Significance
The pneumococcus accounts for 25% of deaths in children under 5 y of age in developing countries. One of the most important virulence factors expressed by this pathogen is the pore-forming toxin, pneumolysin (Ply), an example of a Gram-positive cholesterol-dependent cytolysin (CDC). We show that Ply interacts with the Lewis histo-blood group antigen sialyl LewisX and that blocking this interaction can protect RBCs from lysis. We also identify glycan receptors on RBCs for the CDC streptolysin O from group A streptococcus. Our study supports the emerging paradigm shift that CDCs have cellular receptors other than cholesterol that define target cell tropism.
Keywords: Streptococcus pneumoniae, pneumolysin, cholesterol-dependent cytolysin, glycan binding, streptolysin O
Abstract
The cholesterol-dependent cytolysin (CDC) pneumolysin (Ply) is a key virulence factor of Streptococcus pneumoniae. Membrane cholesterol is required for the cytolytic activity of this toxin, but it is not clear whether cholesterol is the only cellular receptor. Analysis of Ply binding to a glycan microarray revealed that Ply has lectin activity and binds glycans, including the Lewis histo-blood group antigens. Surface plasmon resonance analysis showed that Ply has the highest affinity for the sialyl LewisX (sLeX) structure, with a Kd of 1.88 × 10−5 M. Ply hemolytic activity against human RBCs showed dose-dependent inhibition by sLeX. Flow cytometric analysis and Western blots showed that blocking binding of Ply to the sLeX glycolipid on RBCs prevents deposition of the toxin in the membrane. The lectin domain responsible for sLeX binding is in domain 4 of Ply, which contains candidate carbohydrate-binding sites. Mutagenesis of these predicted carbohydrate-binding residues of Ply resulted in a decrease in hemolytic activity and a reduced affinity for sLeX. This study reveals that this archetypal CDC requires interaction with the sLeX glycolipid cellular receptor as an essential step before membrane insertion. A similar analysis conducted on streptolysin O from Streptococcus pyogenes revealed that this CDC also has glycan-binding properties and that hemolytic activity against RBCs can be blocked with the glycan lacto-N-neotetraose by inhibiting binding to the cell surface. Together, these data support the emerging paradigm shift that pore-forming toxins, including CDCs, have cellular receptors other than cholesterol that define target cell tropism.
Streptococcus pneumoniae is a leading cause of morbidity and mortality worldwide. This bacterial pathogen is responsible for a range of diseases, including pneumonia, meningitis, septicemia, and otitis media. One of the major virulence factors of S. pneumoniae is the multifunctional pore-forming toxin pneumolysin (Ply). Ply is produced by virtually all clinical isolates of S. pneumoniae and is a member of the cholesterol-dependent cytolysin (CDC) family of toxins (1). The key feature of the CDCs, which are expressed by a number of pathogenic Gram-positive bacteria, is the ability to form pores in cholesterol-containing cell membranes. The pore-forming mechanism of the CDCs is a multistep process that involves recognition and binding to the cholesterol-containing membrane by domain 4 of the toxin, oligomerization of ∼34–50 soluble monomers on the target cell membrane to form a large prepore complex (2), and penetration of the prepore structure into the membrane to become a transmembrane β-barrel pore (3–5).
The cytolytic mechanism of the CDCs depends on the presence of cholesterol in the target cell membrane; hence, it was thought that cholesterol served as the cellular receptor for these toxins. The first suggestion of this cholesterol serving as the receptor occurred in the 1970s, when it was found that preincubation of the CDC of Streptococcus pyogenes, streptolysin O (SLO), with free cholesterol inhibited the hemolytic activity of this toxin (6). Later work with the CDC of Listeria monocytogenes, listeriolysin O (LLO), showed that preincubation with cholesterol inhibited hemolytic activity, cytolytic activity, and oligomerization of the toxin in cellular membranes, thereby preventing pore formation. Significantly, however, cholesterol did not interfere with membrane binding or with the induction of cytokine expression in macrophages treated with LLO (7–9). The idea that cholesterol was the cellular receptor for the CDCs was further questioned by the finding that the CDC of Streptococcus intermedius, intermedilysin (ILY), uses human CD59 as its membrane receptor (10). ILY does, however, require cholesterol for the insertion of the prepore complex into the membrane to form the pore. This requirement has also been shown for SLO (11). These findings are supported by the identification of several proteinaceous receptors for membrane lipid-dependent, pore-forming cytotoxins from Staphylococcus aureus (12). Taken together, these reports suggest that although membrane cholesterol is required for the CDCs to form a complete transmembrane pore, the cellular receptor that dictates cell tropism remains to be identified for some of the CDCs, including, most importantly, Ply. In this study, we investigated the glycan-binding properties of Ply to identify candidate cellular receptors for this toxin.
Results
Ply Binds to the Lewis Histo-Blood Group Antigens LewisX and Sialyl LewisX, and Domain 4 Is Required for This Activity.
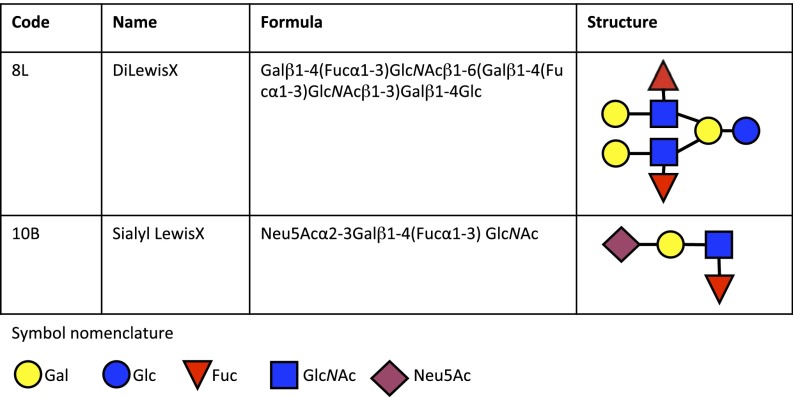
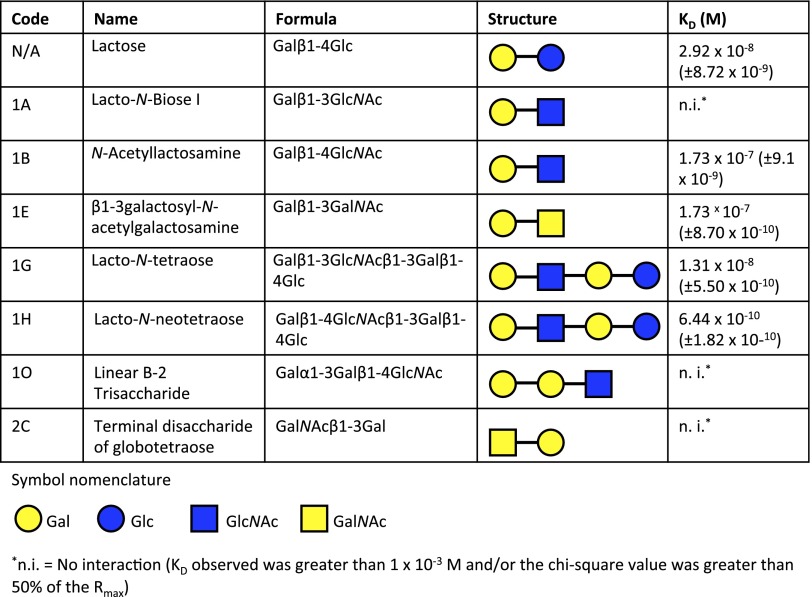
Although it has been shown that preincubation of Ply with cholesterol inhibits hemolytic activity against human RBCs (13, 14), whether cholesterol is the actual cellular receptor for Ply has not been determined. Using glycan array analysis, we investigated the glycan-binding specificities of Ply. Purified recombinant His6-tagged Ply from strain D39 was incubated with a glycan array of 120 distinct glycan structures (15) (Table S1) and revealed significant binding to the fucosylated glycan divalent-LewisX (LeX) [8L; Galβ1–4(Fucα1–3)GlcNAcβ1–6(Galβ1–4(Fucα1–3)GlcNAcβ1–3)Galβ1–4Glc] and the sialylated fucosylated glycan sialyl LewisX (sLeX) [10B; Neu5Acα2–3Galβ1–4(Fucα1–3)GlcNAc] (Fig. 1), as well as the high-molecular weight glycosaminoglycan 1.6-MDa hyaluronan (HA) [14I; (GlcAβ1–3GlcNAcβ1–4)n]. No binding of Ply to any of the smaller molecular weight HA structures was detected, including large polymers up to 222 kDa; therefore, the interaction between Ply and 1.6-MDa HA was considered a weak, polyvalent interaction and was not investigated further.
