Abstract
This paper focuses on the problem of partially matched samples in the presence of confounders. We propose using propensity score matching to adjust for confounding factors for the subset of data with incomplete pairs, followed by integrating the P-values computed from the complete and incomplete paired samples, respectively. Several simulations and a case study on DNA methylation are considered to evaluate the operating characteristics of the proposed method.
Keywords: microarray, confounders, observational studies, full matching, regression
Introduction
The advancement in biotechnologies has revolutionized numerous disciplines including biology and medicine. Several high-throughput platforms including whole genome arrays and the next-generation sequencing instruments are available for profiling large-scale omics data. These cutting edge biotechnologies have spurred rapid biomarker discovery and personalized medicine approach in multiple diseases, in particular, cancer research. In recent years, genomewide profiling utilizing these technologies has been carried out to identify biomarkers associated with cancer development and progression. In this paper, we consider the matched pairs samples for identifying differentially expressed biomarkers between two groups. Matched/paired study design is commonly used in omics/biomarkers profiling because it automatically accounts for confounding factors. Examples of matched/paired designs include profiling n (1) tumor and adjacent normal lesions, (2) pre- and post-drug treatment samples, or (3) one-to-one matching of patients by demographic covariates (eg, age, gender, race, etc.) from the two groups of interest. We will use the tumor versus normal samples hereafter for expository purpose.
Ideally, one expects a total of 2n samples from such matched/paired design. However, in practice, circumstances such as RNA degradation, array failure, or insufficient resources could result in a subset of patients missing in either the tumor or matched normal biomarker profiles. For example, n1(<n) patients have both the tumor and matched normal profiles, whereas n2 and n3 patients have only tumor and normal samples, respectively. Such incomplete or missing paired samples are also known as partially matched samples.
Several methods have been developed to analyze partially matched data generated from the Gaussian distribution.1–4 Recently, Kuan and Huang5 and Yu et al.6 extended the approach to non-parametric setting, which does not require the Gaussian assumption. Specifically in Kuan and Huang,5 we introduced a simple and robust method for analyzing partially matched samples based on the weighted Z-test to combine the P-values computed using (1) paired sample tests (eg, paired t-test or Wilcoxon sign rank test) on the n1 matched pairs and (2) two-sample tests (eg, two-sample t-test or Mann–Whitney test) on the incomplete n2 and n3 pairs. The P-value pooling approach has been shown to achieve good operating characteristics compared to existing methods.
As alluded earlier, matched/paired design is an appealing approach to avoid confounding. However, when a subset of samples has incomplete pairs in partially matched samples scenarios, this can result in unbalanced covariates between the tumor and normal groups. Nonetheless, the above-mentioned methods assume that the confounding factors among the n2 and n3 incomplete matched pairs are absent or negligible. If this assumption does not hold, the conclusions drawn from these methods are no longer valid. In this paper, we introduce an approach to adjust for potential confounders based on propensity score matching method in partially matched samples. Our paper is organized into five sections. In Section 2, we describe the proposed method, followed by Sections 3 and 4, which demonstrate the operating characteristics of the proposed approach in simulations and case study, respectively. We conclude with a discussion in Section 5.
Method
Let (Xi, Yi) be a matched pair for subject i, i = 1, …, n, where Xi and Yi are the tumor and normal measurements, respectively. Without loss of generality, we assume that (Xi, Yi) are complete matched pairs for i = 1, …, n1; Yi’s are missing for i = n1 + 1, …, n1 + n2, and Xi’s are missing for i = n1 + n2 + 1, …, n1 + n2 + n3. That is, n1 patients have both the tumor and matched normal profiles, whereas n2 and n3 patients have only tumor or normal samples, respectively. Let Zi = (Z1i, …, Zpi) denote the p covariates for subject i, for instance, Z1i = age, Z2i = gender, etc. The first step is to create pseudo-pairs between the n2 and n3 incomplete pairs by matching the covariate information. We will use propensity score method to accomplish this step. To simplify the notation, we introduce subscript j to denote sample j, j = 1, …, n2 + n3 among the incomplete pairs, and let Zj denote the corresponding covariate information. Let Oj denote the measurement for subject j. Note that Oj = Xj for j = 1, …, n2 and Oj = Yj for j = n2 + 1, …, n2 + n3. We also let Gj denote the group indicator for sample j, ie, Gj = 1 and 2 for normal and tumor samples, respectively.
Propensity score method
The propensity score method, introduced by Rosenbaum and Rubin,7 is a popular approach in observational studies to create balance in multiple confounding covariates between the two groups. The propensity score is defined as
There are several approaches for estimating ej, including logistic regression and machine learning techniques such as boosted regression,8 classification trees (CART), and random forests. A comparison of these methods is provided in Lee et al.9
There are four main methods for removing confounding effects based on ej, namely (1) propensity score matching, (2) stratification on propensity score, (3) covariate adjustment using propensity score, and (4) inverse probability weighting by propensity score. We refer the readers to Austin10 for a review on these different approaches. In this paper, we consider two approaches based on propensity scores to account for confounding effects. The first approach is covariate adjustment using propensity score via a linear model
| (1) |
where εj ~ N(0, 1) if the biomarker measurements are approximately Gaussian distributed after appropriate normalization and transformation. Otherwise, one can use the generalized linear model11 with appropriate link function for non-Gaussian data. One can then evaluate if the expression of tumor is significantly different from normal by testing the hypothesis H0: βG = 0.
