Significance
The microbial production of ethanol (bioethanol) is a massive commercialized technology. Though alcohols with longer carbon chains are chemically much better suited for current transportation needs, their biotechnological production remains challenging. Here we have engineered the model hyperthermophile Pyrococcus furiosus to produce various alcohols from their corresponding organic acids by constructing a synthetic route termed the AOR/AdhA pathway. Our study is also the first example, to our knowledge, of significant alcohol formation in an archaeon, emphasizing the biotechnological potential of novel microorganisms. Moreover, we show that carbon monoxide and hydrogen (syngas) can be used as the driving forces for alcohol production. The application of the AOR/AdhA pathway in syngas-fermenting microorganisms is potentially a game-changing platform technology for the production of longer bioalcohols.
Keywords: Archaea, metabolic engineering, hyperthermophile, carbon monoxide, aldehydes
Abstract
Bioethanol production is achieved by only two metabolic pathways and only at moderate temperatures. Herein a fundamentally different synthetic pathway for bioalcohol production at 70 °C was constructed by insertion of the gene for bacterial alcohol dehydrogenase (AdhA) into the archaeon Pyrococcus furiosus. The engineered strain converted glucose to ethanol via acetate and acetaldehyde, catalyzed by the host-encoded aldehyde ferredoxin oxidoreductase (AOR) and heterologously expressed AdhA, in an energy-conserving, redox-balanced pathway. Furthermore, the AOR/AdhA pathway also converted exogenously added aliphatic and aromatic carboxylic acids to the corresponding alcohol using glucose, pyruvate, and/or hydrogen as the source of reductant. By heterologous coexpression of a membrane-bound carbon monoxide dehydrogenase, CO was used as a reductant for converting carboxylic acids to alcohols. Redirecting the fermentative metabolism of P. furiosus through strategic insertion of foreign genes creates unprecedented opportunities for thermophilic bioalcohol production. Moreover, the AOR/AdhA pathway is a potentially game-changing strategy for syngas fermentation, especially in combination with carbon chain elongation pathways.
Production of alcohol-based biofuels from renewable feedstocks is currently achieved by only a very limited number of metabolic pathways (1, 2). The US bioethanol industry depends on glucose conversion by yeast wherein pyruvate (C3) is decarboxylated to acetaldehyde and then reduced to ethanol (C2) by a monofunctional alcohol dehydrogenase. The other major pathway is found in some anaerobic bacteria, wherein glucose-derived pyruvate is oxidized to acetyl-CoA, and this is further reduced to ethanol by a bifunctional alcohol dehydrogenase (AdhE) (3, 4). Recently, there has been increasing interest in microorganisms that produce longer-chain alcohols (>C2), which have superior characteristics as fuel molecules compared with ethanol, to replace fossil fuels (1, 2). In this case, glucose conversion requires microbial strains engineered to produce one specific alcohol at a time. For example, the acetone–butanol–ethanol fermentation pathway, found in some Clostridia, has been adapted in yeast, Escherichia coli, and a few other bacteria (2, 5) to produce isopropanol or l-butanol. Similarly, n-propanol, 2-methyl-1-butanol, 3-methyl-1-butanol, and 1-butanol are side products of amino acid fermentation by yeast (2), and modified pathways have been expressed in E. coli to produce a specific alcohol (6). In addition, isopentanol can be produced by a variation of the isoprenoid biosynthesis pathway in engineered E. coli (2).
Production of bioalcohols at temperatures above 70 °C has several advantages over ambient-temperature processes, including lower risk of microbial contamination, higher diffusion rates, and lower cooling and distillation costs (7). However, very few microorganisms able to grow at such temperatures are able to generate ethanol from sugar (8–10), and no bacterium growing above 70 °C produces an alcohol other than ethanol. In addition, no member of the domain Archaea is known to produce any alcohol as a major product, regardless of growth temperature. Herein, we describe the metabolic engineering of an archaeon to produce not only ethanol but a range of alcohols at 70−80 °C via a synthetic pathway not known in nature and fundamentally different from those previously known.