Fig. 1.
Selection of glycans bound by Ply in glycan array analysis. The code corresponds to the glycan code used in Table S1.
To validate the glycan array results and to characterize the interaction of Ply with sLeX and LeX, we performed surface plasmon resonance (SPR) analysis. The LewisB (LeB) glycan and lactose were included as nonbinding controls. Recombinant Ply was immobilized on the sensor chip, and free glycan was flowed over the immobilized protein. Ply bound to LeX, but with a higher affinity binding interaction, was observed with the sLeX glycan (Table 1).
Table 1.
Kd values of Ply and Ply mutants with glycans determined by SPR
| Glycan | Ply | Ply306 | PlyL460D | PlyW433F | PlyS | PlyQ374A | PlyY376A |
| LeX | 3.17 (±1.70) | 2.89 (±1.56) | n.i. | 8.25 (±0.444)* | 2.62 (±2.15) | 2.24 (±0.204) | 3.41 (±0.471) |
| sLeX | 1.88 (±0.423) | 35.0 (±56.3) | n.i. | 13.9 (±2.09)* | 4.30 (±0.543) | 13.7 (±2.03)* | 19.4 (±2.69)* |
Mean Kd values of at least three runs ± 1 SD are reported as 10−5 M. Lactose and LeB were included in SPR analysis. No interactions were observed with these glycans and any Ply sample. n.i., no interaction (Kd observed was greater than 1 × 10−3 M and/or the χ2 value was greater than 50% of the maximum response).
Significant difference in mean Kd value compared with WT Ply with the same glycan (P < 0.05).
The SPR analysis was conducted with a series of Ply mutants with amino acid substitutions and truncations to investigate the impact on glycan binding (Table 1). A number of clinical isolates of S. pneumoniae, such as the serotype 1 ST306 clone, express a variant of Ply (Ply306) that is unable to form pores (16–18). The Ply306 variant bound to LeX with a similar affinity to the WT Ply and to sLeX with a lower affinity compared with the WT Ply. The toxoid PlyL460D mutant is unable to bind to the cell surface or form pores and has no detectable hemolytic activity (19). Binding to LeX and sLeX was abolished in the PlyL460D mutant protein. The toxoid PlyW433F has <1% native hemolytic activity (20). The PlyW433F mutant protein had a significantly lower affinity for LeX and sLeX compared with the WT Ply.
We next investigated the ability of truncated versions of Ply to bind to LeX and sLeX (Table 1). These truncated proteins consisted of domains 1–3 of Ply (PlyL) and domains 1–3 of Ply306 (PlyL306), as well as domain 4 of Ply (PlyS) and domain 4 of PlyL460D (PlySL460D). PlyS and domain 4 of Ply306 share 100% amino acid sequence identity, and PlyL and domains 1–3 of PlyL460D share 100% amino acid sequence identity (21). It was found that PlyS bound to both LeX and sLeX, whereas the PlySL460D mutant, PlyL, and PlyL306 showed no binding to LeX or sLeX. These results suggest that binding of Ply to sLeX is mediated via domain 4 and that residue 460 is critical for this interaction.
sLeX Glycan Can Inhibit Ply Hemolytic Activity by Blocking Binding of the Toxin to the RBC Surface.
LeX and sLeX are members of the Lewis system histo-blood group antigens, which were originally identified on RBCs, where they are acquired from the plasma as glycolipids (22). A common method for assessing Ply activity is by determining the toxin’s ability to lyse RBCs in hemolytic assays. Therefore, we investigated whether the LeX and sLeX glycans could inhibit Ply hemolytic activity against human group O RBCs. The hemolytic activity of 53 ng/mL Ply (1 nM) was determined in the presence of 10 mM Lewis glycan sLeX, LeX, LeB, or Lewis Y (LeY) and compared with Ply with PBS only (Fig. 2A). The presence of the free sLeX glycan significantly decreased Ply hemolytic activity, whereas only minimal inhibition of activity was achieved with LeX. The inhibition observed with LeX was not statistically significant. The hemolytic activity of Ply was unaffected by LeB or LeY. When the hemolytic activity of a range of Ply concentrations in the presence of either the sLeX or LeX glycan was examined, only the free sLeX glycan significantly decreased Ply hemolytic activity (Fig. 2B). Similar inhibition experiments were performed with primary human polymorphonuclear leukocytes (PMNs) and human peripheral blood mononucleated cells (PBMCs) to determine if the sLeX glycan could inhibit Ply cytotoxicity against these immune cells. No inhibition of Ply cytotoxicity against either cell type was observed (Fig. S1 A–D). Inhibition of Ply cytotoxicity against the human alveolar basal epithelial cell line, A549 cells, with the free sLeX glycan was also attempted. We found that 20 mM sLeX could protect A549 cells against Ply cytotoxicity, whereas the presence of 20 mM lactose had no effect (Fig. S2).
Fig. 2.
Phenotypic analysis of Ply hemolytic activity and RBC binding. (A) Hemolytic activity of 53 ng/mL (1 nM) Ply against 1% (vol/vol) human group O RBCs at 37 °C for 30 min was determined after preincubation with 10 mM sLeX, LeX, LeB, or LeY. Results are presented as the mean of triplicate assays ± 1 SD. Significant differences are indicated: ****P < 0.0001. (B) Hemolytic activity of a range of Ply concentrations (conc; 424 ng/mL, 212 ng/mL, 106 ng/mL, and 53 ng/mL) against 1% (vol/vol) human group O RBCs at 37 °C for 30 min was determined after preincubation with PBS only (Ply), 10 mM sLeX, or 10 mM LeX. Results are presented as the mean of triplicate assays ± 1 SD. (C and D) Flow cytometric analysis of Ply binding to human RBCs. Human RBCs were incubated with 50 ng/mL Ply (Ply), 50 ng/mL Ply preincubated with 10 mM sLeX (Ply + sLeX), 50 ng/mL Ply preincubated with lactose (Ply + lac), or PBS only (no Ply) on ice for 30 min. Binding of Ply to the RBC surface was detected with anti-Ply polyclonal mouse serum and preconjugated rabbit anti-mouse Alexa Fluor 488 and goat anti-rabbit Alexa Fluor 488 antibodies. FL, fluorescence parameter; Max, maximum.
Ply mediates hemolysis of RBCs by binding to the cell membrane, where it oligomerizes to form the prepore complex, which then inserts into the membrane to form a transmembrane pore. To investigate whether the presence of sLeX was preventing Ply binding to the RBC membrane or was interfering with Ply oligomerization and subsequent pore formation, we treated human RBCs with 50 ng/mL Ply, a concentration of Ply that resulted in less than 100% lysis of RBCs, in the presence or absence of 10 mM sLeX glycan. The amount of Ply bound to the unlysed RBCs was analyzed by Western blotting using anti-Ply monoclonal antibody. No Ply was detected from the unlysed RBCs treated with Ply in the presence of sLeX at 37 °C, whereas very little was detected from unlysed RBCs treated with Ply in the presence of sLeX at 4 °C (Fig. S3). Having established that the presence of sLeX inhibits the binding of Ply to RBCs, we then used flow cytometry to assess the deposition of Ply onto the RBC surface. Incubation with 10 mM sLeX glycan inhibited the binding of Ply to the RBC surface (Fig. 2C), whereas the presence of a control carbohydrate, lactose, had no effect on the binding of Ply to the RBC surface (Fig. 2D). These results suggest that the interaction between Ply and sLeX blocks the binding of this toxin to the RBC membrane and that sLeX is a cellular receptor on human RBCs for this toxin.
Monoclonal Antibodies Specific for sLeX and LeX Can Block Ply Hemolytic Activity.