The second approach is based on Mahalanobis distance on covariate ranks with propensity score caliper12 for matching the covariates between the n2 tumor and n3 normal samples from incomplete pairs. Let rj be the vectors of covariate ranks for sample j. The Mahalanobis distance between sample j in the tumor group and sample k in the normal group is defined as
where is the estimated pooled covariance matrix for the ranks. On the other hand, the propensity score caliper c is defined as the maximum propensity score distance between sample j and k allowed within a match. In other words,
The choice of caliper width is related to bias–variance trade-off where small caliper width results in bias reduction but at the expense of increasing variance, and vice versa.13 A few studies have been conducted to investigate the optimal caliper width in propensity score matching, including the work of Austin13 and Wang et al.14 Based on these works and our own experience in propensity score matching, we recommend using caliper width equal to 0.2 of the standard deviation of the logit of the propensity score, which tends to have better performance, ie,
where is the variance of logit of the propensity score in the Gth group. The samples are matched using the optimal full matching algorithm.15–17 Matching algorithm aims to group tumor and normal samples that have similar covariates, ie, small djk. Optimal full matching subdivides the samples into collection of matched sets S, where each set consists of a tumor with any number of normal samples or a normal sample with any number of tumors by minimizing the net discrepancy Σj,k∈Sdjk 15,17 The Olsen’s algorithm is used to create optimal matching (see Hansen15 and Hansen and Klopfer16 for details).
Test statistics for matched set
The tumor and normal samples within each matched set tend to be correlated since they have comparable baseline covariates.7,18 For one-to-one pairing, one usually uses paired sample t-test or Wilcoxon signed-rank test to test if the expression level of tumors is significantly different from the normal samples. However, in the full matching scenario, each tumor is paired with several normal samples and vice versa; thus, the paired sample t-test or Wilcoxon signed-rank test needs to be generalized to such one-to-many pairing. In this paper, we consider a generalization of the paired sample t-test under the scenario that the biomarker measurements are approximately Gaussian distributed. Following Rosner,19 a generalized paired sample t-test can be derived based on a one-way random effects ANOVA model given by
where α is the overall within-pair mean difference between tumor and normal samples, is the random effect for the sth pairing, is the random error, and S is the total number of matched sets. In addition, σs = σ2 (1/m1s + 1/m2s) where m1s and m2s are the number of tumors and normals in matched set s, respectively. The hypothesis for testing if the expression of tumor is different from normal samples translates into testing α = 0. The test statistic is given by
where
and
σ2 is estimated using the usual unbiased estimator, whereas α and are estimated using numerical methods (see Rosner19 for details). For large samples, the P-value of can be obtained from the asymptotic distribution N(0, 1). For small samples, the P-value can be computed from the permutation test, by permuting the labels of tumor and normal samples within each matched set. Suppose there are m1s tumors and a total of Ns samples in matched set s, then the total number of possible permutations is .
On the other hand, the generalized non-parametric test for one-to-many pairing can be carried out via the aligned rank test of Hodges and Lehmann20 if the data are non-Gaussian. We refer the readers to Hodges and Lehmann,20 and Heller et al.21 for additional details on implementing the aligned rank test.
P-values pooling
We follow the idea of our earlier work in Kuan and Huang5 to test if the biomarker is significantly up or down regulated in tumor compared to normal samples by pooling the P-values from the n1 complete and (n2, n3) incomplete pairs. The P-value for the n1 complete matched pairs is computed using either the paired sample t-test or Wilcoxon signed-rank test, denoted as p1. On the other hand, the P-value for the incomplete pairs p2 is computed based on the linear model using propensity score as covariate (equation (1) of Section 2.1), the generalized t-test, or aligned signed rank test (Section 2.2). The next step is to pool the two P-values by borrowing the idea of meta-analysis. Several methods are available for pooling P-values including the inverse normal and Fisher’s methods. In Kuan and Huang,5 we showed that pooling P-values based on weighted Z-test has good operating characteristics compared to other methods. The weighted Z-test for combining the P-values is based on transforming the P-values into Z-score Za = Φ−1(1 − pk), k = 1, 2. The combined P-value by the weighted Z-test5,22 is given by
| (2) |
where wk’s are the corresponding weights. Although different choices of weights have been proposed in the literature, Kuan and Huang,5 Zaykin23 showed that setting the weights as the square root of the sample sizes works well in practice. Thus, we set and . In addition, pooling P-values is only meaningful if p1 and p2 are computed from one-sided hypothesis tests to avoid directional conflict. One can obtain a two-sided combined P-value as follows. Let p1 and p2 be the one-sided P-value for the same alternative (eg, “greater”) hypothesis, and pc be the combined one-sided P-value from equation (2). The two-sided P-value is given by
Simulation
We carry out simulation to evaluate the performance of propensity score method to adjust for potential confounders in partially matched samples. n paired sample measurements (Xij, Yij) of a biomarker i for the tumor and matched normal group are generated from bivariate Gaussian distribution,
where Z1 and Z2 are confounders, and μX and μY are the true mean expressions for tumor and normal groups, respectively. We consider (n1, n2, n3) = (70, 15, 15), (50, 25, 25), and (30, 35, 35) and set σX = σY = 1, whereas ρ ~ U(0, 1) to capture various degrees of correlation between tumor and normal matched pairs. In addition, we set μY = 0 and μX = 0, 0.1, 0.2, …, 0.5 for different effect sizes, and β = 0, 0.5, 1, and 2 for zero, moderate, strong, and very strong confounding effects. To simulate unbalanced confounders arising from incomplete matched pairs, we generate Z1, Z2 ~ N(0, 1) for i = 1, …, n1, Z1 ~ N(−0.2, 1), Z2 ~ N(0.2, 1) for i = n1 + 1, …, n1 + n2, and Z1 ~ (0.2, 1), Z2 ~ N(−0.2, 1) for i = n1 + n2 + 1, …, n1 + n2 + n3.
