Abstract
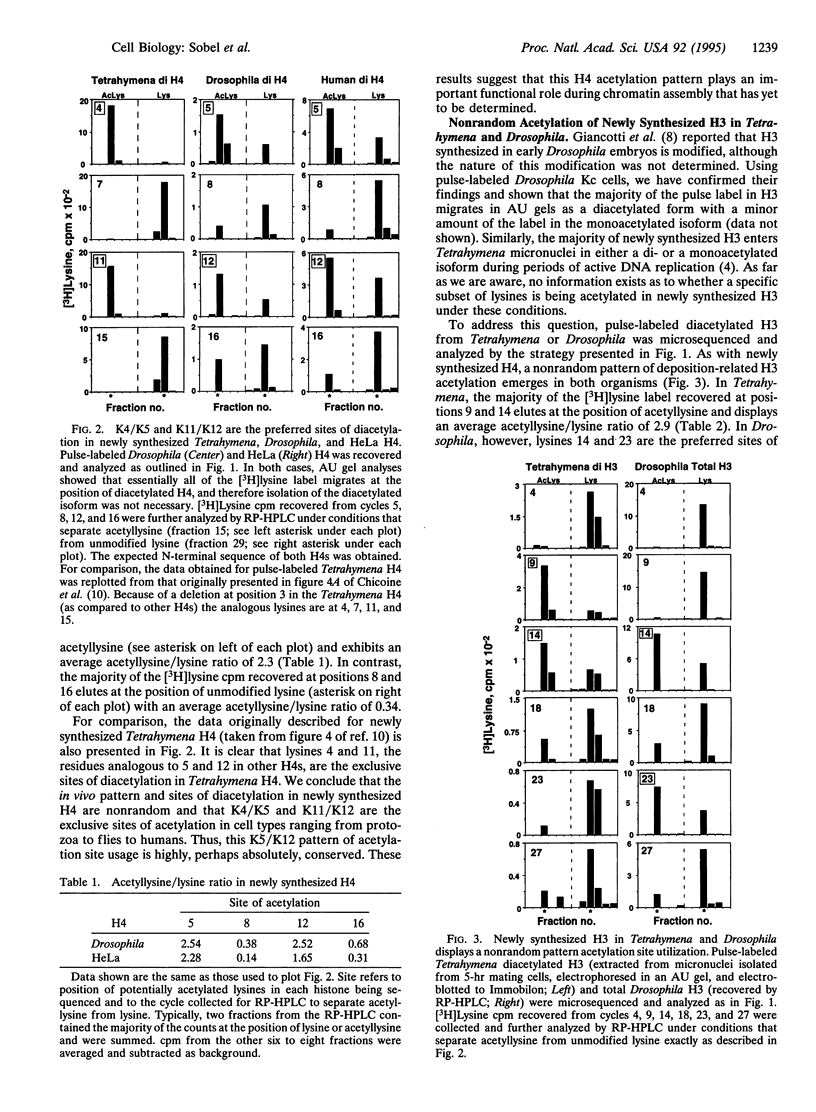
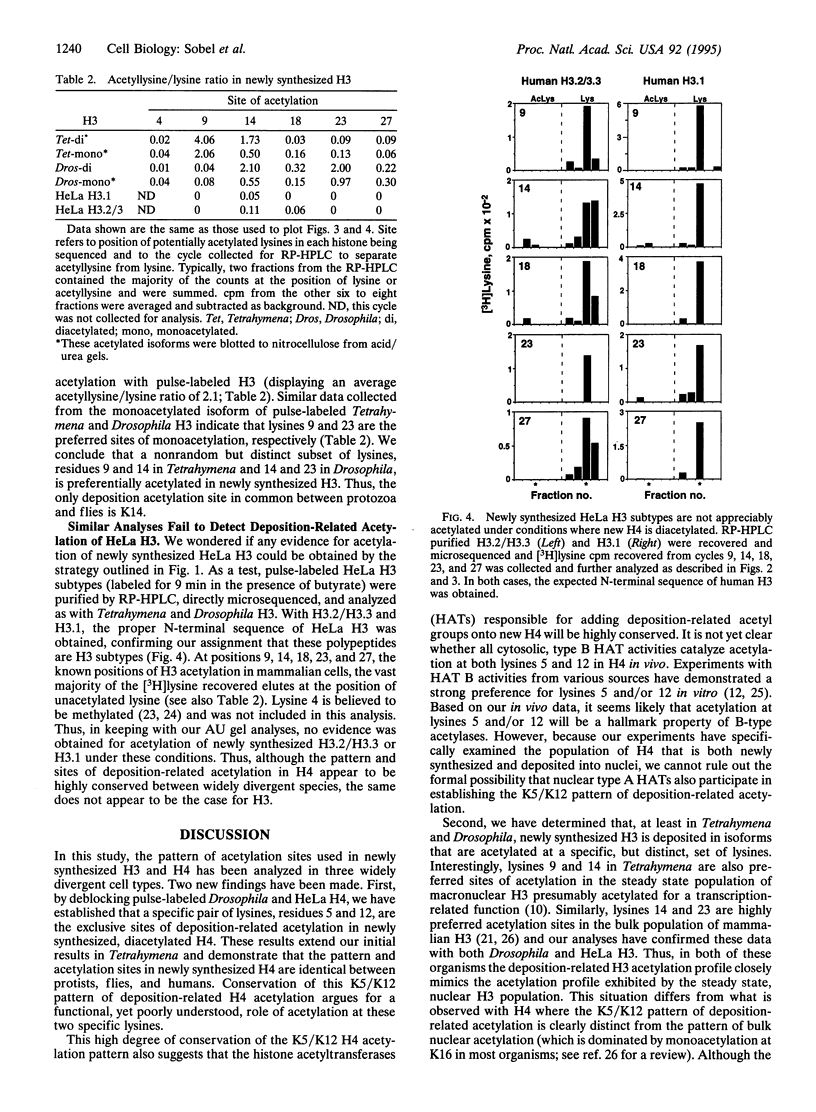
Newly synthesized histone H4 is deposited in a diacetylated isoform in a wide variety of organisms. In Tetrahymena a specific pair of residues, lysines 4 and 11, have been shown to undergo this modification in vivo. In this report, we demonstrate that the analogous residues, lysines 5 and 12, are acetylated in Drosophila and HeLa H4. These data strongly suggest that deposition-related acetylation sites in H4 have been highly, perhaps absolutely, conserved. In Tetrahymena and Drosophila newly synthesized histone H3 is also deposited in several modified forms. Using pulse-labeled H3 we have determined that, like H4, a specific, but distinct, subset of lysines is acetylated in these organisms. In Tetrahymena, lysines 9 and 14 are highly preferred sites of acetylation in new H3 while in Drosophila, lysines 14 and 23 are strongly preferred. No evidence has been obtained for acetylation of newly synthesized H3 in HeLa cells. Thus, although the pattern and sites of deposition-related acetylation appear to be highly conserved in H4, the same does not appear to be the case for histone H3.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allis C. D., Chicoine L. G., Richman R., Schulman I. G. Deposition-related histone acetylation in micronuclei of conjugating Tetrahymena. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8048–8052. doi: 10.1073/pnas.82.23.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis C. D., Dennison D. K. Identification and purification of young macronuclear anlagen from conjugating cells of Tetrahymena thermophila. Dev Biol. 1982 Oct;93(2):519–533. doi: 10.1016/0012-1606(82)90139-7. [DOI] [PubMed] [Google Scholar]
- Annunziato A. T., Seale R. L. Histone deacetylation is required for the maturation of newly replicated chromatin. J Biol Chem. 1983 Oct 25;258(20):12675–12684. [PubMed] [Google Scholar]
- Chambers S. A., Shaw B. R. Levels of histone H4 diacetylation decrease dramatically during sea urchin embryonic development and correlate with cell doubling rate. J Biol Chem. 1984 Nov 10;259(21):13458–13463. [PubMed] [Google Scholar]
- Chicoine L. G., Schulman I. G., Richman R., Cook R. G., Allis C. D. Nonrandom utilization of acetylation sites in histones isolated from Tetrahymena. Evidence for functionally distinct H4 acetylation sites. J Biol Chem. 1986 Jan 25;261(3):1071–1076. [PubMed] [Google Scholar]
- Cousens L. S., Alberts B. M. Accessibility of newly synthesized chromatin to histone acetylase. J Biol Chem. 1982 Apr 10;257(7):3945–3949. [PubMed] [Google Scholar]
- Cousens L. S., Gallwitz D., Alberts B. M. Different accessibilities in chromatin to histone acetylase. J Biol Chem. 1979 Mar 10;254(5):1716–1723. [PubMed] [Google Scholar]