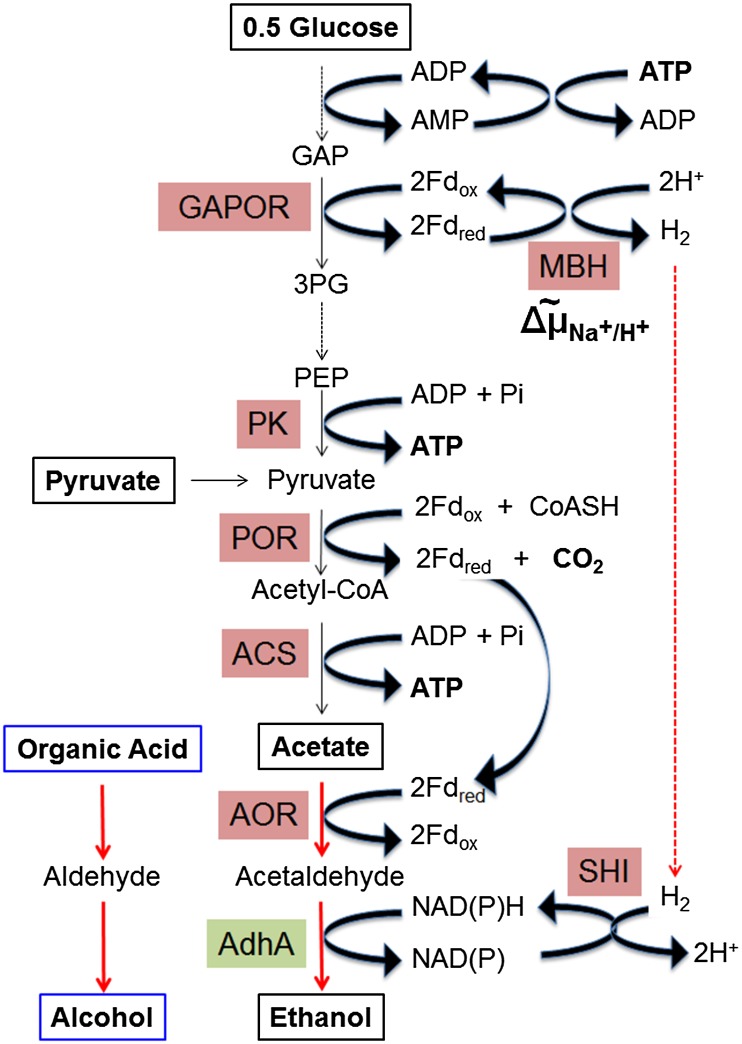
The archaeon Pyrococcus furiosus grows optimally near 100 °C (11) by fermenting simple and complex sugars to acetate, carbon dioxide, and hydrogen gas (12). P. furiosus has an unusual Emden–Meyerhof pathway for the conversion of glucose to pyruvate because reductant is channeled not to NADH but to the redox protein ferredoxin (Fd; Fig. 1) by glyceraldehyde-3-phosphate (GAP) Fd oxidoreductase (GAPOR). Reduced Fd is reoxidized by a membrane-bound, energy-conserving H2-evolving hydrogenase (12). Pyruvate produced by glycolysis is subsequently oxidized to acetyl-CoA by pyruvate Fd oxidoreductase (POR), and acetyl-CoA is converted by ATP-forming acetyl-CoA synthetase (ACS) to acetate. P. furiosus was recently metabolically engineered to generate end products other than acetate in a temperature-controlled manner without the need for chemical inducers. Lactate was produced from glucose, and 3-hydroxypropionate was produced from carbon dioxide and glucose, using heterologously expressed enzymes encoded by foreign genes obtained from microbes that grow near 75 °C (13, 14). At 98 °C, the foreign enzymes were inactive and the engineered P. furiosus strains generated acetate, but near 70 °C, the engineered strains produced either lactate or 3-hydroxypropionate instead.
Fig. 1.
Sugar fermentation coupled to alcohol production by P. furiosus strain A. Glucose from sugars is oxidized to acetate and CO2 and ferredoxin is reduced by GAPOR and POR. In the engineered strain A, acetate is reduced to acetaldehyde by AOR and then reduced to ethanol by the heterologously expressed AdhA from Thermoanaerobacter strain X514. The redox balance is maintained by the production of H2 by the energy-conserving, membrane-bound hydrogenase (MBH) and H2 oxidation by SHI. Organic acids added exogenously are reduced to the corresponding aldehyde and alcohol by AOR and AdhA, respectively, using reductant generated by glucose oxidation.
The primary goal here was to engineer P. furiosus to produce ethanol near 70 °C, applying a similar approach. We found that, unexpectedly, the insertion of a primary alcohol dehydrogenase, AdhA, led to the production of not only ethanol but also of a variety of bioalcohols from their corresponding organic acids. We used gene deletion analysis and 13C labeling to elucidate the biochemical pathway, and hypothesize it might be a remnant of an ancient energy-conservation mechanism. Furthermore, a P. furiosus strain A/Codh was developed that expresses a multisubunit carbon monoxide dehydrogenase. That strain used carbon monoxide as the electron donor for organic acid reduction to bioalcohols, emphasizing the biotechnological versatility and potential of the new synthetic pathway.
Results and Discussion
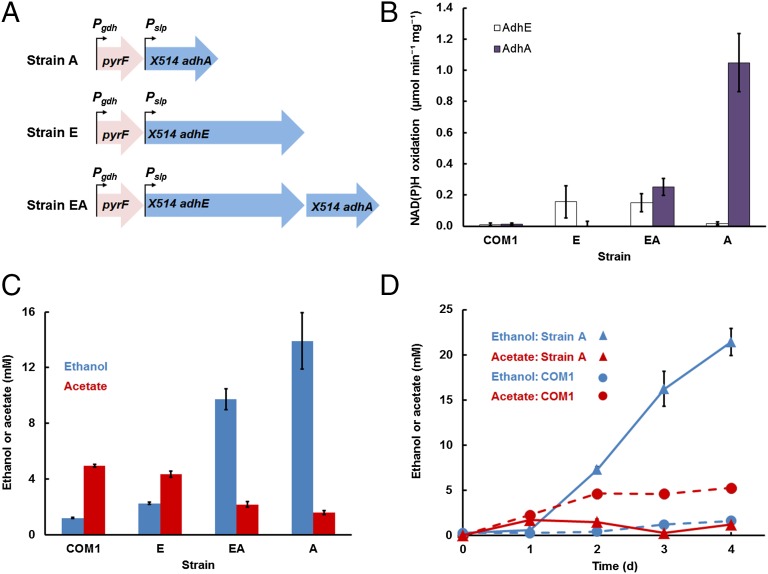
For ethanol production, the foreign genes to be inserted into P. furiosus encoded the bifunctional AdhE and the monofunctional AdhA enzymes, which generate ethanol from acetyl-CoA and acetaldehyde, respectively (9, 15). The genes were obtained from the thermophilic bacterium Thermoanaerobacter strain X514, which grows near 70 °C (16). These genes were inserted, individually and in combination, into the P. furiosus genome (17), yielding strain E (containing adhE), strain A (containing adhA), and strain EA (containing adhE and adhA; Fig. 2A and SI Appendix, Fig. S1 and Table S1). As expected, when grown at 98 °C, no AdhA or AdhE activity could be detected in cell extracts of any strain, although both activities were measured when the strains were grown at 72 °C (Fig. 2B). The activity of AdhA was lower in cell extracts of strain EA compared with those of strain A, possibly due to a lower expression level of adhA, because it is the second gene in the synthetic operon inserted into strain EA.
Fig. 2.
