from MEND to synaptic plasticity (24), is currently emerging. This investigation highlights one means by which DHHC5 substrates are recruited to the enzyme active site—via its extended disordered C-tail. DHHC5 palmitoylation of PLM is supported by and requires direct interaction of DHHC5 with PLM, but it does not require PDZ protein interaction. DHHC5 binding partners identified to date (GRIP1 and PSD-95) bind DHHC5 through its PDZ binding motif, not the disordered C-tail (2, 25). PLM, to our knowledge, is the first DHHC5 substrate found to physically associate with a different region of DHHC5. We propose the region between N218 and T334 of DHHC5 interacts with the carboxyl terminus of PLM (from S37 to R72), because both are located cytoplasmically. Notably, palmitoylation of flotillin 2, which does not physically associate with DHHC5, requires the same region of DHHC5 as PLM. The failure of flotillin 2 to coimmunoprecipitate with DHHC5 may reflect a lower affinity interaction than PLM or a different means of recruiting this particular substrate—possibly indirectly via an adaptor protein, much as is the case for protein kinases. Macromolecular complex assembly as a means of lending specificity to posttranslational modifications is well characterized in the biology of protein kinases (with A-kinase anchoring proteins perhaps the best example in the heart) (26), but remains uninvestigated for DHHCs.
Understanding the mechanisms by which DHHCs recruit substrates is clearly key to understanding the spatial and temporal control of protein palmitoylation. Disorder predictions for all human DHHCs (Fig. S3) indicate that the majority of family members consist of a core-ordered cytosolic cysteine-rich domain with relatively disordered intracellular N and C termini. DHHCs 2 and 3 possess notably little disorder, which may account for their tractability in cell-free systems (20). In some isoforms the region of C-tail disorder is relatively large (for example, DHHCs 1, 5, 8, and 14). Our experimental data together with these disorder predictions are consistent with previous reports that regions outside the cysteine-rich domain are responsible for substrate recruitment to DHHCs (27–29). We suggest that disordered domain interactions underlie substrate recognition by (and hence substrate specificity of) some DHHCs. It is notable that DHHC2, the only isoform other than DHHC5 at the cell surface, does not coimmunoprecipitate with PLM and is predicted to be highly ordered. DHHC2, like DHHC5, is abundantly expressed in cardiac muscle and reported to functionally couple to the Na pump in the heart (8), but our data suggest this is achieved by a mechanism either independent of PLM or without the formation of a stable complex.
PLM Palmitoylation Sites.
We have previously speculated that given the orientation of PLM C42 toward the pump α-subunit, palmitoylation at this site rather than PLM C40 mediates pump inhibition by palmitoylated PLM (10). That this is unequivocally not the case is one conclusion of the present investigation: PLM-mediated inhibition of the Na pump absolutely requires its palmitoylation at C40. PLM is principally palmitoylated at C40 in HEK cells, whereas we estimate less than 10% of PLM C42 is palmitoylated. Our PEGylation assay confirms that PLM is predominantly singly palmitoylated in myocytes. Because we find both PLM C40 and C42 are palmitoylated by DHHC5, it is reasonable to infer that DHHC5 exhibits the same preference for PLM C40 in cardiac myocytes as it does in HEK cells. Site-specific reagents to determine palmitoylation site occupancy in cardiac tissue have resisted development in our hands, and we acknowledge the limitation of extrapolating the DHHC5 preference for C40 in HEK cells to myocytes. The molecular basis for this preference remains undetermined, but may reflect the relative inaccessibility of PLM C42 to DHHC5 when complexed with the Na pump (Fig. 4D).
Functional Implications of DHHC5-Mediated Pump Regulation.
DHHC5 regulates Na pump density at the cell surface via MEND (8). Consistent with our previous investigation (9) we found no influence of PLM or its palmitoylation status on cell surface expression of the pump α-subunit (Fig. S2). Hence we propose that palmitoylation of PLM C40 by DHHC5 has a direct inhibitory effect on pump enzymatic activity at the cell surface and therefore regulates cardiac contractility through the functional coupling of the Na pump and NCX (12). DHHC5 therefore inhibits the pump through both palmitoylation of PLM and the MEND pathway. The convergence of multiple signaling pathways on PLM highlights the key role of this regulatory protein in control of intracellular Na in cardiac myocytes (30). Intracellular Na influences a vast array of cellular processes in the heart (12), including contractility via NCX and mitochondrial energetics (31). The steep reliance of NCX activity on the transmembrane Na gradient means that even small changes in intracellular Na have a substantial influence on systolic Ca. Remarkably, intracellular Na exerts a greater influence on peak systolic Ca than the activity of any of the cardiac Ca transporters (32), so the change in Na pump activity caused by palmitoylation of PLM C40 by DHHC5 is likely to be functionally significant in the heart.