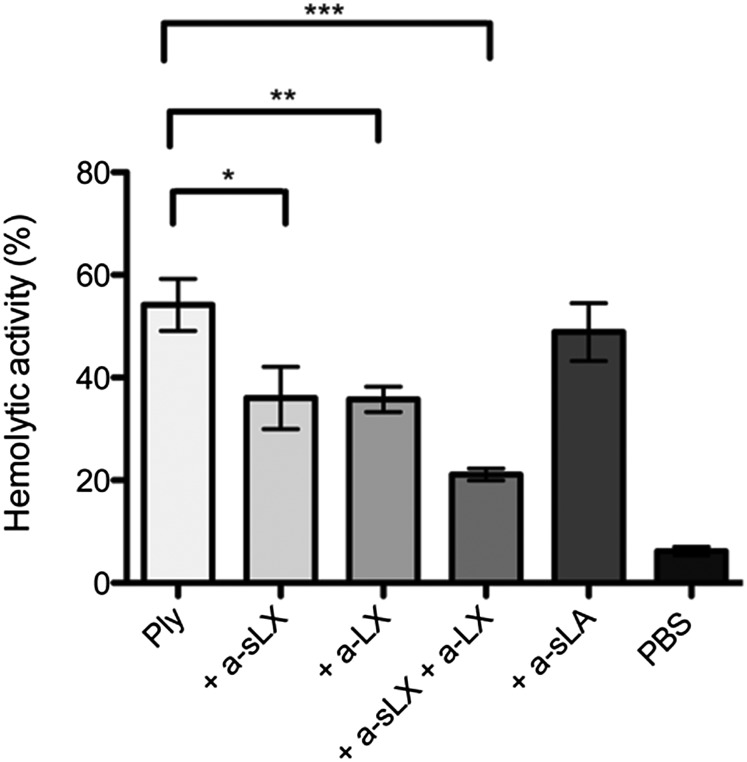
To confirm further that the LeX/sLeX glycans are receptors for Ply to mediate lysis of RBCs, RBCs were preincubated with anti-sLeX and anti-LeX monoclonal antibodies before the addition of a hemolytic dose for 50% lysis (HD50) of Ply. Each antibody significantly reduced Ply hemolytic activity, whereas a combination of both monoclonal antibodies caused an even greater reduction in Ply hemolytic activity (Fig. 3). Preincubation of RBCs with an equivalent concentration of anti-sialyl Lewis A monoclonal antibody that binds to a distinct structure had no effect on Ply hemolytic activity, indicating that specifically blocking access of Ply to the LeX glycans on the RBC surface inhibits hemolytic activity. These data further suggest that the LeX and sLeX glycans are cellular receptors for Ply on human RBCs.
Fig. 3.
Monoclonal antibodies against sLeX and LeX can inhibit Ply hemolytic activity against human group O RBCs. One percent (vol/vol) human group O RBCs was preincubated with 50 μg/mL monoclonal antibodies specific for sLeX (+ a-sLX), LeX (+ a-LX), sLeX and LeX (+ a-sLX + a-LX), or siayl Lewis A (+ a-sLA) before the addition of the HD50 of Ply (∼20 ng/mL) and was incubated at 16 °C for 60 min. Results are presented as the mean of triplicate assays ± 1 SD. Significant differences are indicated: *P < 0.05; **P < 0.005; ***P < 0.0005.
Protein/Carbohydrate-Binding Site Prediction in Domain 4 of Ply and Other CDCs.
Our collective data suggest that domain 4 of Ply binds LeX and sLeX glycans; however, the exact binding site is unknown. In silico analysis was used along with 3D models to predict the Ply LeX/sLeX-binding site. Currently, there are four structures of CDCs in the Protein Data Bank (PDB) database; perfringolysin (PFO; PDB ID code 1PFO), SLO (PDB ID code 4HSC), suilysin (SLY; PDB ID code 3HVN), and ILY (PDB ID code 1S3R). Four separate Ply models were built based on each known CDC structure using SWISS-MODEL (23, 24). Each model was then uploaded to the in silico molecular biology laboratory protein/carbohydrate-binding site prediction software (25). The whole Ply protein was used for the prediction to verify that carbohydrate-binding sites should only be predicted in domain 4 and not in domains 1–3.
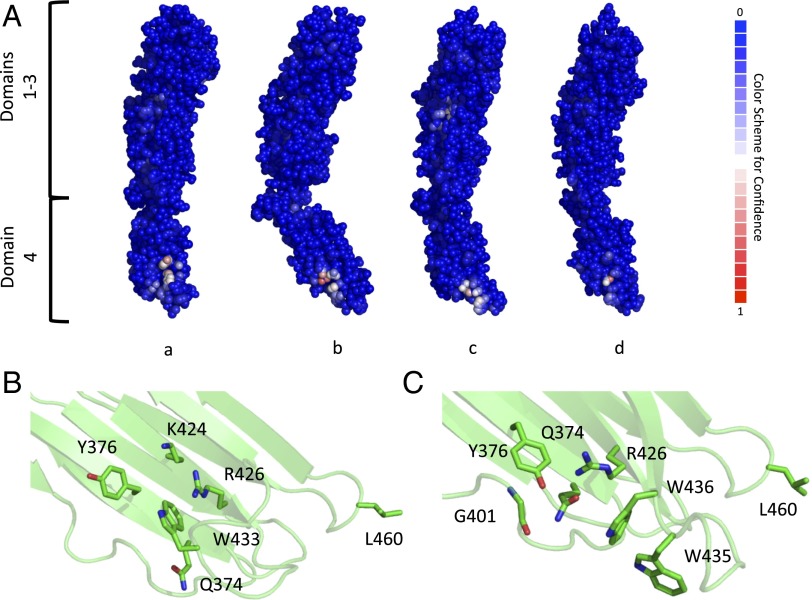
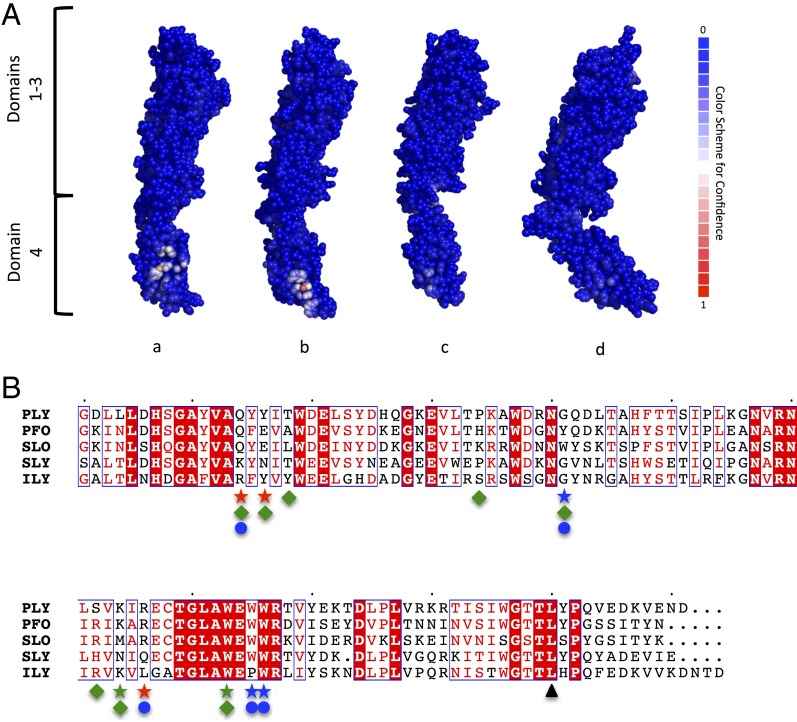
A carbohydrate-binding site located in domain 4 was predicted for all Ply models (Fig. 4A). Each putative binding site had a different patch score, or prediction confidence level, where a higher value represents a greater confidence level. The putative binding site for the Ply model based on the PFO structure had a patch score of 5.337, whereas for the Ply models based on ILY, SLO, and SLY, the patch scores were 3.2431, 1.7969, and 0.9176, respectively. When looking more closely at the predicted binding site between the four models, it appears to have two possible arrangements. The first is from the PFO-based model, where the main contributing residues would be Q374, Y376, K424, R426, and W433 (Fig. 4B). The other three Ply models follow the second binding site configuration that uses Q374, Y376, G401, R426, W435, and W436 as the central putative binding residues (Fig. 4C). It is noteworthy that both predicted carbohydrate-binding sites overlap an undecapeptide region (E427–R437 in Ply) that is strongly conserved among CDCs and is known to be important for hemolytic activity (26) and that Ply with a W433F mutation has markedly reduced hemolytic activity and sLeX-binding affinity (Table 1).