Formation of ethanol from sugars by engineered P. furiosus strains. (A) Genetic constructs with Thermoanaerobacter strain X514 adhE and/or adhA for genome insertion into P. furiosus strain COM1. (B) Specific activities of AdhE (open bars) and AdhA (solid bars) in cell-free extracts of P. furiosus strains EA, E, and A, and parent strain COM1 grown at 72 °C. (C) Ethanol (blue bars) and acetate (red bars) produced after 4 d incubation at 72 °C with maltose (5 g⋅L−1) as the carbon source. (D) Time course of ethanol (blue) and acetate (red) production in strain A (▲) and COM1 (●) at 72 °C with cellobiose (5 g⋅L−1) as the carbon source. After 4 d, ∼35% of the cellobiose was converted to ethanol. Experimental data represent the average of three independently prepared cell extracts or cultures (n = 3; ±SD).
Surprisingly, however, at 72 °C, strain E produced very little ethanol, only slightly more than the trace amounts produced by the parent strain (Fig. 2C). This result might be explained by high activity of the P. furiosus enzyme ATP-forming ACS, which competes with AdhE for the substrate acetyl-CoA (18). Even more unexpected was that strain A generated very high amounts of ethanol (>20 mM), even more than that produced by strain EA (Fig. 2C) with very little acetate (<2 mM; Fig. 2D).
Because AdhA can generate ethanol only from acetaldehyde, and P. furiosus strain A does not contain bifunctional AdhE activity (Fig. 2B), acetyl-CoA is not likely to be the source of this acetaldehyde for ethanol production. Acetaldehyde could arise in P. furiosus from the decarboxylation of pyruvate, which was previously shown to be a significant side reaction of POR (19). Alternatively, it could arise by the reduction of acetate by P. furiosus aldehyde Fd oxidoreductase (AOR). This enzyme is highly expressed in P. furiosus when grown on sugars or peptides, and it has been shown to catalyze the reverse reaction in vitro, the oxidation of various aldehydes to their corresponding acid. It is thought that the in vivo function of AOR is to oxidize toxic aldehydes generated from the 2-ketoacids that are produced during sugar and peptide fermentation. However, this hypothesis has not been experimentally verified (20).
To distinguish between pyruvate or acetate as the source of acetaldehyde, 13C-labeled acetate was added to P. furiosus strain A growing at 72 °C on sugar (the disaccharide maltose), and the isotopic composition of the ethanol produced was analyzed. Approximately 50% of the ethanol formed after 40 h incubation contained the 13C label (SI Appendix, Fig. S2); this can only occur if the acetaldehyde for ethanol production was derived from the added labeled acetate, which was subsequently diluted by unlabeled acetate produced from maltose degradation. To prove that AOR was responsible for reducing the acetate to acetaldehyde, the gene encoding AOR (PF0346) was deleted in strain A. As expected, the new P. furiosus strain A/Δaor, containing Thermoanaerobacter strain X514 AdhA but lacking the host’s AOR, generated only trace amounts of ethanol from maltose, similar to that of the original parent strain (SI Appendix, Fig. S3). It is not clear if AOR normally generates any acetaldehyde from acetate in wild-type P. furiosus (lacking AdhA) and, if so, how that the acetaldehyde is further metabolized. In any event, the properties of strain A call into question the previously proposed role of AOR (20).
The proposed synthetic pathway for ethanol production in P. furiosus strain A is shown in Fig. 1. Acetate generated from glucose oxidation is reduced by AOR, and the acetaldehyde produced is reduced to ethanol by heterologously expressed AdhA. As indicated in Fig. 1, ethanol production from glucose is redox balanced. Reduced Fd for acetate reduction by AOR is supplied by POR and GAPOR (Fig. 1), whereas the NADPH for ethanol production by AdhA must also be generated from reduced Fd; this could occur either by ferredoxin NAD(P) oxidoreductase or from H2 via the cytoplasmic hydrogenase (SHI) of P. furiosus. In addition, energy is conserved in the form of ATP by the ACS reaction. Consequently, this synthetic pathway theoretically converts 0.5 mol of glucose to 1 mol of ethanol and 1 mol of CO2, according to Eq. 1:
| [1] |
Because the synthetic pathway converted the added 13C-labeled acetate to ethanol, we investigated whether other exogenously supplied organic acids would similarly be converted to their corresponding alcohol by the AOR/AdhA pathway; this would seem likely because AOR has a very broad substrate specificity—it oxidizes the decarboxylated forms of keto acids derived from the transaminated derivatives of virtually all 20 amino acids (21). Hence, when 40 mM butyrate was added to a culture of P. furiosus strain A at 72 °C, almost 30 mM butanol was generated (Fig. 3A and SI Appendix, Fig. S4) with the reductant supplied by glucose, according to Eq. 2:
| [2] |
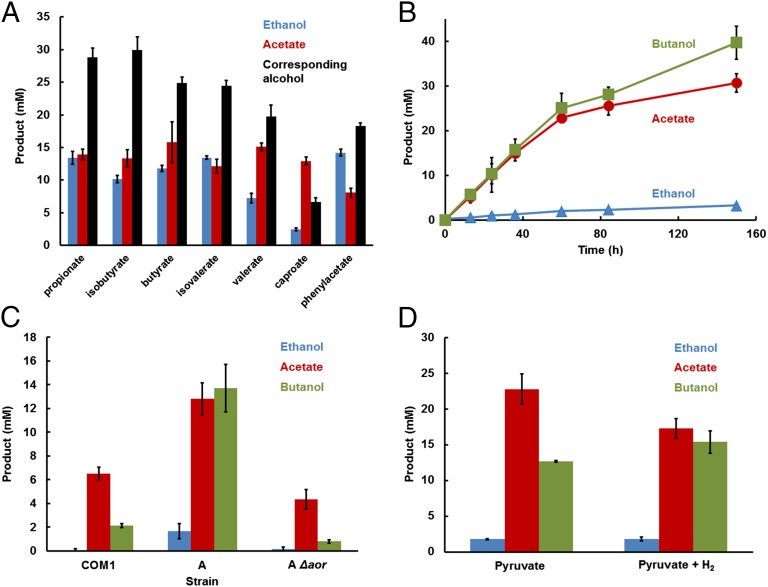
Similar results were obtained when propionate, isobutyrate, valerate, isovalerate, caproate, or phenylacetate were added to P. furious strain A, generating propanol, isobutanol, 1-pentanol, isoamylalcohol, 1-hexanol, and phenylethanol, respectively (Fig. 3A). When butyrate was added to strain A/Δaor, insignificant amounts of butanol were formed (Fig. 3C), once more demonstrating the essential role of AOR in alcohol formation. P. furiosus strain A must, therefore, metabolize the sugar (maltose) to provide reductant for the conversion of the added acid to the corresponding alcohol (Fig. 1 and Eq. 2). Consequently, one would expect acetate to be also generated as the oxidized end product, and this was the case (Fig. 3A). Furthermore, ethanol was produced by the reduction of the acetate generated from sugar oxidation according to Eq. 1 (Fig. 3A). As shown in Fig. 3B, with butyrate as the added acid, acetate and butanol were produced in a 1:1 ratio (Eq. 2). Because butyrate was provided in great excess (100 mM), only minimal amounts of ethanol were produced. Under growth conditions, almost 40 mM (3 g⋅L−1) butanol was generated at a rate of 0.34 mmol⋅h−1⋅g of protein.