We compare the performance of the following methods in our simulation studies:
Gold standard (Gold-std): The P-value was computed from paired sample t-test on n1 + n2 + n3 original matched pairs assuming complete data set. This is the reference test.
Paired only: The P-value was computed from paired sample t-test on the n1 complete matched pairs only, and discarding the n2 and n3 incomplete pairs.
Two sample: Combining the P-value from paired sample t-test on the complete n1 matched pairs and the P-value from two-sample t-test on the incomplete n2 and n3 samples using the weighted Z-test approach.5
Propensity score with full matching (FM-PS): Combining the P-value from paired sample t-test on the complete n1 matched pairs and the P-value from generalized t-test on full matched data by Mahalanobis distance with propensity score caliper c = 0.2 on the incomplete n2 and n3 samples.
Propensity score with regression adjustment (Reg-PS): Combining the P-value from paired sample t-test on the complete n1 matched pairs and the P-value from linear regression model using propensity score as covariate on the incomplete n2 and n3 samples.
Single biomarker
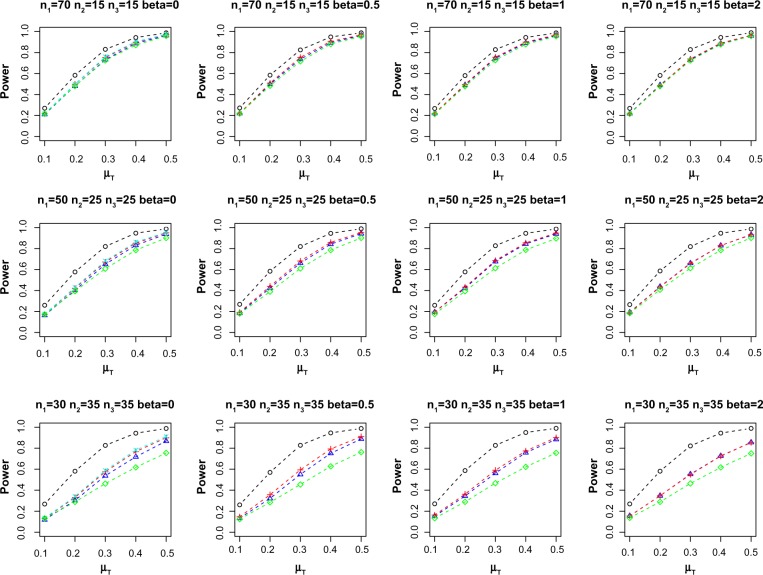
We first evaluate the performance of propensity score methods in adjusting for unbalanced covariates in single biomarker setting. Table 1 reports the average empirical Type I error at nominal α = 0.05 over 10,000 replications. When there is no confounding effect, ie, β = 0, all the methods control the Type I error. However, for β ≠ 0, the two-sample method exhibits the largest Type I error inflation. On the other hand, paired only, FM-PS, and Reg-PS methods control the Type I error under all the scenarios considered in the simulation. Figure 1 shows the average power for different combinations of n1, n2, n3, and β for methods that control the Type I error (empirical Type I error ≤ 0.055). As expected, missing samples in incomplete matched pairs reduce the power compared to complete data set (Gold-std). However, incorporating the incomplete matched pairs with proper adjustment for confounders via FM-PS or Reg-PS methods exhibit increased statistical power compared to using only the n1 paired samples when n2 and n3 are substantially large. When n2 and n3 are small relative to n1, using only the n1 paired samples is comparable to methods that incorporate n2 and n3. Both FM-PS and Reg-PS methods show comparable performance in this simulation study.
Table 1.
Average empirical Type I error at nominal α = 0.05. Italicized values indicate that the empirical Type I error is greater than 0.055.
| METHOD | β= 0 | β= 0.5 | β= 1 | β= 2 |
|---|---|---|---|---|
| n1 = 70, n2 = 15, n3 = 15 | ||||
| Gold-std | 0.0455 | 0.0473 | 0.0503 | 0.0501 |
| Paired only | 0.0479 | 0.0494 | 0.0490 | 0.0518 |
| Two-sample | 0.0475 | 0.0643 | 0.0794 | 0.0922 |
| FM-PS | 0.0462 | 0.0485 | 0.0469 | 0.0476 |
| Reg-PS | 0.0480 | 0.0502 | 0.0441 | 0.0442 |
| n1 = 50, n2 = 25, n3 = 25 | ||||
| Gold-std | 0.0527 | 0.0541 | 0.0495 | 0.0541 |
| Paired only | 0.0486 | 0.0536 | 0.0534 | 0.0531 |
| Two-sample | 0.0476 | 0.1044 | 0.1460 | 0.1809 |
| FM-PS | 0.0483 | 0.0467 | 0.0465 | 0.0485 |
| Reg-PS | 0.0494 | 0.0476 | 0.0443 | 0.0416 |
| n1 = 30, n2 = 35, n3 = 35 | ||||
| Gold-std | 0.0522 | 0.0543 | 0.0483 | 0.0489 |
| Paired only | 0.0507 | 0.0494 | 0.0499 | 0.0465 |
| Two-sample | 0.0522 | 0.1712 | 0.2805 | 0.3663 |
| FM-PS | 0.0449 | 0.0454 | 0.0422 | 0.0460 |
| Reg-PS | 0.0524 | 0.0479 | 0.0452 | 0.0446 |
Figure 1.