Fig. 4.
Protein/carbohydrate-binding site prediction for Ply. (A) Atom-based carbohydrate-binding site predictions are shown for each Ply model based on PFO (a), ILY (b), SLO (c), and SLY (d). Residues are shown as spheres with a color code based on the carbohydrate-binding confidence level, where blue has a low confidence and red has a high confidence. The potential glycan-binding sites for the Ply model based on PFO (B) and the Ply model based on ILY (C) are shown. Residues are shown in stick representation, with carbon atoms presented in green, nitrogen presented in blue, and oxygen presented in red.
This site in domain 4 was the only predicted carbohydrate-binding site for Ply modeled with PFO and ILY. However, there was a second predicted binding site located in domain 3 for Ply modeled from SLO and SLY with patch scores of 0.4422 and 0.6753, respectively. Due to the low patch scores and the fact that Ply domains 1–3 did not show any binding to the LeX glycans, this site is presumed to be a false-positive prediction and is not further discussed.
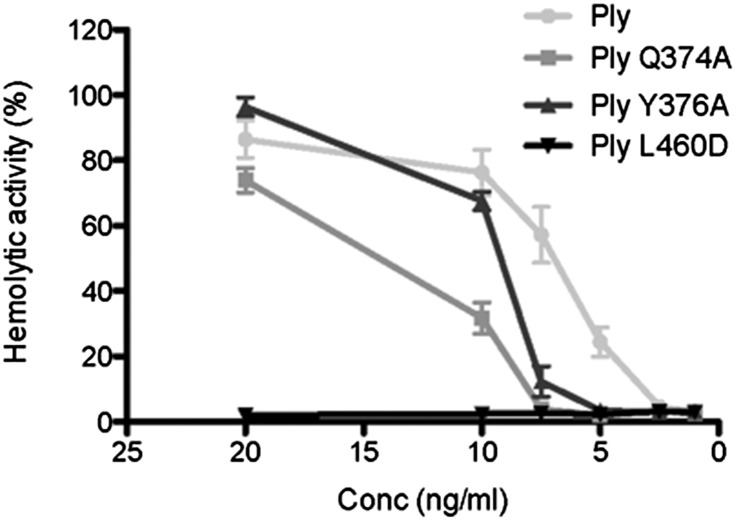
To investigate the validity of the predicted carbohydrate-binding sites further, site-directed mutagenesis was performed on two additional putative binding residues to give the mutant proteins PlyQ374A and PlyY376A. Like PlyW433F, both of these mutant proteins had reduced hemolytic activity against human RBCs relative to the WT protein (Fig. 5). Furthermore, SPR analysis of glycan binding showed that both had a significantly reduced affinity for sLeX compared with WT Ply, whereas the affinities of the mutant proteins for LeX were not significantly different from the affinity of the WT protein (Table 1).
Fig. 5.
Hemolytic activity of Ply mutant proteins with substitutions of predicted carbohydrate-binding residues is reduced compared with the WT Ply protein. The hemolytic activity of WT Ply and Ply mutant proteins at a range of concentrations (20 ng/mL, 10 ng/mL, 7.5 ng/mL, 5 ng/mL, 2.5 ng/mL, and 1 ng/mL) against 1% (vol/vol) human group O RBCs at 37 °C for 30 min was determined. Results are presented as the mean of triplicate assays ± 1 SD.
Our study is the first, to our knowledge, of a CDC domain 4 binding a carbohydrate ligand; thus, it is possible that other CDCs will also bind carbohydrates. Because the carbohydrate-binding site prediction software was able to predict a binding site in domain 4 of Ply, this same software was used to predict carbohydrate-binding sites for the other CDCs with known structures. In fact, the software predicted a carbohydrate-binding site for PFO and SLO in domain 4 (patch scores of 3.3631 and 3.1449, respectively) (Fig. 6A). The putative residues contributing to the binding site for PFO are Q405, E407, A409, H425, Y432, R453, K455, and W464, which align with the residues from the first predicted Ply binding site arrangement (Fig. 6B). In contrast, the possible SLO binding site residues are Q476, W503, W537, and W538, which mirror the residues from the second putative binding site configuration of Ply (Fig. 6B). Of note, the only binding site prediction for ILY is located in domain 3, with a patch score of 0.7493, whereas there were no binding sites predicted for SLY.
Fig. 6.
Protein/carbohydrate-binding site predictions for other CDCs. (A) Atom-based carbohydrate-binding site predictions for the four CDCs PFO (a; PDB ID code 1PFO), SLO (b; PDB ID code 4HSC), SLY (c; PDB ID code 3HVN), and ILY (d; PDB ID code1S3R). Residues are represented as spheres and color-coded by carbohydrate-binding confidence level, where blue is low confidence and red is high confidence. (B) Amino acid sequence alignment of domain 4 for CDCs. Amino acids highlighted in red indicate fully conserved residues, whereas residues boxed in blue show similar residues across the groups, with the red characters displaying the similarity within the group. Green, blue, and red stars indicate residues identified as potential glycan-interacting residues for the binding sites from the Ply model derived from PFO, ILY, or both models, respectively. Green diamonds and blue circles show putative binding site residues for PFO and SLY, respectively, whereas ▲ indicates L460.
SLO Also Binds to Glycans, and Glycan Binding Is Required for Hemolytic Activity and Deposition on the RBC Surface.
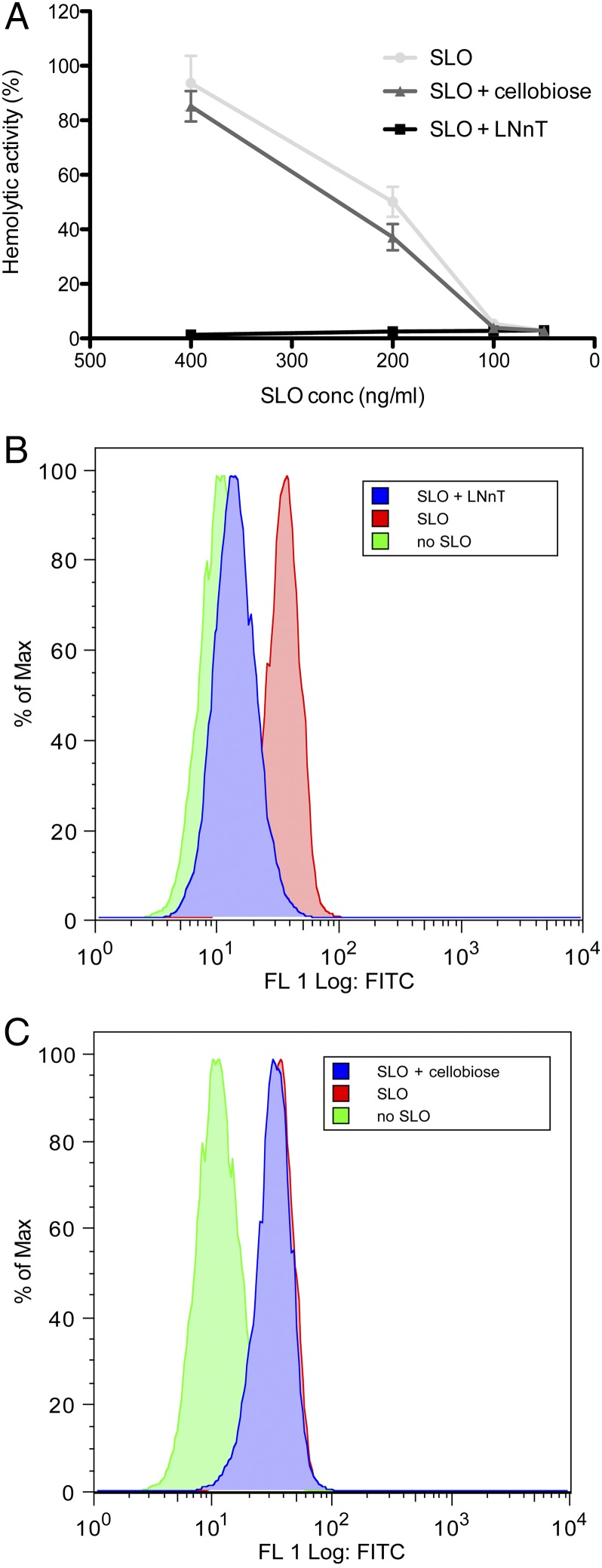
Because carbohydrate-binding sites were predicted in domain 4 of SLO, the glycan-binding profile of this toxin was investigated by glycan array analysis. SLO displayed significant binding to 47 glycan structures. These glycan structures included terminal Gal, terminal GlcNAc, terminal GalNAc, terminal Glc, fucosyl, and sialylated structures (Table S1). SPR analysis was used to characterize further and verify a selection of these glycan interactions. High-affinity interactions with Kds in the nanomolar range were detected with a number of these glycans (Fig. 7). To determine if the glycan-binding function of SLO contributed to its ability to lyse human RBCs, hemolysis assays were performed with SLO in the presence of the glycan with the highest affinity as determined by SPR analysis, lacto-N-neotetraose (LNnT). The presence of 2 mM free LNnT significantly reduced SLO hemolytic activity at a range of toxin concentrations, whereas the presence of the disaccharide d-cellobiose [Glcβ(1→4)Glc] did not significantly reduce SLO-mediated hemolytic activity at any of the concentrations tested (Fig. 8A). Furthermore, using flow cytometric analysis, it was found that free LNnT blocked SLO binding to the RBC surface (Fig. 8B), whereas the presence of d-cellobiose at the same concentration had no effect on the binding of SLO to the RBC surface (Fig. 8C).