Fig. 3.
Reduction of organic acids to alcohols by P. furiosus strain A. (A) Various organic acids were reduced to the corresponding alcohols (black bars) with concomitant production of acetate (red bars) and ethanol (blue bars) during incubation of strain A with maltose for 5 d at 72 °C. (B) Equimolar formation of butanol (green squares) from butyrate and acetate (red circles) from maltose by a 10-fold concentrated cell suspension, with only minor amounts of ethanol (blue triangles) formed. (C) Effect of aor deletion on butanol formation in strain A. (D) Effect of hydrogen on the oxidation of pyruvate to acetate and reduction of butyrate to butanol by a 10-fold concentrated cell suspension of strain A. All experimental data represent the average of three independent cultures (n = 3; ±SD).
P. furiosus can also grow with pyruvate as a carbon and energy source, and so pyruvate should be able replace maltose and supply reductant via POR for butyrate reduction, and this should also result in the formation of ATP (Fig. 1 and Eq. 3). As shown in Fig. 3D, this proved to be the case. The acetate:butanol ratio is predicted to be 2:1 for redox balance (Eq. 3), and this was confirmed experimentally (Fig. 3D). Hydrogen gas (H2) could also be used as a source of reductant in addition to pyruvate (Fig. 1). Use of H2 is predicted to result in the production of equimolar amounts of butanol and acetate (Eq. 4), and this was also demonstrated in vivo (Fig. 3D).
| [3] |
| [4] |
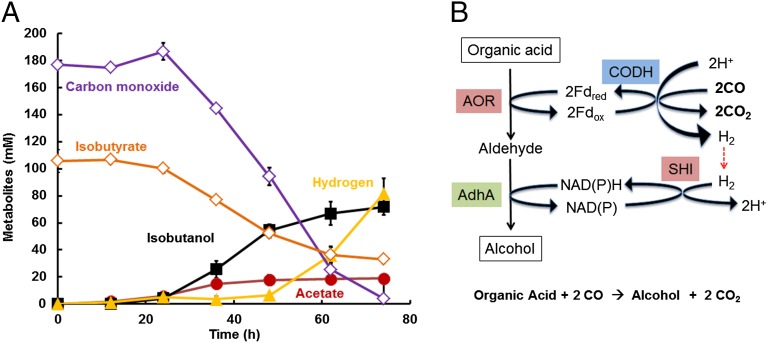
Hence, exogenous acid to alcohol conversion by P. furiosus strain A can be driven by the oxidation of glucose, pyruvate, or pyruvate plus H2. Hydrogen gas cannot be used as the sole source of reductant for alcohol production, however, because its redox potential [H2/H+, E0′ = −414 mV (pH 7.0)] is low enough to reduce NADP (E0′ = −320 mV) but not low enough to drive the reduction of P. furiosus Fd (E0′ = −480 mV) (22) for the AOR reaction. In contrast, carbon monoxide (CO) oxidation is a very low potential reaction (CO/CO2, E0′ = −558 mV) that could potentially be coupled to Fd reduction for the AOR reaction, but P. furiosus does not metabolize CO (or any other C1 compound). Using a bacterial artificial chromosome, we genetically inserted into the chromosome of P. furiosus strain A the 16-gene operon encoding the complete carbon monoxide dehydrogenase/membrane-bound hydrogenase complex (CODH; SI Appendix, Fig. S5) of the carboxydotrophic thermophile Thermococcus onnurineus, which oxidizes CO to H2 and CO2 at 80 °C (23, 24). Remarkably, engineered P. furiosus strain A/Codh was able to use the strong reducing power of CO to produce high concentrations of the alcohol from the corresponding acid at 72 °C. For example, the CO-dependent production of isobutanol (70 mM) from isobutyrate (105 mM) by strain A/Codh is shown in Fig. 4A.
Fig. 4.
CO as source of reductant for conversion of organic acids to alcohols by P. furiosus strain A/Codh. (A) Isobutanol formation (black squares) from isobutyrate (orange diamonds) in the presence of CO (purple diamonds) by cultures of strain A/Codh grown with maltose as the carbon source at 72 °C (n = 2; ±SD). (B) CO oxidation linked to organic acid reduction by P. furiosus strain A/Codh. The CODH complex oxidizes CO with the production of H2 and also reduces ferredoxin to provide low potential electrons to the AOR reaction. NADPH for the AdhA reaction is supplied by H2 oxidation by SHI.