Average power at nominal α = 0.05. Sample size (n1, n2, n3) and effect of confounding β are indicated in the header of each plot. Power curves for methods that did not control Type I error (ie, empirical Type I error > 0.055) are not shown.
Notes: ○: Gold-std, ∆: FM-PS, +: Reg-PS, ×: two-sample, and ⋄: paired only.
Multiple biomarkers
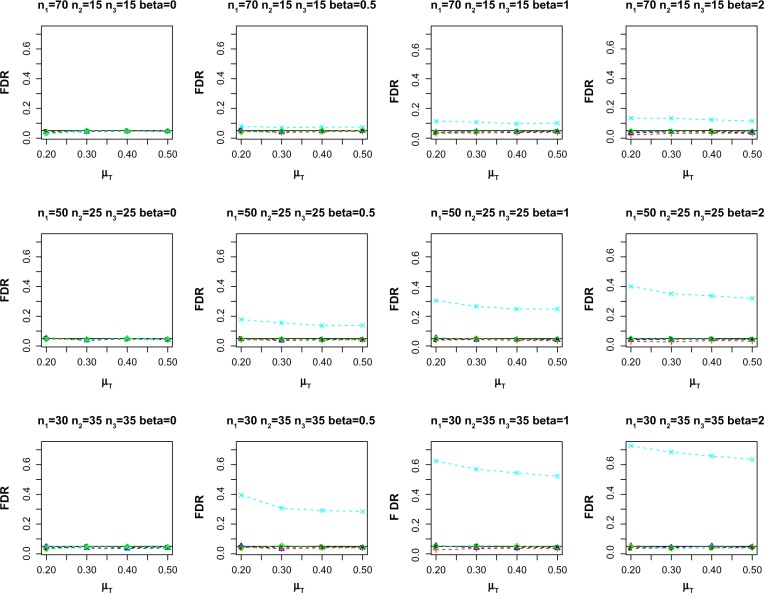
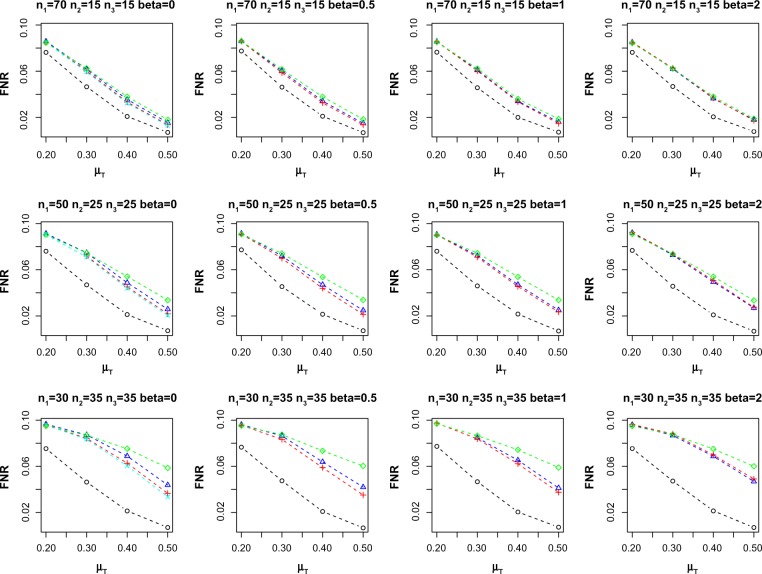
As omics data involve testing multiple biomarkers simultaneously (within a multiple hypothesis testing framework), we also simulate the observations from multiple biomarkers setting. We consider 1000 biomarkers and repeat each simulation setting over 100 replications. We use the false discovery rate (FDR) procedure of Benjamini and Hochberg24 to adjust for multiple hypothesis testing. Figure 2 reports the average empirical FDR for the different methods at nominal FDR = 0.05. Similar to the single biomarker case, two-sample method exhibits the largest inflated empirical FDR for β ≠ 0. On the other hand, FM-PS, Reg-PS, and paired only methods control the FDR across the different scenarios. Figure 3 shows the empirical false nondiscovery rate (FNR) for the methods under comparison. FNR is an analog of Type II error in multiple hypothesis testing settings, and is defined as the proportion of false negatives among the total number of non-rejection. Empirical FNR is large when the number of incomplete matched pairs is large. On the other hand, both FM-PS and Reg-PS methods result in lower FNR compared to paired only method.
Figure 2.
Average empirical FDR at nominal FDR = 0.05. Sample size (n1, n2, n3) and effect of confounding β are indicated in the header of each plot.
Notes: ○: Gold-std, ∆: FM-PS, +: Reg-PS, ×: two-sample, and ⋄: paired only.
Figure 3.
Average empirical FNR at nominal FDR = 0.05. Sample size (n1, n2, n3) and effect of confounding β are indicated in the header of each plot. FNR values for methods that did not control FDR (ie, empirical FDR > 0.055) are not shown.
Notes: ○: Gold-std, ∆: FM-PS, +: Reg-PS, ×: two-sample, and ⋄: paired only.