Fig. 7.
Selection of glycans bound by SLO in glycan array analysis. The code corresponds to the glycan code used in Table S1. The mean Kd values of at least three runs ± 1 SD are reported.
Fig. 8.
Phenotypic analysis of SLO function showing that hemolytic activity against human RBCs can be inhibited with LNnT by blocking binding of the toxin to the RBC membrane. (A) Hemolytic activity of a range of SLO concentrations (400 ng/mL, 200 ng/mL, 100 ng/mL, and 50 ng/mL) against 1% (vol/vol) human group O RBCs at 37 °C for 30 min was determined after preincubation with PBS only (SLO), 2 mM LNnT, or 2 mM d-cellobiose. Results are presented as the mean of triplicate assays ± 1 SD. (B and C) Flow cytometric analysis of SLO binding to human RBCs. Human RBCs were incubated with 50 ng/mL SLO (SLO), 50 ng/mL SLO preincubated with 2 mM LNnT (SLO + LNnT), 50 ng/mL SLO preincubated with d-cellobiose (SLO + cellobiose), or PBS only (no SLO) on ice for 15 min. Binding of SLO to the RBC surface was detected with anti-SLO polyclonal mouse serum and preconjugated rabbit anti-mouse Alexa Fluor 488 and goat anti-rabbit Alexa Fluor 488 antibodies.
Discussion
Although it is known that the CDC of S. pneumoniae, Ply, requires membrane cholesterol for its cytolytic effects, it has not been definitively shown that cholesterol functions as the cellular receptor. We performed a comprehensive screen to analyze the glycan-binding specificity of purified WT Ply and found that this toxin binds to the Lewis histo-blood group antigens divalent LeX and sLeX. We have shown that Ply has a higher affinity for sLeX than LeX and that the sLeX glycan may function as a receptor for this toxin on human RBCs.
The noncytolytic Ply306 variant expressed by the serotype 1 ST306 clonal group bound to LeX with a comparable affinity to WT Ply and to sLeX with a lower affinity, which, when taken with previous reports, suggests that this variant’s sLeX- and LeX-binding affinities are sufficient to retain binding to the RBC surface (17). However, it is difficult to interpret this phenotype in terms of distinct domain functions, because this mutant Ply contains multiple amino acid changes throughout the protein. In contrast, the single amino acid change toxoid PlyL460D mutant was unable to bind to either sLeX or LeX, which is consistent with its inability to bind to the cell surface (19). Furthermore, we discovered that domain 4 is responsible for binding to sLeX and LeX, and, in particular, that mutation of the L460 residue for an Asp is sufficient to abrogate binding completely, supporting previous studies that have highlighted the importance of the L1 loop in the initial engagement of CDCs with the cell membrane (19).
The Lewis system antigens are structurally related to the determinants of the ABO blood group system. These carbohydrate structures consist of two major antigenic cores: type 1 (Galβ1–3GlcNAc) and type 2 (Galβ1–4GlcNAc). Fucosylation of the type 1 core produces Lewis A (LeA) and LeB, whereas fucosylation of type 2 produces Lewis X (LeX) and LeY (27). The Lewis antigens were originally identified on human erythrocytes, in plasma, and in mucous secretions, where they were assumed to be glycoproteins. It was later shown that RBCs passively acquire Lewis antigens from plasma, predominantly as glycosphingolipids, and incorporate them into the erythrocyte membrane (22). The type 1 Lewis antigens, LeA and LeB, are important histo-blood group antigens, whereas the type 2 Lewis antigens, LeX and LeY, are expressed at relatively low levels in normal tissues but are found to be overexpressed on the surface of human tumor cells from various sites (27).
The sialylated derivative of LeX, sLeX, is an important carbohydrate moiety for humans. The sLeX antigen has been detected on the surface of multiple cell types, including granulocytes (particularly neutrophils), monocytes (28, 29), platelets (30), natural killer cells (31), activated lymphocytes (32), and helper memory T cells (33), where it may be present on glycoproteins or glycolipids. The sLeX antigen has a critical role in the inflammation process (34). Expression of sLeX is up-regulated during inflammation on the surface of leukocytes, where it serves as an essential component of the ligands for the three major types of selectins, P-, L-, and E-selectins. These selectins initiate the “tethering and rolling” of leukocytes to vascular endothelial cells or platelets during inflammation (35, 36). P-selectin is expressed by activated platelets and endothelial cells, and it binds to P-selectin glycoprotein ligand-1 (PSGL-1), which contains an O-linked sLeX glycan on a Thr residue near a sulfated Tyr trio, and is expressed on myeloid cells and subsets of lymphocytes (37). L-selectin is found on most leukocytes and binds to glycoproteins carrying an O-linked sLeX with a sulfate on the 6-hydroxyl of the GlcNAc residue, such as glycosylation-dependent cell adhesion molecule 1 (38). E-selectin is found on activated endothelium and appears to bind sLeX on sialylated glycosphingolipids on human neutrophils (39). It has also been reported that Mac1 (CD11b/CD18) of human neutrophils, a major membrane protein decorated with sLeX, binds to E-selectin and that the sLeX moieties are critical for this interaction (40). Attempts were made to inhibit Ply cytotoxicity against human PMNs and PBMCs with sLeX, but no inhibition was observed. The failure of sLeX to inhibit Ply cytotoxicity against these cells may be due to the overload of sLeX-containing glycoproteins and glycolipids on the surface of human PMNs, particularly neutrophils (39, 40), or to the presence of alternative receptors for Ply on human leukocytes. However, sLeX did inhibit cytotoxicity of Ply for A549 (human alveolar) cells, suggesting that this glycan present on glycolipids and/or glycoproteins may serve as a receptor for Ply on cell types other than RBCs. Susceptibility of lung cells to Ply is consistent with the known importance of the toxin in the pathogenesis of pneumonia (1).
In silico modeling and carbohydrate-binding site predictions show a possible glycan-binding site within the Ply domain 4, consistent with array and SPR data. Whereas the putative binding sites for all four of the constructed Ply models were located in a similar position on domain 4, with respect to the entire protein, there appears to be two possible binding site configurations dependent on which CDC structure was used to model the Ply 3D structure (Fig. 4 A and B). Site-directed mutagenesis of two of the predicted carbohydrate-binding site residues resulted in a decrease in the affinity of these mutant proteins for sLeX and a decrease in hemolytic activity against human RBCs compared with the WT Ply protein, suggesting that these residues are involved in glycan binding and that glycan binding is required for native levels of hemolytic activity against human RBCs. The SPR data revealed that an L460D mutation in Ply domain 4 abolished sLeX and LeX binding, suggesting that this residue may also be involved in glycan binding. Surprisingly, this residue does not appear to be in either of the predicted carbohydrate-binding site conformations (Fig. 4 A and B). Because the L460D mutation is not a conservative mutation, it is unclear if the alteration causes a conformational change that could affect the predicted binding sites. Nevertheless, our SPR data also show that the W433F mutation in Ply significantly reduces binding to both LeX and sLeX. This conservative mutation is known to result in >99% loss of Ply hemolytic activity, and the W433F toxoid has been used extensively as an experimental vaccine antigen (20). It is interesting that the PlyQ374A and PlyY376A mutant proteins are still able to bind LeX and retain greater hemolytic activity than the PlyW433F toxoid, suggesting that LeX can still serve as a receptor for Ply to mediate hemolytic activity on human RBCs. W433 is predicted to be an important carbohydrate-binding residue in the PFO-based model, but not in the alternative configuration predicted for the other Ply models. Thus, although further mutational analyses are required to pinpoint the exact sLeX-binding site, our data suggest that the site predicted by the PFO-based model is the more accurate reflection of the true situation in native Ply. Previous studies have proposed that the conserved undecapeptide region of Ply, and W433 in particular, is critically important for the toxin’s capacity to bind membrane cholesterol (41). However, our data now suggest that this region may be more important for the glycan interactions that precede cholesterol-dependent toxin oligomerization and membrane insertion.