CO oxidation is not coupled to the production of acetate or any other organic compound (Fig. 4B), in contrast to the use of glucose or pyruvate to drive alcohol production by P. furiosus (SI Appendix, Fig. S6). A cell suspension of strain A/Codh used CO as the only electron source to drive the AOR/AdhA pathway and convert isobutyrate to isobutanol, with no other products (except for CO2 from CO oxidation; SI Appendix, Fig. S7). Organic acids are therefore converted to the corresponding alcohol with minimal input of the host’s energy metabolism (Fig. 4B). The CODH complex is thought to convert CO to H2 and CO2 without the involvement of intermediate electron carriers like ferredoxin in T. onnurineus, but this cannot be the case in P. furiosus. Because H2 cannot be the sole source of reductant for organic acid production, T. onnurineus CODH expressed in P. furiosus must also reduce Fd directly, thereby allowed the resulting P. furiosus strain A/Codh to use CO as reductant for the reduction of organic acids by the AOR/AdhA pathway (Fig. 4B). CO-dependent conversion of acids to alcohols also results in H2 production (SI Appendix, Fig. S7); therefore, though CODH reduces P. furiosus Fd directly for the AOR reaction, the NADPH for the AdhA reaction is supplied at least in part via H2 and SHI (Fig. 4B). However, no net H2 production was observed until isobutanol production slowed down (Fig. 4A). The use of CO to provide the reducing equivalents to convert organic acids to their corresponding alcohols has great potential for using industrial syngas (CO and H2) as both an energy source and a carbon source in microbial fermentations to convert organic acids generated from H2 and CO2 (25) to the corresponding alcohol.
Hence, remarkably, with the introduction of a single foreign enzyme, encoded by adhA from Thermoanaerobacter strain X514, P. furiosus can convert glucose to ethanol as well as various organic acids to the corresponding alcohol. Moreover, with the introduction of second enzyme, CODH, CO can serve as the sole source of reductant for the reduction of structurally diverse acids, including aliphatic (C2–C6) and aromatic (phenyl acetate) derivatives. Accordingly, we found that recombinant AdhA (produced in P. furiosus) was able to reduce C2–C6 aldehydes and phenyl acetaldehyde to the corresponding alcohol, and thus it has a broad substrate spectrum, matching that of AOR (21) (SI Appendix, Fig. S8). AdhA also has high affinities for acetaldehyde, butyraldehyde, and NADPH, with apparent Michaelis constant (Km) values of 63, 166, and 31 µM, respectively (SI Appendix, Fig. S9), and so it is able to efficiently reduce the aldehydes generated by AOR. Maximal activity of AdhA was produced in P. furiosus when cells were grown in the 70 to 77 °C range, representing the optimal temperature for production and folding of the AdhA polypeptide (SI Appendix, Fig. S10A); this correlates well with the optimum temperature for in vivo production of butanol from butyrate (70−80 °C; SI Appendix, Fig. S10B). In fact, some butanol was still produced at 94 °C, which corresponds to the upper limit for AdhA activity (SI Appendix, Fig. S8).
Conversion of organic acids to the corresponding alcohols has been reported using the mesophilic anaerobic bacterium Clostridium ljungdahlii (26) grown with CO as the energy source. Although the mechanism and pathway of carbon and electron flow has not been demonstrated, it likely proceeds via activation of organic acids to their CoA ester and CO-derived reducing equivalents are used to form alcohols from the acyl-CoA esters. In contrast, the synthetic AOR/AdhA pathway of P. furiosus for CO-dependent acid-to-alcohol conversion does not involve CoA derivatives (Fig. 4B). There are also reports suggesting that cell-free extracts and/or cell suspensions of the anaerobic bacteria Moorella thermoacetica and Clostridium formicaceticum catalyze a “through reduction” of acids to alcohols (27–29). These reactions were performed using CO, formate or H2 as electron donors in the presence of artificial viologen dyes as electron carriers. However, the fermentation of glucose to ethanol via an AOR/AdhA-type pathway has not been shown previously. Moreover, direct involvement of AOR in microbial alcohol production from organic acids has not been previously demonstrated. It is the low potential fermentative pathway of P. furiosus, where ferredoxin is the sole electron acceptor of sugar oxidation that fuels the AOR reaction and alcohol production.
Conversion of organic acids to alcohols might have a primordial origin, because the synthesis of organic acids from CO2 has been shown experimentally using metal catalysts (30). Furthermore, C2–C6 carboxylic acids have been postulated to be the dominant carbon species in early earth hydrothermal vents based on thermodynamic considerations (31). P. furiosus was isolated from a hot marine vent system (11), and archaea in general are considered by some as the most primitive of all life forms. AOR might, therefore, be a remnant of an ancient pathway for energy conservation in a reducing early earth environment, where geochemically formed organic acids could have served as electron acceptors with carbon monoxide as the potential electron donor.
Materials and Methods
Transformation of P. furiosus.
Escherichia coli XL1 Blue-MRF′ (Agilent Technologies) was used to amplify plasmid DNA. Plasmid DNA purification was performed using the StrataPrep Plasmid Miniprep Kit (Agilent). Extraction of DNA from P. furiosus, transformation of P. furiosus, screening of transformants, and strain purification were performed as previously described, except that the defined medium contained maltose (5 g⋅L−1) instead of cellobiose as the sole growth substrate. The DNA sequence modification of isolated P. furiosus strains was verified by sequencing as previously described (17). Primers used to construct the strains, all plasmids, and all strains are listed in SI Appendix, Fig. S1 and Tables S1 and S2.
Construction of adhA-Containing Strains A, E, EA, and A/Codh.