Case Study
We illustrate the proposed propensity score adjustment for partially matched samples on a publicly available DNA methylation data from Selamat et al.25 (downloaded from Gene Expression Omnibus (GEO) under accession number GSE32861). The data set consists of 58 matched pairs of lung adenocarcinoma and adjacent non-tumor lung tissue after removing paired sample 3023_T/N.25 Methylation for these samples was profiled using the Illumina HumanMethylation27 BeadChip, which covers 27,578 CpGs. We use a subset of baseline covariates measured for each sample (ie, age, smoking status, stage, recurrence, KRAS mutation, EGFR mutation, and LKB1 mutation) to illustrate the performance of the different methods. Age and stage are continuous and ordinal variables, respectively, whereas the other covariates are binary variables.
We randomly choose n1 out of 58 matched pairs to be complete matched pairs. Next, we generate 58 − n1 indicator variables, ie, δk, k = 1, …, 58 − n1, where
and
after standardizing each covariate. This function generates approximately equal number of 0s and 1s on average. Among the remaining 58 – n1 pairs, we set n2 pairs to be missing in non-tumor lung tissue corresponding to those with δk = 1, and the remaining to be missing in lung adenocarcinoma. We consider n1 = 10, 20, 30, 40, and for each n1, the process is repeated 50 times.
We apply FM-PS, Reg-PS, two-sample, and paired only methods on the logit-transformed methylation β-values, ie, log(β/(1 − β))26,27 of each CpG. Since CpGs that are truly differentially methylated between lung adenocarcinoma and non-tumor lung tissues are unknown in the case study, we use the results from paired sample t-test on the full 58 matched pairs as Gold-std. We define the true positive CpGs as the subset of CpGs that are significant at the Benjamini and Hochberg24 FDR of 0.05 for the Gold-std method. We compare the list of significant CpGs identified by FM-PS, Reg-PS, two-sample, and paired only methods at FDR = 0.05 to the true positive CpGs. In Table 2, we report the average empirical FDR, FNR, and average true positive (ATP) CpGs identified by each method. The ATP CpG is also the number of overlapping CpGs identified by each method and the Gold-std method. Two-sample method declares a larger number of false positives as indicated by the inflated empirical FDR. In this case study, the effect of confounding is moderate; thus, FM-PS, Reg-PS, and paired only methods are able to control the FDR. However, both FM-PS and Reg-PS methods have lower FNR and larger ATP compared to paired only method. This shows that the propensity score method for partially matched samples is able to adjust for confounders and improve the power of detecting differentially methylated CpGs.
Table 2.
Average empirical FDR, FNR, and ATP at nominal FDR = 0.05.
| METHOD | PAIRED ONLY | TWO-SAMPLE | FM-PS | Reg-PS |
|---|---|---|---|---|
| n1 = 10, n2 = 24, n3 = 24 | ||||
| FDR | 0.0304 | 0.0822 | 0.0285 | 0.0256 |
| FNR | 0.5439 | 0.2121 | 0.4649 | 0.4478 |
| ATP | 3457 | 13722 | 6908 | 7527 |
| n1 = 20, n2 = 19, n3 = 19 | ||||
| FDR | 0.0322 | 0.0625 | 0.0256 | 0.0247 |
| FNR | 0.4443 | 0.1897 | 0.4053 | 0.3810 |
| ATP | 7690 | 14014 | 8941 | 9568 |
| n1 = 30, n2 = 14, n3 = 14 | ||||
| FDR | 0.0352 | 0.0537 | 0.0393 | 0.0333 |
| FNR | 0.3512 | 0.1632 | 0.3241 | 0.2948 |
| ATP | 10422 | 14395 | 11183 | 11715 |
| n1 = 40, n2 = 9, n3 = 9 | ||||
| FDR | 0.0354 | 0.0648 | 0.0404 | 0.0534 |
| FNR | 0.2404 | 0.1193 | 0.2269 | 0.1803 |
| ATP | 12955 | 15028 | 13201 | 13973 |
We carry out a gene ontology (GO) analysis to provide biological insights into the list of significant CpGs at FDR = 0.05 identified from the Gold-std method using the Bioconductor package topGO.28 We consider both the elim Fisher’s exact test (elim.Fisher) and elim Kolmogorov–Smirnov test (elim.KS) implemented in topGO based on Alexa et al.29 The elim method has been shown to improve interpretation of the GO analysis by integrating GO graph topology and iteratively removing genes that map to significant GO terms from a higher level GO terms.29 The P-values from each of the test are adjusted via the Benjamini–Hochberg method.24 Tables 3–5 report the GO terms corresponding to biological process (BP), molecular function (MF), and cellular component (CC) that exhibit adjusted P-values <0.05 by both the elim.Fisher and elim.KS test, respectively. For example, the BP GO analysis identifies a GO term related to positive regulation of ERK1 and ERK2 cascade, which has been shown to be implicated in lung adenocarcinomas.30,31
Table 3.