Putative carbohydrate-binding sites were also identified within domain 4 of two other CDCs, SLO and PFO, but not for ILY and SLY. Glycan array analysis of SLO found that this toxin binds to a range of glycan structures and that the binding of SLO to the RBC membrane can be inhibited with the glycan LNnT. The ability of LNnT to inhibit SLO binding to the RBC membrane suggests that the interaction of SLO with a glycan is an essential initial step preceding pore formation in the RBC membrane and that a glycan may also serve as a receptor target for this toxin. LNnT is found on human erythrocytes as the glycosphingolipid paragloboside, also known as lacto-N-neotetraosyl ceramide. Paragloboside is an intermediate in the biosynthesis of the erythrocyte blood group ABH and P1 glycosphingolipid antigens, and it has also been isolated from human PMNs (42). This glycolipid may be a receptor for SLO on human RBCs. A recent study showed that membrane binding of SLO to A549 cells does not require cholesterol, indicating that cholesterol is not the membrane receptor for this CDC on this particular cell type. A mutant version of SLO, which had particular residues of domain 4 swapped with the homologous residues of PFO, had decreased binding to A549 cell membranes relative to the WT, which was cholesterol-insensitive, and lost all hemolytic activity (43). This swap region contains one of the predicted carbohydrate-binding residues from our in silico analysis and is consistent with our findings that glycan binding by SLO is the initial first step to allow pore formation of this toxin. Furthermore, in support of our findings, it has recently been shown that a specific Gal-binding lectin inhibits the hemolytic effect of SLO (44).
In summary, our data show that the hemolytic activity of the archetypal CDCs, Ply and SLO, require binding to cellular glycan receptors for efficient deposition into the RBC membrane. The examples provided in this study are the first, to our knowledge, of a CDC targeting a glycolipid receptor, because all others have targeted protein or glycoprotein receptors. This study, along with recent studies on other streptococcal (10, 45) and staphylococcal cytolysins (12), supports the emerging paradigm that receptors other than membrane lipid composition define the cellular tropism of pore-forming toxins, including CDCs.
Materials and Methods
Cloning, Expression, and Purification of Full-Length Ply Variants.
Primers were designed to amplify full-length ply from the cytolytic strain D39 (Ply) and the noncytolytic serotype 1 ST306 clinical isolate A0229467 (Ply306), which were cloned into the pQE vector (Qiagen). The PlyL460D mutant was constructed as previously described (19) and cloned into the pQE vector. The PlyQ346A and PlyY376A mutants were constructed as previously described by means of overlap extension PCR as previously described (46) using the primers RHPlyQ374AF (GTT GCC GCG TAT TAT ATT ACT TGG GAT), RHPlyQ374AR (CAT CCC AAG TAA TAT AAT ACG CGG CAA C), RHPlyY376AF (GTT BGCC CAA TAT GCG ATT ACT), RHPlyY376AR (CAT CCC AAG TAA TCG CAT ATT GGG CAA C), RHPlyF(5) (GGT GGT GCT TAT GCT TTG TCG), and RHPlyR(5) (GTG GGC AAT GAC AAA GGA TGT G), and they were also cloned into the pQE vector. The resultant His6-tagged constructs were used to transform Escherichia coli BL21 (DE3) lpxM-. Bacterial cultures were pelleted and disrupted at 20 kpsi by a Constant Cell disruptor. Recombinant protein was purified by Ni2+ immobilized metal affinity chromatography (GE Healthcare). Protein concentrations were determined by measuring the A280 with the calculated molar extinction coefficient, 0.07231 cm−1⋅μM−1 (47).
Cloning, Expression, and Purification of Ply Mutants and Domains.
PlyL (domains 1–3) and PlyS (domain 4) cloning and recombinant protein production procedures have been described previously (13). Identical primers were used for PlyL306 and PlySL460D utilizing the appropriate template DNA described above. Bacterial cultures were pelleted and disrupted at 20 kpsi by a Constant Cell disruptor. Recombinant protein was purified by Ni2+ immobilized metal affinity chromatography, followed by buffer exchange into 20 mM Tris (pH 8.0) and 50 mM NaCl. PlyL and PlyL306 were further purified by gel filtration chromatography using a Sephacryl S-200 column (GE Healthcare) equilibrated in 20 mM Tris (pH 8.0) and 50 mM NaCl. Protein concentrations were determined as above, with calculated molar extinction coefficients, 0.03037 and 0.04194 cm−1⋅μM−1 for PlyL and PlyS, respectively.
Cloning, Expression, and Purification of SLO.
Recombinant SLO was expressed and purified from a pET15b construct expressing full-length SLO without the signal sequence (48) transformed into E. coli BL21 Star (Life Technologies). Recombinant protein expression was induced at an OD600 of 0.6 with 0.5 mM isopropyl 1-thio-d-galactopyranoside and maintained at 28 °C for 4 h. Cultures were pelleted, resuspended in a buffer of 300 mM NaCl (pH 8.0) and 100 mM Tris supplemented with 5 mM imidazole, 50% (vol/vol) glycerol, 1 mg/mL lysozyme, 100 mg/mL DNase, and the protease inhibitors 2 mM PMSF and Complete Protease Inhibitor Mixture (Roche), before cell disruption by sonication. Soluble His6-tagged SLO was purified by Ni2+ immobilized metal affinity chromatography and purified protein stored in a low-salt buffer of 150 mM NaCl and 50 mM Tris. Protein concentrations were determined by measuring the A280 using the molar extinction coefficient 0.06429 cm−1⋅μM−1 calculated using the ProtParam platform at web.expasy.org/protparam.
Glycan Array.
Glycan array slides were printed on SuperEpoxy 2(ArrayIt)-activated substrates as previously described (15) using the list of glycans described in Table S1 and methods as described in SI Materials and Methods.
SPR Analysis.
The interactions between the Ply proteins or domains thereof or SLO and test glycans were analyzed using SPR as described in SI Materials and Methods.
Hemolysis Assays.
Individual batches of human group O RBCs were washed three times with PBS and were used in each hemolytic assay at a final concentration of 1% (vol/vol). The total volume of each assay was 100 μL. Purified WT Ply, Ply mutants, or SLO prereduced with 1 mM DTT was diluted in PBS to the appropriate concentration. Assays were incubated at 37 °C for 30 min and were then centrifuged at 1,000 × g for 5 min at 4 °C to remove unlysed RBCs. Fifty microliters of the supernatants containing released Hb was transferred to the wells of a 96-well plate, and the A405 was measured. The 100% lysis assays consisted of 1% RBCs (vol/vol) with 0.5% saponin. Hemolytic activity (%) was calculated as follows:
Results are presented as the mean of triplicate assays ± 1 SD. The activity of toxin against each individual batch of RBCs was determined before being used in hemolytic assays.
Inhibition of Ply/SLO Hemolytic Activity with Free Glycan.
Ply or SLO at the appropriate concentration was preincubated with free glycan in PBS for 15 min at room temperature before the addition of 1% (vol/vol) washed human group O RBCs. Assays were incubated for 37 °C for 30 min. Hemolytic activity was determined as described above. Assays were also performed with each concentration of glycan with 1% (vol/vol) RBCs without toxin. Incubation of RBCs with all of the glycans tested had no effect on cell lysis.
Analysis of RBC Binding by Western Blotting.
Binding of Ply to the RBC membrane was analyzed by Western blotting as described in SI Materials and Methods.