P. furiosus strain COM1 (17) served as the parent strain for genetic manipulations for the heterologous expression of the bifunctional aldehyde/alcohol dehydrogenase AdhE (Teth514 0627; GeneID: 5876124) and the primary alcohol dehydrogenase AdhA (Teth514 0564; GeneID 5877753) from Thermoanaerobacter strain X514 (16). Genomic DNA was isolated according to Zhou et al. (32). adhE was amplified from genomic DNA by PCR using the primer pairs AdhE-Pslp-F/AdhE-R2 (for construction of plasmid pMB303SLP) or AdhE-Pslp-F/AdhE-SphI-R (for construction of plasmid pMB304SLP). AdhA (for construction of plasmid pMB303SLP) was amplified using AdhA-F2/AdhA-SphI-R. The constitutive promoter Pslp was amplified from genomic DNA of P. furiosus with the primer set Pslp-SacII-F/Pslp-adhE-R. Fusion products of Pslp and adhE or adhE and adhA were obtained by overlap PCR. Products from overlap PCR were digested with the restriction enzymes SacII and SphI and ligated into plasmid vector pSPF300 as described previously (13) to make plasmids pMB303SLP (containing adhE and adhA under control of Pslp) and pMB304SLP (containing only adhE under control of Pslp). pMB303SLP contained a ribosomal binding site of the cold-induced protein CipA (PcipA RBS, 16 bases) between adhE and adhA. pMB303SLP and pMB304SLP were used for transformation of P. furiosus strain ΔpdaD.
For transformation of P. furiosus strain COM1, the Pslp-adhE-adhA or Pslp-adhE fusions were amplified from pMB303SLP and pMB304SLP using the primer pairs Pslp-SphI-F/AdhA-AscI-R or Pslp-SphI-F/AdhE-AscI-R, respectively, and additionally introducing the hpyA1 terminator T hpyA1 (17). The resulting PCR products were digested with AscI and SphI, and then ligated into plasmid pGL007 (14) to make plasmids pMB403SLP and pMB404SLP. Plasmid pMB407SLP for construction of strain A (Fig. 2A) is derived from plasmid pMB403SLP. Using the primers AdhA-Pslp-F/SP2.055, everything but the adhE gene was amplified from plasmid pMB403SLP, and the PCR product was assembled to yield plasmid pMB407SLP using a Gibson Assembly Master Mix (NEB). All plasmids were digested with the restriction enzyme NdeI, and the resulting linear DNA was used to transform strain COM1 to yield strains EA, E, and A (Fig. 2A).
A linear DNA construct was used for a knockout of the aor gene (PF0346) in strain A to make strain AΔaor. First, the marker PgdhpyrF was removed from strain A by selection on 5-fluoroacetic acid (17) to yield strain MW610. Then, primer pairs AOR1/AOR2 and AOR3/AOR4 were used to amplify 500-bp regions upstream and downstream of PF0346. AOR5/SP2.037 and SP2.088/AOR6 were used to amplify the marker PgdhpyrF from pGL007 (14). The PCR products were combined by overlap PCR, and the resulting DNA fragment containing the marker PgdhpyrF flanked by 500-bp regions upstream and downstream of PF0346 was used to transform strain A. The deletion was verified by PCR and sequence analysis.
The pGL058 plasmid containing the T. onnurineus Codh was constructed via Gibson Assembly (NEB) of the following fragments: the 8.8-kb backbone BAC vector containing the pyrF genetic marker and flanking homologous recombination regions targeting the intergenic space between convergent genes PF1232 and PF1233, amplified from pGL054 (33) with primers SP.238 and SP.237; the 200-bp mbh1 (PF1423) promoter region of the membrane-bound hydrogenase gene cluster, amplified from P. furiosus genomic DNA with primers SP.239 and SP.243; and the 13.3-kb Codh gene cluster (TON_1017-TON_1031), amplified from T. onnurineus genomic DNA using primers SP.244 and SP.245 (SI Appendix, Fig. S5). The pGL058 plasmid was linearized using the unique PvuI restriction site on the BAC vector backbone before transformation of P. furiosus strain MW610.
Cultivation of the Strains and Alcohol Production Experiments.
Thermoanaerobacter strain X514 was cultivated at 65 °C on complex medium used for cultivation of thermophilic heterotrophic anaerobes (modified DSMZ 516 medium) with 5 g⋅L−1 cellobiose as electron donor (34). P. furiosus (DSM 3638) was routinely grown at the indicated temperatures with 5 g⋅L−1 maltose and 2 g⋅L−1 yeast extract as described previously (13). A total of 20 µM uracil (Sigma Chemical) was added as needed (17). In temperature-switch experiments, cells were grown at 95 °C until mid- to late-exponential growth phase (0.5−1 × 108 cells), then cooled to 72 °C and kept at this temperature for another 20−48 h as described previously (13, 14). Growth was followed by cell counting and determination of cell protein concentration.
Cell Suspension Experiments.
P. furiosus strain A and strain A/Codh were grown at 72 °C for 4 d to reach high cell density (>1 × 108 cells per milliliter), pelleted by centrifugation (6,000 × g) for 10 min, and then resuspended in media (1/10 of the original culture volume to achieve a 10× concentration). Maltose, pyruvate (as the electron donor), and organic acids (as electron acceptors) were added in excess (≥40−100 mM). To test the effect of hydrogen or CO as the electron donor, the argon headspace was replaced by either gas (2 bar).
13C-Acetate Conversion Experiment.
The 10-mL cultures of P. furiosus strain A were supplied with 8-mM double 13C-labeled sodium acetate (sodium acetate-13C2; Sigma-Aldrich) in addition to 5 g⋅L−1 (unlabeled) maltose and incubated at 72 °C. Samples were taken from the cultures over a 4-d time course to study the change in the carbon isotope signature of acetate and ethanol. Samples (100 µL) of spent media in 2-mL glass vials were acidified by addition of 10 µL 2 M H2SO4. Acidified samples were heated on a hot plate until boiling, and 1- to 4-µL samples were removed and analyzed by GC-MS to obtain 13C/12C ratios for acetic acid and ethanol. This ratio was taken to be equal to the ratio of the measured abundances for masses 62 and 60 for acetic acid and masses 45 and 47 for ethanol. Significant amounts of mixed compounds (containing both 13C and 12C) were not detected. A helium mobile phase was used at a head pressure of 12 psi on an Alltech Econo-Cap 30 m × 0.25 mm EC-WAX column (0.25-µM film) using a Hewlett Packard HP5890A GC with a Hewlett Packard 5971A electron ionization MS. The temperature was held at 40 °C for a 3-min solvent delay and then increased to 220 °C at a rate of 15 °C/min where it was held for an additional minute. For acetic acid measurements in samples with high amounts of ethanol, the method was modified to begin at 100 °C to avoid overloading the MS detector with ethanol while still obtaining sufficient signal for acetic acid. Mass spectra were collected at m/z of 40–200 at a scan rate of four scans per second.