Significant BP GO terms for the CpGs identified by the Gold-std method in the lung adenocarcinoma case study. The reported P-values are adjusted via the Benjamini–Hochberg FDR control.24
| BIOLOGICAL PROCESS | ||||||
|---|---|---|---|---|---|---|
| GO ID | TERM | ANNOTATED | SIGNIFICANT | EXPECTED | elim.Fisher | elim.KS |
| GO:0007268 | Synaptic transmission | 1153 | 803 | 685.54 | 2.18e-07 | 2.54e-17 |
| GO:0048704 | Embryonic skeletal system morphogenesis | 159 | 120 | 94.54 | 0.0122 | 1.94e-ll |
| GO:0031424 | Keratinization | 58 | 53 | 34.49 | 0.00019 | 2.97e-ll |
| GO:0007155 | Cell adhesion | 1577 | 1067 | 937.64 | 0.0122 | 7.27e-08 |
| GO:0048265 | Response to pain | 54 | 46 | 32.11 | 0.0139 | 9.22e-08 |
| GO:0009952 | Anterior/posterior pattern specification | 350 | 246 | 208.1 | 0.0283 | 1.21e-07 |
| GO:0007156 | Homophilic cell adhesion | 177 | 133 | 105.24 | 0.0102 | 1.29e-07 |
| GO:0007186 | G-protein coupled receptor signaling pat. | 1035 | 721 | 615.38 | 0.000349 | 6.24e-06 |
| GO:0007193 | Adenylate cyclase-inhibiting G-protein c. | 91 | 73 | 54.11 | 0.0122 | 6.24e-06 |
| GO:0007267 | Cell-cell signaling | 1923 | 1319 | 1143.36 | 0.0122 | 6.24e-06 |
| GO:0070374 | Positive regulation of ERK1 and ERK2 cas. | 0.174 | 128 | 103.46 | 0.0201 | 3.26e-05 |
| GO:0050911 | Detection of chemical stimulus involved. | 19 | 19 | 11.3 | 0.0161 | 4.15e-05 |
| GO:0001755 | Neural crest cell migration | 80 | 65 | 47.57 | 0.0122 | 4.15e-05 |
| GO:0023019 | Signal transduction involved in regulati. | 33 | 30 | 19.62 | 0.0201 | 4.15e-05 |
| GO:0007204 | Elevation of cytosolic calcium ion conce. | 326 | 240 | 193.83 | 0.0322 | 9.08e-05 |
| GO:0048484 | Enteric nervous system development | 30 | 28 | 17.84 | 0.0139 | 9.73e-05 |
| GO:0030198 | Eextracellular matrix organization | 537 | 375 | 319.28 | 0.0102 | 9.73e-05 |
| GO:0021527 | Spinal cord association neuron different. | 27 | 25 | 16.05 | 0.0322 | 0.000155 |
| GO:0042742 | Defense response to bacterium | 209 | 150 | 124.27 | 0.0313 | 0.000266 |
| GO:0006954 | Inflammatory response | 845 | 564 | 502.41 | 0.0217 | 0.000556 |
| GO:0030855 | Epithelial cell differentiation | 905 | 611 | 538.09 | 0.0139 | 0.00112 |
| GO:0045666 | Positive regulation of neuron differenti. | 112 | 86 | 66.59 | 0.0217 | 0.00185 |
| GO:0030335 | Positive regulation of cell migration | 428 | 294 | 254.48 | 0.0139 | 0.00265 |
| GO:0019233 | Sensory perception of pain | 137 | 105 | 81.46 | 0.0122 | 0.00301 |
| GO:0048485 | Sympathetic nervous system development | 45 | 40 | 26.76 | 0.0122 | 0.00496 |
| GO:0045165 | Cell fate commitment | 417 | 299 | 247.94 | 0.034 | 0.00999 |
| GO:0007215 | Glutamate receptor signaling pathway | 88 | 75 | 52.32 | 0.0122 | 0.0181 |
| GO:0021846 | Cell proliferation in forebrain | 43 | 38 | 25.57 | 0.0139 | 0.0194 |
| GO:0007631 | Feeding behavior | 155 | 117 | 92.16 | 0.0122 | 0.0273 |
Table 5.
Significant CC GO terms for the CpGs identified by the Gold-std method in the lung adenocarcinoma case study. The reported P-values are adjusted via the Benjamini–Hochberg FDR control.24
| CELLULAR COMPONENT | ||||||
|---|---|---|---|---|---|---|
| GO ID | TERM | ANNOTATED | SIGNIFICANT | EXPECTED | elim.Fisher | elim.KS |
| GO:0005887 | Integral to plasma membrane | 2079 | 1443 | 1242.77 | 3.14e-14 | 3.64e-35 |
| GO:0005576 | Extracellular region | 3134 | 2161 | 1873.41 | 1.35e-09 | 1.82e-27 |
| GO:0005615 | Extracellular space | 1398 | 964 | 835.68 | 5.49e-ll | 4.19e-25 |
| GO:0005886 | Plasma membrane | 6289 | 4079 | 3759.38 | 2.08e-05 | 2.84e-14 |
| GO:0005578 | Proteinaceous extracellular matrix | 547 | 402 | 326.98 | 1.32e-07 | 1.39e-13 |
| GO:0016021 | Integral to membrane | 6898 | 4421 | 4123.42 | 0.000363 | 1.86e-ll |
| GO:0 04 5211 | Postsynaptic membrane | 294 | 209 | 175.74 | 0.00296 | 4.46e-10 |
| GO:0008076 | Voltage-gated potassium channel complex | 117 | 92 | 69.94 | 0.0012 | 7.64e-10 |
| GO:0030054 | Cell junction | 11 9 7 | 776 | 715.53 | 0.0105 | 2.57e-06 |
| GO:0034774 | Secretory granule lumen | 105 | 79 | 62.77 | 0.0 419 | 0.00149 |
Discussion