It is widely reported that PLM reduces the pump’s Na affinity in both intact cells (12) and following reconstitution in proteoliposomes (33). Molecular dynamics simulations suggest an interaction between the PLM carboxyl terminus (R65, R66, and R70) and the pump N domain may be responsible for at least part of this functional inhibition (33). The structural imposition of a membrane-anchoring fatty acid on PLM C40 may influence the interaction between elements of the PLM carboxyl terminus and the pump intracellular domains, but it is more probable that PLM palmitoylation influences the pump within the membrane bilayer itself, which is also a site of physical contact between these proteins (34, 35). Palmitoylation is well established as regulating protein partitioning to membrane microdomains (36). Phospholipid interactions with the pump α-subunit can directly regulate the pump (37), and reconstitution experiments suggest that the phospholipid composition of a particular membrane bilayer modifies the regulatory influence of PLM (33, 38). Therefore, we suggest that palmitoylation of PLM C40 modifies regulatory phospholipid interactions to inhibit the pump.
Conclusions
By combining truncation analysis with coimmunoprecipitation and Acyl-RAC, we demonstrate that recruitment and palmitoylation of the DHHC5 substrate PLM requires a region of the DHHC5 intracellular C-tail.
Methods
All experiments involving animals were approved by the University of Dundee Welfare and Ethical Use of Animals Committee.
Materials and Antibodies.
Unless indicated otherwise, all reagents were obtained from Sigma-Aldrich and were of the highest grade available. Antibodies were as previously described (9). Anti-PLM and anti-HADHA were from Abcam, anti-DHHC5 was from Sigma, anti-clathrin heavy chain was from BD Biosciences, anti-ras was from Millipore, and anti-hemagglutinin A (HA) was from Roche.
Plasmids, Cell Lines, Transfection, and Silencing.
Plasmids encoding HA-tagged murine DHHC isoforms were generously provided by Masaki Fukata, Division of Membrane Physiology, National Institute for Physiological Sciences, Okazaki, Aichi, Japan.
HEK-derived FT-293 cells expressing canine PLM–YFP under the control of a tetracycline (tet)-sensitive promoter are described elsewhere (9). FT-293 cells expressing tet-inducible PLM–YFP C40A and PLM–YFP C42A were generated in an identical manner using the Invitrogen Flip-In T-REx system. An additional FT-293 line constitutively expressing untagged human PLM was also generated using the Invitrogen Flip-In system.
All transfections of plasmid DNA used Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
siRNA targeted to human DHHC5 was predesigned and supplied by Qiagen. Knockdown used two siRNAs (20 nM each), which were delivered using Trans-IT X2 (Cambridge Bioscience), according to the manufacturer’s instructions. Cells were harvested 72 h after transfection.
Coimmunoprecipitation.
Proteins were solubilized in 2 mg/mL C12E10 in PBS supplemented with protease inhibitors, and targets were captured overnight at 4 °C using antibodies preimmobilized on protein G Sepharose beads (GE Life Sciences) or anti-HA beads (Roche) as appropriate, as described previously (17).
Purification of Palmitoylated Proteins.
Acylated proteins were captured on preequilibrated thiopropyl Sepharose (GE Life Sciences) in the presence of 200 mM hydroxylamine following blockade of free thiols with methyl methanethiosulfonate, as described in detail elsewhere (17). We routinely assess palmitoylation stoichiometry by comparing target protein abundance in the starting (unfractionated) cell lysate to that captured during the Acyl-RAC procedure. The resin-captured fraction is reduced in volume fivefold compared with the unfractionated cell lysate, so stoichiometrically palmitoylated proteins are fivefold enriched, and enrichment less than fivefold indicates substoichiometric palmitoylation (17).
Adult Rat Ventricular Myocytes.
Calcium-tolerant adult rat ventricular myocytes (ARVMs) were isolated by retrograde perfusion of collagenase in the Langendorff mode.
PEG Switch.