Preparation of Cell Extracts and Enzyme Assays.
P. furiosus cells were harvested by centrifugation for 10 min at 6,000 × g. Cells were lysed under anoxic conditions by osmotic shock in 50 mM Tris⋅HCl (pH 8.0) and 2 mM sodium dithionite, and additionally by a short sonication treatment (30 s, maximum 36 W). The lysis buffer contained 0.5 µg/mL DNase I (Sigma) to decrease the viscosity of the protein extract. Particles were removed by centrifugation at 30,000 × g for 10 min to yield the whole-cell extracts (S30). The supernatant of the whole-cell extract subjected to ultracentrifugation at 100,000 × g for 1 h yielded the cytoplasmatic protein fraction (S100). The protein content was determined using a standard Bradford assay. Whole-cell extracts and S100 were kept anoxic at all times, and enzyme activity assays were performed under reducing conditions in anoxic 50 mM Mops (pH 7.5) plus 2 mM DTT at 70 °C. Unless noted otherwise, aldehyde dehydrogenase (E.C. 1.2.1.3) was determined by oxidation of NADH (0.2 mM) with acetaldehyde (1 mM) as the substrate, and alcohol dehydrogenase was determined by the oxidation of NADPH (0.2 mM) with butyraldehyde (1 mM) as the substrate. Absorption of both NADH and NADPH was measured at 340 nm (ε = 6.22 M−1⋅cm−1), and NAD(P)H oxidation activities are given in micromoles per minute per milligram. Aldehyde ferredoxin oxidoreductase was measured by the oxidation of butyraldehyde (1 mM) with benzyl viologen (1 mM) as electron acceptor as described previously (35). Vmax and Km values of AdhA were calculated using nonlinear regression (nls function) in R (36). Standard Gibbs free energies ΔG°′ were calculated from the free energies of formation ΔGf°, which were taken from Thauer et al. (37).
Chemical Analyses.
Alcohols and organic acids were measured using an Agilent 7890A GC equipped with a Carbowax/20m column and an FID detector. Ethanol and organic acids were also determined using the Megazyme Ethanol Assay Kit (Megazyme) and using a Waters HPLC model 2690 equipped with an Aminex HPX-87H column (300 × 7.8 mm; Bio-Rad) and a photodiode array detector (model 996; Waters), respectively. Hydrogen and CO were determined on a GC-8A gas chromatograph (Shimadzu) equipped with a thermal conductivity detector and a molecular sieve column (model 5A 80/100; Alltech) with argon as the carrier gas.
Supplementary Material
Acknowledgments
We are indebted to Amanda M. Rhaesa for technical assistance and Sanjeev K. Chandrayan, Matthew Keller, R. Chris Hopkins, Irina Kataeva, and Angeli L. Menon for helpful discussions. This work was supported by Bioenergy Science Center, a US Department of Energy (DOE) Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science, Grant DE-PS02-06ER64304; ARPA-E Electrofuels Program of DOE Grant DE-AR0000081; and National Science Foundation Grant CBET-1264052/CBET-1264053.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 17352.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413789111/-/DCSupplemental.
References
- 1.Nielsen J, Larsson C, van Maris A, Pronk J. Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol. 2013;24(3):398–404. doi: 10.1016/j.copbio.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 2.Peralta-Yahya PP, Keasling JD. Advanced biofuel production in microbes. Biotechnol J. 2010;5(2):147–162. doi: 10.1002/biot.200900220. [DOI] [PubMed] [Google Scholar]
- 3.Carere CR, et al. Linking genome content to biofuel production yields: A meta-analysis of major catabolic pathways among select H2 and ethanol-producing bacteria. BMC Microbiol. 2012;12:295. doi: 10.1186/1471-2180-12-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang T, Yao S. Thermophilic, lignocellulolytic bacteria for ethanol production: Current state and perspectives. Appl Microbiol Biotechnol. 2011;92(1):13–27. doi: 10.1007/s00253-011-3456-3. [DOI] [PubMed] [Google Scholar]
- 5.Branduardi P, de Ferra F, Longo V, Porro D. Microbial n-butanol production from Clostridia to non-Clostridial hosts. Eng Life Sci. 2014;14(1):16–26. [Google Scholar]
- 6.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 7.Taylor MP, et al. Thermophilic ethanologenesis: Future prospects for second-generation bioethanol production. Trends Biotechnol. 2009;27(7):398–405. doi: 10.1016/j.tibtech.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Wiegel J, Ljungdahl LG. Thermoanaerobacter ethanolicus gen. nov., spec. nov., a new extreme thermophilic anaerobic bacterium. Arch Microbiol. 1981;128(4):343–348. [Google Scholar]
- 9.Yao S, Mikkelsen MJ. Identification and overexpression of a bifunctional aldehyde/alcohol dehydrogenase responsible for ethanol production in Thermoanaerobacter mathranii. J Mol Microbiol Biotechnol. 2010;19(3):123–133. doi: 10.1159/000321498. [DOI] [PubMed] [Google Scholar]
- 10.Svetlitchnyi VA, et al. Single-step ethanol production from lignocellulose using novel extremely thermophilic bacteria. Biotechnol Biofuels. 2013;6(1):31. doi: 10.1186/1754-6834-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiala G, Stetter KO. Pyrococcus furiosus sp.nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100 °C. Arch Microbiol. 1986;145(1):56–61. [Google Scholar]