Analysis of RBC Surface Binding by Flow Cytometry.
For analysis of Ply binding, 1% (vol/vol) washed human RBCs in a final volume of 50 μL of ice-cold PBS were incubated on ice for 30 min with 50 ng/mL Ply, 50 ng/mL Ply preincubated for 15 min at room temperature with 10 mM sLeX or 10 mM lactose, PBS only (no Ply), 10 mM sLeX, or 10 mM lactose. For analysis of SLO binding, 1% (vol/vol) washed human RBCs in a final volume of 50 μL of ice-cold PBS were incubated on ice for 30 min with 50 ng/mL SLO, 50 ng/mL SLO preincubated for 15 min at room temperature with 2 mM LNnT or 2 mM d-cellobiose, PBS only (no SLO), 2 mM LNnT, or 2 mM d-cellobiose. RBCs were harvested at 1,000 × g and then washed twice with 500 μL of ice-cold PBS containing 2.5% (wt/vol) BSA. RBCs were incubated on ice for 45 min with either anti-Ply polyclonal mouse serum (1:300, generated in this study by immunizing mice with PlyL460D) or anti-SLO polyclonal mouse serum (1:100) diluted in ice-cold PBS containing 2.5% (wt/vol) BSA. RBCs were pelleted and washed twice with 500 μL of ice-cold PBS containing 2.5% (wt/vol) BSA. RBCs were incubated on ice for 45 min with preconjugated rabbit anti-mouse Alexa Fluor 488 and goat anti-rabbit Alexa Fluor 488 antibodies (1:600 and 1:1,200, respectively; Life Technologies) diluted in ice-cold PBS containing 2.5% BSA. RBCs were pelleted and washed twice with 500 μL of ice-cold PBS containing 2.5% BSA and were then fixed for 15 min on ice using 0.25% glutaraldehyde in PBS. RBCs were pelleted and washed twice with 500 μL of ice-cold PBS containing 2.5% BSA and then resuspended in 1 mL of ice-cold PBS containing 2.5% BSA and analyzed using a CyAn ADP Analyzer (Beckman Coulter) utilizing the 488-nm laser for excitation and the fluorescence parameter 1 (FL1)/FITC emission filter and FlowJo 8.7 software (TreeStar, Inc.). A total of 104 gated events were collected per sample. Incubation of RBCs with 10 mM sLeX, 10 mM lactose, 2 mM LNnT, or 2 mM d-cellobiose showed no difference in fluorescence compared with RBCs with PBS only.
Analysis of Ply Cytotoxicity Against PMNs and PBMCs.
Cytotoxicity of Ply against human PMNs and PBMCs was determined as described in SI Materials and Methods.
Analysis of Ply Cytotoxicity Against A549 Cells.
Cytotoxicity of Ply against the human alveolar epithelial cell line, A549 cells, was determined as described in SI Materials and Methods.
Inhibition of Ply Hemolytic Activity with Anti-sLeX and Anti-LeX Monoclonal Antibodies.
Before performing monoclonal antibody inhibition assays, the HD50 for Ply against washed 1% (vol/vol) human group O RBCs at 16 °C for 60 min was determined. Washed 1% (vol/vol) RBCs were preincubated with 50 μg/mL monoclonal antibodies to sLeX (clones 258–12,767) and LeX (clone AHN1.1) diluted in PBS for 30 min at room temperature before the addition of 20 ng/mL Ply, which was determined to be approximately the HD50 of Ply against that particular batch of RBCs. RBCs were also preincubated with 50 μg/mL monoclonal antibody to sialyl Lewis A (121SLE) and PBS only to serve as negative controls. Assays were incubated at 16 °C for 60 min. Hemolytic activity was determined as described. Whole-cell ELISA was performed against RBCs using each monoclonal antibody to determine the titer. No significant differences between the titers of each of the monoclonal antibodies against the RBCs were observed, suggesting that the monoclonal antibodies bound to the RBCs with equal affinities. The monoclonal antibodies used in the inhibition assay did not cause agglutination of the RBCs at the concentration used.
Statistical Analysis.
Data were analyzed for statistical significance with Prism 5 software (GraphPad Software, Inc.) using a one-tailed unpaired Student t test. P values < 0.05 were considered significant.
Supplementary Material
Acknowledgments
This work was supported by Program Grant 565526 from the National Health and Medical Research Council of Australia (NHMRC) (to M.J.W., A.W.P., J.C.P., and M.P.J.). M.A.H. was supported by a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship. C.J.D and L.E.H.-T. were supported by a Smart Futures Fund Research Partnerships Program Grant. J.C.P. is an NHMRC Senior Principal Research Fellow, A.W.P. is an Australian Research Council Discovery Outstanding Researcher Award Fellow, and M.J.W. is an NHMRC Principal Research Fellow. D.B.A.J. was supported in part by Public Health Service Institutional Research Training Award T32AI007180. V.J.T. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412703111/-/DCSupplemental.
References
- 1.Paton JC, Andrew PW, Boulnois GJ, Mitchell TJ. Molecular analysis of the pathogenicity of Streptococcus pneumoniae: The role of pneumococcal proteins. Annu Rev Microbiol. 1993;47:89–115. doi: 10.1146/annurev.mi.47.100193.000513. [DOI] [PubMed] [Google Scholar]
- 2.Ramachandran R, Tweten RK, Johnson AE. Membrane-dependent conformational changes initiate cholesterol-dependent cytolysin oligomerization and intersubunit beta-strand alignment. Nat Struct Mol Biol. 2004;11(8):697–705. doi: 10.1038/nsmb793. [DOI] [PubMed] [Google Scholar]
- 3.Shepard LA, et al. Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: An alpha-helical to beta-sheet transition identified by fluorescence spectroscopy. Biochemistry. 1998;37(41):14563–14574. doi: 10.1021/bi981452f. [DOI] [PubMed] [Google Scholar]
- 4.Dunstone MA, Tweten RK. Packing a punch: The mechanism of pore formation by cholesterol dependent cytolysins and membrane attack complex/perforin-like proteins. Curr Opin Struct Biol. 2012;22(3):342–349. doi: 10.1016/j.sbi.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shatursky O, et al. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: A novel paradigm for pore-forming toxins. Cell. 1999;99(3):293–299. doi: 10.1016/s0092-8674(00)81660-8. [DOI] [PubMed] [Google Scholar]
- 6.Watson KC, Rose TP, Kerr EJ. Some factors influencing the effect of cholesterol on streptolysin O activity. J Clin Pathol. 1972;25(10):885–891. doi: 10.1136/jcp.25.10.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishibori T, Xiong H, Kawamura I, Arakawa M, Mitsuyama M. Induction of cytokine gene expression by listeriolysin O and roles of macrophages and NK cells. Infect Immun. 1996;64(8):3188–3195. doi: 10.1128/iai.64.8.3188-3195.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshikawa H, et al. Membrane damage and interleukin-1 production in murine macrophages exposed to listeriolysin O. Infect Immun. 1993;61(4):1334–1339. doi: 10.1128/iai.61.4.1334-1339.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs T, et al. Listeriolysin O: Cholesterol inhibits cytolysis but not binding to cellular membranes. Mol Microbiol. 1998;28(6):1081–1089. doi: 10.1046/j.1365-2958.1998.00858.x. [DOI] [PubMed] [Google Scholar]
- 10.Giddings KS, Zhao J, Sims PJ, Tweten RK. Human CD59 is a receptor for the cholesterol-dependent cytolysin intermedilysin. Nat Struct Mol Biol. 2004;11(12):1173–1178. doi: 10.1038/nsmb862. [DOI] [PubMed] [Google Scholar]
- 11.Giddings KS, Johnson AE, Tweten RK. Redefining cholesterol’s role in the mechanism of the cholesterol-dependent cytolysins. Proc Natl Acad Sci USA. 2003;100(20):11315–11320. doi: 10.1073/pnas.2033520100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DuMont AL, Torres VJ. Cell targeting by the Staphylococcus aureus pore-forming toxins: It’s not just about lipids. Trends Microbiol. 2014;22(1):21–27. doi: 10.1016/j.tim.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim JE, Park SA, Bong SM, Chi YM, Lee KS. Characterization of pneumolysin from Streptococcus pneumoniae, interacting with carbohydrate moiety and cholesterol as a component of cell membrane. Biochem Biophys Res Commun. 2013;430(2):659–663. doi: 10.1016/j.bbrc.2012.11.095. [DOI] [PubMed] [Google Scholar]