- Giancotti V., Russo E., de Cristini F., Graziosi G., Micali F., Crane-Robinson C. Histone modification in early and late Drosophila embryos. Biochem J. 1984 Mar 1;218(2):321–329. doi: 10.1042/bj2180321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- Harisanova N. T., Ralchev K. H. Histones and histone acetylation during the embryonic development of Drosophila hydei. Cell Differ. 1986 Sep;19(2):115–124. doi: 10.1016/0045-6039(86)90068-0. [DOI] [PubMed] [Google Scholar]
- Jackson V., Shires A., Tanphaichitr N., Chalkley R. Modifications to histones immediately after synthesis. J Mol Biol. 1976 Jun 25;104(2):471–483. doi: 10.1016/0022-2836(76)90282-5. [DOI] [PubMed] [Google Scholar]
- Johnson L. M., Fisher-Adams G., Grunstein M. Identification of a non-basic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J. 1992 Jun;11(6):2201–2209. doi: 10.1002/j.1460-2075.1992.tb05279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijima M., Yoshida M., Sugita K., Horinouchi S., Beppu T. Trapoxin, an antitumor cyclic tetrapeptide, is an irreversible inhibitor of mammalian histone deacetylase. J Biol Chem. 1993 Oct 25;268(30):22429–22435. [PubMed] [Google Scholar]
- Kim U. J., Han M., Kayne P., Grunstein M. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 1988 Jul;7(7):2211–2219. doi: 10.1002/j.1460-2075.1988.tb03060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. Y., Hayes J. J., Pruss D., Wolffe A. P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993 Jan 15;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Madireddi M. T., Davis M. C., Allis C. D. Identification of a novel polypeptide involved in the formation of DNA-containing vesicles during macronuclear development in Tetrahymena. Dev Biol. 1994 Oct;165(2):418–431. doi: 10.1006/dbio.1994.1264. [DOI] [PubMed] [Google Scholar]
- Martindale D. W., Allis C. D., Bruns P. J. Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp Cell Res. 1982 Jul;140(1):227–236. doi: 10.1016/0014-4827(82)90172-0. [DOI] [PubMed] [Google Scholar]
- Marvin K. W., Yau P., Bradbury E. M. Isolation and characterization of acetylated histones H3 and H4 and their assembly into nucleosomes. J Biol Chem. 1990 Nov 15;265(32):19839–19847. [PubMed] [Google Scholar]
- Megee P. C., Morgan B. A., Mittman B. A., Smith M. M. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science. 1990 Feb 16;247(4944):841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- Mingarro I., Sendra R., Salvador M. L., Franco L. Site specificity of pea histone acetyltransferase B in vitro. J Biol Chem. 1993 Jun 25;268(18):13248–13252. [PubMed] [Google Scholar]