Partially matched samples could give rise to unbalanced covariate distribution among the incomplete matched pairs in large-scale matched pair omics studies. This paper extends the P-value pooling method of Kuan and Huang5 to a framework based on propensity score for adjusting unbalanced covariate distribution among the incomplete matched pairs. We consider two approaches using propensity score, namely, (1) full matching followed by generalized t-test (FM-PS) and (2) propensity score as covariate in regression model (Reg-PS). Both methods are able to reduce the number of false positives by accounting for the confounders. Currently, we use the full matching approach based on Mahalanobis distance with propensity score calipers15–17 and the one-way random effects ANOVA model19 for deriving the generalized paired t-test. One can also use other matching algorithms based on propensity score.10
In this paper, we assume that the biomarker measurements are properly transformed such that they are approximately Gaussian distributed. If Gaussian assumption is violated, one can replace the generalized paired t-test with the generalized non-parametric aligned rank test of Hodges and Lehmann20 and Heller et al.21 in FM-PS method, and replace regular linear regression with generalized linear models.11 For instance, in our case study on DNA methylation, the analysis is carried out on the logit transformed beta values (also known as M values). An alternative approach is to analyze the untransformed beta values using beta regression in the Reg-PS method. The choice of analyzing DNA methylation data on either beta values or M values is an ongoing active research.32,33
Both the FM-PS and Reg-PS methods exhibit comparable performance in both our simulations and case study. In this paper, we assume a linear propensity score–outcome relationship that enables us to apply direct adjustment with a linear propensity score term in Reg-PS. In such cases, Reg-PS method is computationally more efficient and easier to implement compared to FM-PS method. However, if the propensity score–outcome relationship is non-linear, one will need to consider more complicated models, for instance, the generalized additive model (GAM) as proposed in Myers and Louis.34 In such cases, the FM-PS method may be a better alternative as this approach does not require specification of the propensity score–outcome relationship. Thus, we recommend that the users compare the results from both Reg-PS and FM-PS methods in practice.
Table 4.
Significant MF GO terms for the CpGs identified by the Gold-std method in the lung adenocarcinoma case study. The reported P-values are adjusted via the Benjamini–Hochberg FDR control.24
| MOLECULAR FUNCTION | ||||||
|---|---|---|---|---|---|---|
| GO ID | TERM | ANNOTATED | SIGNIFICANT | EXPECTED | elim.Fisher | elim.KS |
| GO:0004930 | G-protein coupled receptor activity | 741 | 566 | 440.86 | 2.51e-10 | 1.67e-22 |
| GO:0004984 | Olfactory receptor activity | 67 | 62 | 39.86 | 7.72e-07 | 2.26e-15 |
| GO:0005509 | Calcium ion binding | 977 | 654 | 581.27 | 0.000243 | 1.29e-13 |
| GO:0043565 | Sequence-specific DNA binding | 1129 | 735 | 671.71 | 0.00888 | 5.34e-12 |
| GO:0005201 | Extracellular matrix structural constitu. | 123 | 95 | 73.18 | 0.00634 | 1.81e-08 |
| GO:0005234 | Extracellular-glutamate-gated ion channe. | 29 | 26 | 17.25 | 0.0283 | 6.5e-07 |
| GO:0005125 | Cytokine activity | 332 | 234 | 197.53 | 0.00604 | 1.33e-05 |
| GO:0005230 | Extracellular ligand-gated ion channel a. | 122 | 99 | 72.58 | 0.0147 | 3.79e-05 |
| GO:0004890 | GABA-A receptor activity | 33 | 29 | 19.63 | 0.0283 | 6e-05 |
| GO:0015269 | Calcium-activated potassium channel acti. | 27 | 25 | 16.06 | 0.0192 | 0.000132 |
| GO:0004888 | Transmembrane signaling receptor activit. | 1333 | 979 | 793.08 | 0.0363 | 0.000591 |
| GO:0005540 | Hyaluronic acid binding | 32 | 29 | 19.04 | 0.0179 | 0.00185 |
| GO:0015293 | Symporter activity | 213 | 152 | 126.73 | 0.0216 | 0.00232 |
| GO:0020037 | Heme binding | 201 | 144 | 119.59 | 0.0216 | 0.00558 |
| GO:0005506 | Iron ion binding | 238 | 169 | 141.6 | 0.0192 | 0.00558 |
| GO:0016918 | Retinal binding | 28 | 25 | 16.66 | 0.0363 | 0.0104 |
| GO:0005242 | Inward rectifier potassium channel activ. | 37 | 32 | 22.01 | 0.0283 | 0.0126 |
| GO:0015279 | Store-operated calcium channel activity | 16 | 16 | 9.52 | 0.0229 | 0.0212 |
| GO:0008227 | G-protein coupled amine receptor activit. | 67 | 58 | 39.86 | 0.0216 | 0.0315 |
Footnotes
Author Contributions
Conceived and designed the experiments: PK. Analyzed the data: PK. Wrote the first draft of the manuscript: PK. Made critical revisions: PK. The author reviewed and approved of the final manuscript.
FUNDING: Author discloses no funding sources.