- 14.Nöllmann M, Gilbert R, Mitchell T, Sferrazza M, Byron O. The role of cholesterol in the activity of pneumolysin, a bacterial protein toxin. Biophys J. 2004;86(5):3141–3151. doi: 10.1016/S0006-3495(04)74362-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Day CJ, et al. Differential carbohydrate recognition by Campylobacter jejuni strain 11168: Influences of temperature and growth conditions. PLoS ONE. 2009;4(3):e4927. doi: 10.1371/journal.pone.0004927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jefferies JM, et al. Presence of nonhemolytic pneumolysin in serotypes of Streptococcus pneumoniae associated with disease outbreaks. J Infect Dis. 2007;196(6):936–944. doi: 10.1086/520091. [DOI] [PubMed] [Google Scholar]
- 17.Kirkham LA, et al. Identification of invasive serotype 1 pneumococcal isolates that express nonhemolytic pneumolysin. J Clin Microbiol. 2006;44(1):151–159. doi: 10.1128/JCM.44.1.151-159.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lock RA, Zhang QY, Berry AM, Paton JC. Sequence variation in the Streptococcus pneumoniae pneumolysin gene affecting haemolytic activity and electrophoretic mobility of the toxin. Microb Pathog. 1996;21(2):71–83. doi: 10.1006/mpat.1996.0044. [DOI] [PubMed] [Google Scholar]
- 19.Farrand AJ, LaChapelle S, Hotze EM, Johnson AE, Tweten RK. Only two amino acids are essential for cytolytic toxin recognition of cholesterol at the membrane surface. Proc Natl Acad Sci USA. 2010;107(9):4341–4346. doi: 10.1073/pnas.0911581107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paton JC, et al. Purification and immunogenicity of genetically obtained pneumolysin toxoids and their conjugation to Streptococcus pneumoniae type 19F polysaccharide. Infect Immun. 1991;59(7):2297–2304. doi: 10.1128/iai.59.7.2297-2304.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvey RM, Ogunniyi AD, Chen AY, Paton JC. Pneumolysin with low hemolytic activity confers an early growth advantage to Streptococcus pneumoniae in the blood. Infect Immun. 2011;79(10):4122–4130. doi: 10.1128/IAI.05418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcus DM, Cass LE. Glycosphingolipids with Lewis blood group activity: Uptake by human erythrocytes. Science. 1969;164(3879):553–555. doi: 10.1126/science.164.3879.553. [DOI] [PubMed] [Google Scholar]
- 23.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 24.Bordoli L, et al. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4(1):1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 25.Tsai KC, et al. Prediction of carbohydrate binding sites on protein surfaces with 3-dimensional probability density distributions of interacting atoms. PLoS ONE. 2012;7(7):e40846. doi: 10.1371/journal.pone.0040846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boulnois GJ, Paton JC, Mitchell TJ, Andrew PW. Structure and function of pneumolysin, the multifunctional, thiol-activated toxin of Streptococcus pneumoniae. Mol Microbiol. 1991;5(11):2611–2616. doi: 10.1111/j.1365-2958.1991.tb01969.x. [DOI] [PubMed] [Google Scholar]
- 27.Yuriev E, Farrugia W, Scott AM, Ramsland PA. Three-dimensional structures of carbohydrate determinants of Lewis system antigens: Implications for effective antibody targeting of cancer. Immunol Cell Biol. 2005;83(6):709–717. doi: 10.1111/j.1440-1711.2005.01374.x. [DOI] [PubMed] [Google Scholar]
- 28.Fukuda M, Spooncer E, Oates JE, Dell A, Klock JC. Structure of sialylated fucosyl lactosaminoglycan isolated from human granulocytes. J Biol Chem. 1984;259(17):10925–10935. [PubMed] [Google Scholar]
- 29.Spooncer E, Fukuda M, Klock JC, Oates JE, Dell A. Isolation and characterization of polyfucosylated lactosaminoglycan from human granulocytes. J Biol Chem. 1984;259(8):4792–4801. [PubMed] [Google Scholar]
- 30.Cooling LL, Zhang DS, Walker KE, Koerner TA. Detection in human blood platelets of sialyl Lewis X gangliosides, potential ligands for CD62 and other selectins. Glycobiology. 1995;5(6):571–581. doi: 10.1093/glycob/5.6.571. [DOI] [PubMed] [Google Scholar]
- 31.Ohmori K, et al. Sialyl SSEA-1 antigen as a carbohydrate marker of human natural killer cells and immature lymphoid cells. Blood. 1989;74(1):255–261. [PubMed] [Google Scholar]
- 32.Stoolman LM. Adhesion molecules controlling lymphocyte migration. Cell. 1989;56(6):907–910. doi: 10.1016/0092-8674(89)90620-x. [DOI] [PubMed] [Google Scholar]
- 33.Ohmori K, et al. A distinct type of sialyl Lewis X antigen defined by a novel monoclonal antibody is selectively expressed on helper memory T cells. Blood. 1993;82(9):2797–2805. [PubMed] [Google Scholar]
- 34.McEver RP. Selectin-carbohydrate interactions during inflammation and metastasis. Glycoconj J. 1997;14(5):585–591. doi: 10.1023/a:1018584425879. [DOI] [PubMed] [Google Scholar]
- 35.Lasky LA. Selectins: Interpreters of cell-specific carbohydrate information during inflammation. Science. 1992;258(5084):964–969. doi: 10.1126/science.1439808. [DOI] [PubMed] [Google Scholar]
- 36.Varki A. Selectin ligands. Proc Natl Acad Sci USA. 1994;91(16):7390–7397. doi: 10.1073/pnas.91.16.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leppänen A, White SP, Helin J, McEver RP, Cummings RD. Binding of glycosulfopeptides to P-selectin requires stereospecific contributions of individual tyrosine sulfate and sugar residues. J Biol Chem. 2000;275(50):39569–39578. doi: 10.1074/jbc.M005005200. [DOI] [PubMed] [Google Scholar]
- 38.Rosen SD. Ligands for L-selectin: Homing, inflammation, and beyond. Annu Rev Immunol. 2004;22:129–156. doi: 10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- 39.Nimrichter L, et al. E-selectin receptors on human leukocytes. Blood. 2008;112(9):3744–3752. doi: 10.1182/blood-2008-04-149641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zen K, Cui LB, Zhang CY, Liu Y. Critical role of mac-1 sialyl lewis x moieties in regulating neutrophil degranulation and transmigration. J Mol Biol. 2007;374(1):54–63. doi: 10.1016/j.jmb.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 41.Rossjohn J, et al. The molecular mechanism of pneumolysin, a virulence factor from Streptococcus pneumoniae. J Mol Biol. 1998;284(2):449–461. doi: 10.1006/jmbi.1998.2167. [DOI] [PubMed] [Google Scholar]
- 42.Schwarting GA, Marcus DM. The reactions of antibodies to paragloboside (lacto-N-neotetraosyl ceramide) with human erythrocytes and lymphocytes. J Immunol. 1977;118(4):1415–1419. [PubMed] [Google Scholar]
- 43.Mozola CC, Magassa N, Caparon MG. A novel cholesterol-insensitive mode of membrane binding promotes cytolysin-mediated translocation by Streptolysin O. Mol Microbiol. 2014;94(3):675–687. doi: 10.1111/mmi.12786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hasan I, et al. A galactose-binding lectin isolated from Aplysia kurodai (sea hare) eggs inhibits streptolysin-induced hemolysis. Molecules. 2014;19(9):13990–14003. doi: 10.3390/molecules190913990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feil SC, et al. Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity. Structure. 2012;20(2):248–258. doi: 10.1016/j.str.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harvey RM, et al. The impact of pneumolysin on the macrophage response to Streptococcus pneumoniae is strain-dependent. PLoS ONE. 2014;9(8):e103625. doi: 10.1371/journal.pone.0103625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gasteiger E, et al. The Proteomics Protocols Handbook. Humana Press; Totowa, NJ: 2005. [Google Scholar]
- 48.Timmer AM, et al. Streptolysin O promotes group A Streptococcus immune evasion by accelerated macrophage apoptosis. J Biol Chem. 2009;284(2):862–871. doi: 10.1074/jbc.M804632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.