Cells were lysed in 2.5% (wt/vol) SDS, 100 mM Hepes, 1 mM EDTA pH 7.4, and free cysteines alkylated by addition of 100 mM maleimide and incubation at 40 °C for 4 h. Excess unreacted maleimide was removed by acetone precipitation, and protein pellets were extensively washed with 70% (vol/vol) acetone, dried, then resolubilized in 1% (wt/vol) SDS, 100 mM Hepes, and 1 mM EDTA pH 7.4. Acylated proteins were PEGylated using 2 mM 5 kDa or 10 kDa PEG maleimide (Sigma) in the presence of 200 mM hydroxylamine (ha, pH 7.4) for 2 h at room temperature. An identical reaction in which ha was replaced with 200 mM NaCl served as a negative control.
Molecular Models of Na Pump/PLM Complex.
We used our existing model of PLM and the Na pump, based on the structure of the pig enzyme in the E2 state (12).
86Rb Uptake Measurements.
Na pump activity was measured as ouabain-sensitive 86Rb uptake by cells cultured on poly-l-lysine–coated 12-well multiwell dishes. All 86Rb uptake measurements were made in the presence of the Na ionophore monensin (Sigma) to ensure consistent intracellular Na between groups, and the Na/K/2Cl cotransporter inhibitor bumetanide (Sigma) to reduce background uptake of 86Rb. Cells were equilibrated in 20 mM NaCl, 120 mM N-Methyl-D-glucamine (NMDG)-Cl, 1 mM MgCl2, 1 mM CaCl2, 5 mM KCl, 10 mM Hepes, 10 mM glucose, 10 µg/mL monensin, and 100 µM bumetanide pH 7.4 for 20–30 min at 37 °C, then preincubated ± ouabain (100 µM) at 37 °C for 5 min before initiating 86Rb uptake by the addition of 1 µCi/mL 86Rb per well. Cells were then incubated for 15 min at 37 °C before 86Rb uptake was stopped by rapidly washing multiwell dishes by successive immersion in three baths of ice-cold PBS. Cells were lysed in 1% (vol/vol) Triton X-100 in PBS, the 86Rb content of lysates measured in a liquid scintillation counter, and the protein content of lysates measured using a Bradford assay with BSA as standard.
Preliminary experiments showed linear ouabain sensitive and insensitive 86Rb uptake between 5 and 20 min under these conditions. In experiments in which NMDG-Cl was replaced with NaCl in the extracellular buffer (i.e., extracellular NaCl 140 mM), ouabain-sensitive 86Rb uptake rate approximately doubled, confirming that the presence of 10 µg/mL monensin in this buffer was sufficient to match intracellular Na to extracellular Na, and also indicating that in the presence of 20 mM extracellular NaCl, Na pump activity was being measured close to the Km for Na. Ouabain-sensitive 86Rb uptake was typically >90% of total 86Rb uptake.
mRNA Preparation and Quantitative PCR.
mRNA was isolated from rat hearts that were briefly perfused in the Langendorff mode to remove contaminating blood, then snap frozen in liquid nitrogen. mRNA was prepared using the SV Total RNA Isolation system (Promega) according to the manufacturer’s instructions. Briefly, hearts were powdered by grinding in liquid nitrogen, and RNA was prepared from 60 mg of pulverized tissue. cDNA was prepared from 200 ng RNA using the GoScript Reverse Transcription system (Promega) according to the manufacturer’s instructions.
The mRNA expression of each DHHC was quantified relative to the geometric mean of β-actin and GAPDH using Fast Start Universal SYBR Green Mastermix (Roche) in a 25-µL reaction using an ABIPrism 7000 Real-Time PCR machine. Reactions were performed in 25-µL volumes with 0.2-µM primers. cDNA from four independent heart samples (55–115 ng of cDNA) was used per reaction. Cycling conditions were 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. All of the primers were previously validated with efficiencies calculated to be within 0.1 of the control using the equation e = 10(−1/slope) − 1. The internal reference controls were endogenous β-actin and GAPDH detected using Qiagen primer sets Rn_Actb_1_SG QT00193473 and Rn_Gapd_1_SG QT00199633. DHHC primers were designed and validated in house except for DHHCs 6 (Rn_Zdhhc6_1_SG QT02382807), 19 (Rn_Zdhhc19_1_SG QT01575462), and 25 (RnZdhhc25_1_SG QT00436163), which were purchased from Qiagen.
Sucrose Gradient Fractionation.
Caveolin-enriched buoyant membranes were prepared from freshly isolated ARVMs using a discontinuous sucrose gradient as described previously (17).
Site-Directed Mutagenesis.
