Abstract
Background
Clostridium difficile infection (CDI) is mediated by potent extracellular toxins and is spread largely via bacterial spores. We and others have shown that some antibiotics stimulate C. difficile toxin production in a strain-specific manner; however, the effects of newer anti-C. difficile antibiotics on this process remain to be investigated.
Methods
The effects of the protein synthesis inhibitor tigecycline on sporulation and toxin A and toxin B production were compared in historical (strain 9689) and hypervirulent BI/NAP1/027 (strain 5325) isolates of C. difficile in vitro.
Results
Tigecycline at 1/4× MIC stimulated an increased and earlier toxin A and/or B gene expression in both the historical and the hypervirulent strains, although a commensurate increase in toxin protein production was observed only in the 9689 strain. In fact, in the hypervirulent 5325 strain, toxin production was dramatically suppressed. By comparison, subinhibitory concentrations of vancomycin and metronidazole also stimulated increased protein toxin production by the historical, but not the hypervirulent, strain. In addition, tigecycline dose-dependently reduced viable spore production by both the 9689 and 5325 strains. Vancomycin treatment also suppressed spore formation in both C. difficile strains; however, metronidazole, while reducing spore formation in the 9689 strain, stimulated a near 2 log increase in spore production by the 5325 isolate.
Conclusions
In summary, these findings suggest that the treatment of CDI patients with tigecycline could effectively both control disease progression and limit its spread by disrupting sporulation.
Keywords: antibiotics, human disease, pathogenic bacteria, infectious diseases
Introduction
Clostridium difficile is the most common cause of infective nosocomial diarrhoea in adults worldwide. In the USA, >500 000 new cases of C. difficile emerge each year, resulting in longer hospital stays and billions of dollars of additional healthcare costs.1,2 Recently, the frequency and severity of C. difficile infections (CDIs) have dramatically increased due to the emergence of the hypervirulent BI/NAP1/027 (NAP1) strain. NAP1 strains are characterized by increases in cytotoxin [toxin A (TcdA) and toxin B (TcdB)] production, sporulation and antimicrobial resistance.1–4
Current antibiotic treatment guidelines for severe CDIs recommend the oral administration of vancomycin or metronidazole. However, the therapeutic options for recurrent or refractory CDIs are limited. Recently, several new anti-C. difficile antibiotics such as rifaximin, fidaxomicin, nitazoxanide, ramoplanin and tigecycline have emerged to manage severe and reoccurring cases of CDI.5 Tigecycline (Tygacil®), a broad-spectrum protein synthesis inhibitor antibiotic of the glycylcycline class, has strong antimicrobial activity against C. difficile. Clinically, tigecycline has been used as a successful alternative for treating severe cases of CDI,6,7 including recurrent infections that are refractory to current therapy.8
Previous in vitro studies by our laboratory and others have shown that antimicrobials that target the bacterial cell wall or DNA replication mechanisms increase TcdA and TcdB production by both historical9,10 and hypervirulent (BI/NAP1/027) strains of C. difficile.11 The present study investigated whether, at subinhibitory concentrations, tigecycline has similar effects, and whether this response was similar to that of other antibiotics used to treat CDI. Our findings demonstrate that, overall, tigecycline stimulates increased cytotoxin- and sporulation-related gene expression but blocks their production, thereby lowering TcdA/B levels and preventing sporulation in two distinct strains of C. difficile. These studies suggest that tigecycline may limit both toxin-mediated tissue damage and the transmission of C. difficile in healthcare institutions. Since bacterial spores contribute to CDI recurrence,12 this activity may also account for tigecycline's ability to interfere with the reinfection cycle.
Materials and methods
C. difficile strains
Two strains of C. difficile were studied. ATCC strain 9689 is a historical non-NAP1 clinical CDI isolate determined as toxinotype 0 by PFGE. Strain 5325 is a hypervirulent NAP1 clinical isolate collected in 1993 (a kind gift from Dr Stuart Johnson, VA Chicago Health Care System, Chicago, IL, USA) and identified as group BI by restriction endonuclease analysis.
MICs
Analytical grade tigecycline was provided by Pfizer. Vancomycin and metronidazole were purchased from Sigma–Aldrich (St Louis, MO, USA). The MICs of all antibiotics were determined for the C. difficile ATCC 5325 and 9689 strains by microbroth dilution assay according to CLSI guidelines for such testing in anaerobes13 and as we have previously described.11 In brief, 200 μL of an overnight culture was inoculated into fresh pre-reduced brain heart infusion (BHI) broth (20 mL). Cultures were grown anaerobically to an OD630 of 0.08–0.1 (∼1 × 106 cfu/mL) and added to triplicate wells of a 96-well plate containing serially diluted (2–0.5 mg/L) tigecycline. The plates were incubated anaerobically at 37°C for 48 h and growth (turbidity) was assessed by a microplate reader (OD630). MICs were defined as the lowest antibiotic concentrations that inhibited measurable bacterial growth (i.e. OD630 equal to wells containing no bacteria).