- Morgan B. A., Mittman B. A., Smith M. M. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol Cell Biol. 1991 Aug;11(8):4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry C. A., Annunziato A. T. Influence of histone acetylation on the solubility, H1 content and DNase I sensitivity of newly assembled chromatin. Nucleic Acids Res. 1989 Jun 12;17(11):4275–4291. doi: 10.1093/nar/17.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesis K. H., Matthews H. R. Histone acetylation in replication and transcription: turnover at specific acetylation sites in histone H4 from Physarum polycephalum. Arch Biochem Biophys. 1986 Dec;251(2):665–673. doi: 10.1016/0003-9861(86)90376-0. [DOI] [PubMed] [Google Scholar]
- Riggs M. G., Whittaker R. G., Neumann J. R., Ingram V. M. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature. 1977 Aug 4;268(5619):462–464. doi: 10.1038/268462a0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Wangh L. J., Allfrey V. G. Processing of newly synthesized histone molecules. Science. 1975 Oct 10;190(4210):117–128. doi: 10.1126/science.1166303. [DOI] [PubMed] [Google Scholar]
- Sobel R. E., Cook R. G., Allis C. D. Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology. J Biol Chem. 1994 Jul 15;269(28):18576–18582. [PubMed] [Google Scholar]
- Thorne A. W., Kmiciek D., Mitchelson K., Sautiere P., Crane-Robinson C. Patterns of histone acetylation. Eur J Biochem. 1990 Nov 13;193(3):701–713. doi: 10.1111/j.1432-1033.1990.tb19390.x. [DOI] [PubMed] [Google Scholar]
- Turner B. M. Decoding the nucleosome. Cell. 1993 Oct 8;75(1):5–8. [PubMed] [Google Scholar]
- Wang T., Allis C. D. An abundant high-mobility-group-like protein is targeted to micronuclei in a cell cycle-dependent and developmentally regulated fashion in Tetrahymena thermophila. Mol Cell Biol. 1993 Jan;13(1):163–173. doi: 10.1128/mcb.13.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner D., Panneerselvam C., Horecker B. L. Sequencing of peptides and proteins with blocked N-terminal amino acids: N-acetylserine or N-acetylthreonine. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1947–1949. doi: 10.1073/pnas.87.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P. Implications of DNA replication for eukaryotic gene expression. J Cell Sci. 1991 Jun;99(Pt 2):201–206. doi: 10.1242/jcs.99.2.201. [DOI] [PubMed] [Google Scholar]
- Woodland H. R. The modification of stored histones H3 and H4 during the oogenesis and early development of Xenopus laevis. Dev Biol. 1979 Feb;68(2):360–370. doi: 10.1016/0012-1606(79)90210-0. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Panusz H. T., Hatch C. L., Bonner W. M. Histones and their modifications. CRC Crit Rev Biochem. 1986;20(2):201–263. doi: 10.3109/10409238609083735. [DOI] [PubMed] [Google Scholar]