All mutations were introduced using the QuikChange Site-Directed Mutagenesis kit (Agilent) and confirmed by sequencing. The codon encoding N218 of murine DHHC5 was mutated to a stop codon using the oligonucleotide primers GGCTAGGGGACGCACAACCTGAACAGGTTACAGG and CCTGTAACCTGTTCAGGTTGTGCGTCCCCTAGCC. The codon encoding T334 of murine DHHC5 was mutated to a stop codon using the oligonucleotide primers CGAACACACCTCAGCCTGGCTTAATGAGGATAGCAG and CTGCTATCCTCATTAAGCCAGGCTGAGGTGTGTTCG. The codon encoding R526 of murine DHHC5 was mutated to a stop codon using the oligonucleotide primers CTGGCCCACCACACTGAGAACCCTCACC and GGTGAGGGTTCTCAGTGTGGTGGGCCAG.
Supplementary Material
Acknowledgments
This work was supported by British Heart Foundation Grants RG/12/4/29426 (to M. J. Shattock and W.F.) and PG/12/6/29366 (to W.F. and M.L.J.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.E.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413627111/-/DCSupplemental.
References
- 1.Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47(6):1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, et al. DHHC5 interacts with PDZ domain 3 of post-synaptic density-95 (PSD-95) protein and plays a role in learning and memory. J Biol Chem. 2010;285(17):13022–13031. doi: 10.1074/jbc.M109.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohno Y, Kihara A, Sano T, Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim Biophys Acta. 2006;1761(4):474–483. doi: 10.1016/j.bbalip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2012;9(1):84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang R, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456(7224):904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang MM, Tsou LK, Charron G, Raghavan AS, Hang HC. Tandem fluorescence imaging of dynamic S-acylation and protein turnover. Proc Natl Acad Sci USA. 2010;107(19):8627–8632. doi: 10.1073/pnas.0912306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilgemann DW, Fine M, Linder ME, Jennings BC, Lin MJ. Massive endocytosis triggered by surface membrane palmitoylation under mitochondrial control in BHK fibroblasts. eLife. 2013;2:e01293. doi: 10.7554/eLife.01293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin MJ, et al. Massive palmitoylation-dependent endocytosis during reoxygenation of anoxic cardiac muscle. eLife. 2013;2:e01295. doi: 10.7554/eLife.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tulloch LB, et al. The inhibitory effect of phospholemman on the sodium pump requires its palmitoylation. J Biol Chem. 2011;286(41):36020–36031. doi: 10.1074/jbc.M111.282145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howie J, Tulloch LB, Shattock MJ, Fuller W. Regulation of the cardiac Na(+) pump by palmitoylation of its catalytic and regulatory subunits. Biochem Soc Trans. 2013;41(1):95–100. doi: 10.1042/BST20120269. [DOI] [PubMed] [Google Scholar]
- 11.Fuller W, et al. FXYD1 phosphorylation in vitro and in adult rat cardiac myocytes: Threonine 69 is a novel substrate for protein kinase C. Am J Physiol Cell Physiol. 2009;296(6):C1346–C1355. doi: 10.1152/ajpcell.00523.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller W, et al. Regulation of the cardiac sodium pump. Cell Mol Life Sci. 2013;70(8):1357–1380. doi: 10.1007/s00018-012-1134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bibert S, et al. FXYD proteins reverse inhibition of the Na+-K+ pump mediated by glutathionylation of its beta1 subunit. J Biol Chem. 2011;286(21):18562–18572. doi: 10.1074/jbc.M110.184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahlers BA, et al. Identification of an endogenous inhibitor of the cardiac Na+/Ca2+ exchanger, phospholemman. J Biol Chem. 2005;280(20):19875–19882. doi: 10.1074/jbc.M414703200. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, et al. Phospholemman modulates the gating of cardiac L-type calcium channels. Biophys J. 2010;98(7):1149–1159. doi: 10.1016/j.bpj.2009.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moorman JR, et al. Unitary anion currents through phospholemman channel molecules. Nature. 1995;377(6551):737–740. doi: 10.1038/377737a0. [DOI] [PubMed] [Google Scholar]