- 12.Sapra R, Bagramyan K, Adams MWW. A simple energy-conserving system: Proton reduction coupled to proton translocation. Proc Natl Acad Sci USA. 2003;100(13):7545–7550. doi: 10.1073/pnas.1331436100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basen M, Sun J, Adams MWW. Engineering a hyperthermophilic archaeon for temperature-dependent product formation. MBio. 2012;3(2):e00053–e12. doi: 10.1128/mBio.00053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller MW, et al. Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide. Proc Natl Acad Sci USA. 2013;110(15):5840–5845. doi: 10.1073/pnas.1222607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pei J, et al. Thermoanaerobacter spp. control ethanol pathway via transcriptional regulation and versatility of key enzymes. Metab Eng. 2010;12(5):420–428. doi: 10.1016/j.ymben.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Roh Y, et al. Isolation and characterization of metal-reducing thermoanaerobacter strains from deep subsurface environments of the Piceance Basin, Colorado. Appl Environ Microbiol. 2002;68(12):6013–6020. doi: 10.1128/AEM.68.12.6013-6020.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipscomb GL, et al. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: Construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl Environ Microbiol. 2011;77(7):2232–2238. doi: 10.1128/AEM.02624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorgersen MP, Lipscomb GL, Schut GJ, Kelly RM, Adams MWW. Deletion of acetyl-CoA synthetases I and II increases production of 3-hydroxypropionate by the metabolically-engineered hyperthermophile Pyrococcus furiosus. Metab Eng. 2014;22:83–88. doi: 10.1016/j.ymben.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Ma K, Hutchins A, Sung SJS, Adams MWW. Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase. Proc Natl Acad Sci USA. 1997;94(18):9608–9613. doi: 10.1073/pnas.94.18.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heider J, Ma K, Adams MWW. Purification, characterization, and metabolic function of tungsten-containing aldehyde ferredoxin oxidoreductase from the hyperthermophilic and proteolytic archaeon Thermococcus strain ES-1. J Bacteriol. 1995;177(16):4757–4764. doi: 10.1128/jb.177.16.4757-4764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy R, Adams MWW. Tungsten-dependent aldehyde oxidoreductase: A new family of enzymes containing the pterin cofactor. Met Ions Biol Syst. 2002;39:673–697. [PubMed] [Google Scholar]
- 22.Park JB, Fan CL, Hoffman BM, Adams MWW. Potentiometric and electron nuclear double resonance properties of the two spin forms of the [4Fe-4S]+ cluster in the novel ferredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus. J Biol Chem. 1991;266(29):19351–19356. [PubMed] [Google Scholar]
- 23.Schut GJ, Boyd ES, Peters JW, Adams MWW. The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol Rev. 2013;37(2):182–203. doi: 10.1111/j.1574-6976.2012.00346.x. [DOI] [PubMed] [Google Scholar]
- 24.Yun SH, et al. Proteome analysis of Thermococcus onnurineus NA1 reveals the expression of hydrogen gene cluster under carboxydotrophic growth. J Proteomics. 2011;74(10):1926–1933. doi: 10.1016/j.jprot.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Bengelsdorf FR, Straub M, Dürre P. Bacterial synthesis gas (syngas) fermentation. Environ Technol. 2013;34(13-16):1639–1651. doi: 10.1080/09593330.2013.827747. [DOI] [PubMed] [Google Scholar]
- 26.Perez JM, Richter H, Loftus SE, Angenent LT. Biocatalytic reduction of short-chain carboxylic acids into their corresponding alcohols with syngas fermentation. Biotechnol Bioeng. 2013;110(4):1066–1077. doi: 10.1002/bit.24786. [DOI] [PubMed] [Google Scholar]
- 27.Fraisse L, Simon H. Observations on the reduction of non-activated carboxylates by Clostridium formicoaceticum with carbon monoxide or formate and the influence of various viologens. Arch Microbiol. 1988;150:381–386. [Google Scholar]
- 28.Simon H, White H, Lebertz H, Thanos I. Reduction of 2-enoates and alkanoates with carbon monoxide or formate, viologens, and Clostridium thermoaceticum to saturated acids and unsaturated and saturated alcohols. Angew Chem Int Ed Engl. 1987;26:785–787. [Google Scholar]
- 29.White H, Strobl G, Feicht R, Simon H. Carboxylic acid reductase: A new tungsten enzyme catalyses the reduction of non-activated carboxylic acids to aldehydes. Eur J Biochem. 1989;184(1):89–96. doi: 10.1111/j.1432-1033.1989.tb14993.x. [DOI] [PubMed] [Google Scholar]
- 30.Huber C, Wächtershäuser G. Activated acetic acid by carbon fixation on (Fe,Ni)S under primordial conditions. Science. 1997;276(5310):245–247. doi: 10.1126/science.276.5310.245. [DOI] [PubMed] [Google Scholar]
- 31.Amend JP, LaRowe DE, McCollom TM, Shock EL. The energetics of organic synthesis inside and outside the cell. Phil Trans R Soc B. 2013;368:1622. doi: 10.1098/rstb.2012.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Fries MR, Chee-Sanford JC, Tiedje JM. Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth of toluene and description of Azoarcus tolulyticus sp. nov. Int J Syst Bacteriol. 1995;45(3):500–506. doi: 10.1099/00207713-45-3-500. [DOI] [PubMed] [Google Scholar]
- 33.Lipscomb GL, et al. Engineering hydrogen gas production from formate in a hyperthermophile by heterologous production of an 18-subunit membrane-bound complex. J Biol Chem. 2014;289(5):2873–2879. doi: 10.1074/jbc.M113.530725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang SJ, et al. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl Environ Microbiol. 2009;75(14):4762–4769. doi: 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukund S, Adams MWW. The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an aldehyde ferredoxin oxidoreductase. Evidence for its participation in a unique glycolytic pathway. J Biol Chem. 1991;266(22):14208–14216. [PubMed] [Google Scholar]
- 36.R Development Core Team 2013 R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna). Available at www.R-project.org/
- 37.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41(1):100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.