ACADEMIC EDITOR: JT Efird, Editor in Chief
COMPETING INTERESTS: Author discloses no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
REFERENCES
- 1.Lin P, Stivers L. On differences of means with incomplete data. Biometrika. 1974;61(2):325–34. [Google Scholar]
- 2.Ekbohm G. On comparing means in the paired case with incomplete data on both responses. Biometrika. 1976;63(2):299–304. [Google Scholar]
- 3.Kim B, Kim I, Lee S, Kim S, Rha S, Chung H. Statistical methods of translating microarray data into clinically relevant diagnostic information in colorectal cancer. Bioinformatics. 2004;21(4):517–28. doi: 10.1093/bioinformatics/bti029. [DOI] [PubMed] [Google Scholar]
- 4.Looney S, Jones P. A method for comparing two normal means using combined samples of correlated and uncorrelated data. Stat Med. 2003;22:1601–10. doi: 10.1002/sim.1514. [DOI] [PubMed] [Google Scholar]
- 5.Kuan P, Huang B. A simple and robust method for partially matched samples using the p-values pooling approach. Stat Med. 2013;32(19):3247–59. doi: 10.1002/sim.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu D, Lim J, Liang F, Kim K, Kim B, Jang W. Permutation test for incomplete paired data with application to cDNA microarray data. Comput Stat Data Anal. 2012;56:510–21. [Google Scholar]
- 7.Rosenbaum P, Rubin D. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. [Google Scholar]
- 8.McCaffrey D, Ridgeway G, Morral A. Propensity score estimation with boosted regression for evaluating causal effects in observational studies. Psychol Methods. 2004;9(4):403–25. doi: 10.1037/1082-989X.9.4.403. [DOI] [PubMed] [Google Scholar]
- 9.Lee B, Lessler J, Stuart E. Improving propensity score weighting using machine learning. Stat Med. 2010;29:337–46. doi: 10.1002/sim.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Austin P. An introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivariate Behav Res. 2011;46:399–424. doi: 10.1080/00273171.2011.568786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCullagh P, Nelder J. Generalized Linear Model. London: Chapman and Hall; 1983. [Google Scholar]
- 12.Rosenbaum P, Rubin D. Constructing a control-group using multivariate matched sampling that incorporate the propensity score. Am Stat. 1985;39:33–8. [Google Scholar]
- 13.Austin P. Optimal caliper widths for propensity-score matching when estimating differences in means and differences in proportions in observational studies. Pharm Stat. 2011;10:150–61. doi: 10.1002/pst.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Cai H, Li C, et al. Optimal caliper width for propensity score matching of three treatment groups: a Monte Carlo study. PLoS One. 2013;8(12):e81045. doi: 10.1371/journal.pone.0081045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen B. Full matching in an observational study of coaching for the SAT. J Am Stat Assoc. 2004;99:609–18. [Google Scholar]
- 16.Hansen B, Klopfer S. Optimal full matching and related designs via network flows. J Comput Graph Stat. 2006;15:609–27. [Google Scholar]
- 17.Rosenbaum P. A characterization of optimal designs for observational studies. J R Stat Soc B. 1991;53:597–610. [Google Scholar]
- 18.Austin P. Comparing paired vs non-paired statistical methods of analyses when making inferences about absolute risk reductions in propensity-score matched samples. Stat Med. 2011;30:1292–301. doi: 10.1002/sim.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosner B. A generalization of the paired t-test. J R Stat Soc C. 1982;31(1):9–13. [Google Scholar]
- 20.Hodges J, Lehmann E. Rank methods for combination of independent experiments in analysis of variance. Ann Math Stat. 1962;33:482–97. [Google Scholar]
- 21.Heller R, Manduchi E, Small D. Matching methods for observational microarray studies. Bioinformatics. 2008;25(7):904–9. doi: 10.1093/bioinformatics/btn650. [DOI] [PubMed] [Google Scholar]
- 22.Liptak T. On the combination of independent tests. Magyar Tud Aanyos Akad Aemia Mat Kutatao Int Kozl. 1958;3:171–97. [Google Scholar]
- 23.Zaykin D. Optimally weighted z-test is a powerful method for combining probabilities in meta-analysis. J Evol Biol. 2011;24:1836–41. doi: 10.1111/j.1420-9101.2011.02297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 25.Selamat S, Chung B, Girard L, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuan P, Wang S, Chu H. A statistical framework for Illumina DNA methylation. Bioinformatics. 2010;26(22):2849–55. doi: 10.1093/bioinformatics/btq553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuan P, Chiang D. Integrating prior knowledge in multiple testing under dependence with applications to detecting differential DNA methylation. Biometrics. 2012;68(3):774–83. doi: 10.1111/j.1541-0420.2011.01730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexa A, Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. 2010. (R Package Version 2.14.0). [Google Scholar]
- 29.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating go graph structure. Bioinformatics. 2006;22(13):1600–7. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 30.Moorehead R, Sanchez O, Baldwin M, Khokha R. Transgenic overexpression of IGF-II induces spontaneous lung tumors: a model for human lung adenocarcinoma. Oncogene. 2003;22:853–7. doi: 10.1038/sj.onc.1206188. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Su T, Lv D, et al. Erk1/2 mediates lung adenocarcinoma cell proliferation and autophagy induced by apelin-13. Acta Biochim Biophys Sin (Shanghai) 2014;42(2):100–11. doi: 10.1093/abbs/gmt140. [DOI] [PubMed] [Google Scholar]
- 32.Du P, Zhang X, Huang C, et al. Comparison of beta-value and m-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bock C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 2012;13:705–19. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 34.Myers J, Louis T. Johns Hopkins University Department of Biostatistics Working Papers 203. Baltimore: 2010. Regression adjustment and stratification by propensity score in treatment effect estimation. [Google Scholar]