- 17.Wypijewski KJ, et al. A separate pool of cardiac phospholemman that does not regulate or associate with the sodium pump: Multimers of phospholemman in ventricular muscle. J Biol Chem. 2013;288(19):13808–13820. doi: 10.1074/jbc.M113.460956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian L, McClafferty H, Jeffries O, Shipston MJ. Multiple palmitoyltransferases are required for palmitoylation-dependent regulation of large conductance calcium- and voltage-activated potassium channels. J Biol Chem. 2010;285(31):23954–23962. doi: 10.1074/jbc.M110.137802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swarthout JT, et al. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem. 2005;280(35):31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 20.Jennings BC, Linder ME. DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J Biol Chem. 2012;287(10):7236–7245. doi: 10.1074/jbc.M111.337246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dietzen DJ, Hastings WR, Lublin DM. Caveolin is palmitoylated on multiple cysteine residues. Palmitoylation is not necessary for localization of caveolin to caveolae. J Biol Chem. 1995;270(12):6838–6842. doi: 10.1074/jbc.270.12.6838. [DOI] [PubMed] [Google Scholar]
- 22.Kanai R, Ogawa H, Vilsen B, Cornelius F, Toyoshima C. Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state. Nature. 2013;502(7470):201–206. doi: 10.1038/nature12578. [DOI] [PubMed] [Google Scholar]
- 23.Mahmmoud YA, et al. Regulation of Na,K-ATPase by PLMS, the phospholemman-like protein from shark: Molecular cloning, sequence, expression, cellular distribution, and functional effects of PLMS. J Biol Chem. 2003;278(39):37427–37438. doi: 10.1074/jbc.M305126200. [DOI] [PubMed] [Google Scholar]
- 24.Brigidi GS, et al. Palmitoylation of δ-catenin by DHHC5 mediates activity-induced synapse plasticity. Nat Neurosci. 2014;17(4):522–532. doi: 10.1038/nn.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas GM, Hayashi T, Chiu SL, Chen CM, Huganir RL. Palmitoylation by DHHC5/8 targets GRIP1 to dendritic endosomes to regulate AMPA-R trafficking. Neuron. 2012;73(3):482–496. doi: 10.1016/j.neuron.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McConnachie G, Langeberg LK, Scott JD. AKAP signaling complexes: Getting to the heart of the matter. Trends Mol Med. 2006;12(7):317–323. doi: 10.1016/j.molmed.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Huang K, et al. Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. FASEB J. 2009;23(8):2605–2615. doi: 10.1096/fj.08-127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greaves J, et al. The hydrophobic cysteine-rich domain of SNAP25 couples with downstream residues to mediate membrane interactions and recognition by DHHC palmitoyl transferases. Mol Biol Cell. 2009;20(6):1845–1854. doi: 10.1091/mbc.E08-09-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadolski MJ, Linder ME. Molecular recognition of the palmitoylation substrate Vac8 by its palmitoyltransferase Pfa3. J Biol Chem. 2009;284(26):17720–17730. doi: 10.1074/jbc.M109.005447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boguslavskyi A, et al. Cardiac hypertrophy in mice expressing unphosphorylatable phospholemman. Cardiovasc Res. 2014;104(1):72–82. doi: 10.1093/cvr/cvu182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohlhaas M, et al. Elevated cytosolic Na+ increases mitochondrial formation of reactive oxygen species in failing cardiac myocytes. Circulation. 2010;121(14):1606–1613. doi: 10.1161/CIRCULATIONAHA.109.914911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hilgemann DW. New insights into the molecular and cellular workings of the cardiac Na+/Ca2+ exchanger. Am J Physiol Cell Physiol. 2004;287(5):C1167–C1172. doi: 10.1152/ajpcell.00288.2004. [DOI] [PubMed] [Google Scholar]
- 33.Cirri E, et al. Surface charges of the membrane crucially affect regulation of Na,K-ATPase by phospholemman (FXYD1) J Membr Biol. 2013;246(12):967–979. doi: 10.1007/s00232-013-9600-5. [DOI] [PubMed] [Google Scholar]
- 34.Mishra NK, et al. FXYD proteins stabilize Na,K-ATPase: Amplification of specific phosphatidylserine-protein interactions. J Biol Chem. 2011;286(11):9699–9712. doi: 10.1074/jbc.M110.184234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khafaga M, et al. Na+/K+-ATPase E960 and phospholemman F28 are critical for their functional interaction. Proc Natl Acad Sci USA. 2012;109(50):20756–20761. doi: 10.1073/pnas.1207866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levental I, Lingwood D, Grzybek M, Coskun U, Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci USA. 2010;107(51):22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haviv H, Habeck M, Kanai R, Toyoshima C, Karlish SJ. Neutral phospholipids stimulate Na,K-ATPase activity: A specific lipid-protein interaction. J Biol Chem. 2013;288(14):10073–10081. doi: 10.1074/jbc.M112.446997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cirri E, et al. Phospholemman (FXYD1) raises the affinity of the human α1β1 isoform of Na,K-ATPase for Na ions. Biochemistry. 2011;50(18):3736–3748. doi: 10.1021/bi2001714. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.