Growth curves and RNA isolation
C. difficile isolates were cultured anaerobically to log phase (OD630 0.08–0.1) in BHI broth and then split into four flasks each containing 199 mL. Antibiotic was prepared and diluted in sterile water as 200× stocks. After 3 h of bacterial culture, 1 mL of prepared tigecycline was added to each flask to give final concentrations of 1/4×, 1/8× and 1/16× MIC, respectively. Sterile water (1 mL) served as a negative treatment control. At times −3 h (stock culture split), 0 h (antibiotics added) and 6, 12, 24 and 48 h after antibiotic addition, 10 mL samples were removed from each C. difficile culture and a small aliquot (20 μL) was used to determine viable cfu. The remaining organisms were collected by centrifugation (13 000 g) and used to prepare total RNA as described below for a PCR analysis of gene expression. The resultant supernatants were filter sterilized and frozen at −70°C to assess soluble TcdA/B production by ELISA.
Analysis of gene expression
RNA was isolated from collected bacterial pellets using the RiboPure-Bacteria kit (Ambion, Austin, TX, USA) according to the manufacturer's recommendations. Contaminating genomic DNA was removed by two rounds of DNase treatment (DNA-free kit; Ambion), and the final RNA yield and quality were assessed by ultraviolet absorbance and agarose gel electrophoresis, respectively. Changes in tcdA, tcdB and spo0A gene expression were assessed by real-time RT–PCR using methods we have previously described11 and the primers given in Table 1. cDNA was generated from 1 μg of total RNA and diluted 1 : 40 in sterile water. C. difficile 16S rRNA served as the internal control. Real-time RT–PCR was performed on a 7500 Fast Real-time PCR system (Applied Biosystems, Carlsbad, CA, USA) using the RT2 Real-Time SYBR Green/Rox PCR Master Mix (SuperArray, Frederick, MD, USA) and the following cycles: 10 min at 95°C and then 40 cycles each at 95°C for 15 s and 55°C for 60 s. Relative gene expression was determined using the 2−ΔΔCt method. The sample mean Ct of 16S rRNA (internal control gene) was subtracted from the sample mean Ct of the tcdA, tcdB and spo0A genes (ΔCt). ΔCt of the no treatment control taken at 6 h was subtracted from the mean ΔCt of each experimental sample (ΔΔCt). This 2−ΔΔCt method yields the fold change in gene expression of the gene of interest normalized to the expression of the 16S rRNA internal control and relative to the no treatment control taken at 6 h.
Table 1.
Primer sequences for amplification of C. difficile tcdA, tcdB, spo0A and 16S rRNA gene sequences
| Gene | Primer | Sequence (5′–3′) | Accession number |
|---|---|---|---|
| tcdA | tcdA-F | CAACACCTTAACCCAGCCATA | AM180355 |
| tcdA-R | AGAGTTTTCTGCGGTAGCTGA | AM180355 | |
| tcdB | tcdB-F | ATCTGGAGAATGGAAGGTGGT | AM180355 |
| tcdB-R | TGATGGTGCTGAAAAGAAGTG | AM180355 | |
| spo0a | spo0A-F | AGCGCAATAAATCTAGGAGCA | AM180355 |
| spo0A-R | AGGTTTTGGCTCAACTTGTGT | AM180355 | |
| 16S rRNA | 16S-F | AGCGGTGAAATGCGTAGATAT | AM180355 |
| 16S-R | CAG CGTCAGTTACAGTCCAGA | AM180355 |
Toxin production
Soluble TcdA and TcdB protein levels were measured in combination (i.e. toxin AB) in collected culture supernatant samples using the Wampole Tox A/B II kit (TechLab, Blacksburg, VA, USA) according to the manufacturer's recommendations. TcdB purified from an NAP1 isolate (a kind gift from Dr Jimmy Ballard, University of Oklahoma Health Sciences Center; stock concentration 300 mg/L) was used to construct a standard curve.11,14 Samples were diluted when necessary to obtain readings within the linear range of the standard (7–500 ng/mL). All samples were tested in triplicate.
Sporulation
To determine the number of spores produced by C. difficile strains over the growth cycle, 0.5 mL aliquots were collected from antibiotic-treated and control cultures at 6, 12, 24 and 48 h post-treatment. The collected samples were mixed with 0.5 mL of 100% ethanol for 1 h with rotation at room temperature to kill the vegetative organisms.15 Samples were then pelleted by centrifugation (13 000 g for 5 min) and washed twice in PBS. Following the last wash, the pellets were resuspended in 0.5 mL of PBS. Spores were enumerated by serially diluting the samples in PBS and plating onto BHI agar plates. Plates were incubated anaerobically at 37°C for 72 h and the resultant cfu/mL were deemed to represent the relative numbers of viable spores produced.
Results
MICs
The MIC of tigecycline was determined for both the historical 9689 and the hypervirulent 5325 C. difficile strains by microbroth dilution. Both strains were susceptible to tigecycline, with MIC values of 0.024 and 0.048 mg/L, respectively. These values agree with the MIC90 values reported by Noren et al.16 The MIC values of metronidazole for the 9689 and 5325 strains were 2 mg/L and 8 mg/L, respectively. Both strains displayed an MIC of vancomycin of 4 mg/L.
Effects of tigecycline on the growth of C. difficile strains
There was little difference in the growth dynamics between the two untreated C. difficile strains as both peaked at ∼109 cfu/mL by 6 h and were in the stationary phase by 12 h (Figures 1a and 2a, filled circles). From 12 to 48 h, the viability of strain 9869 remained constant (Figure 1a), whereas that of strain 5325 gradually declined by ∼1 log (Figure 2a).
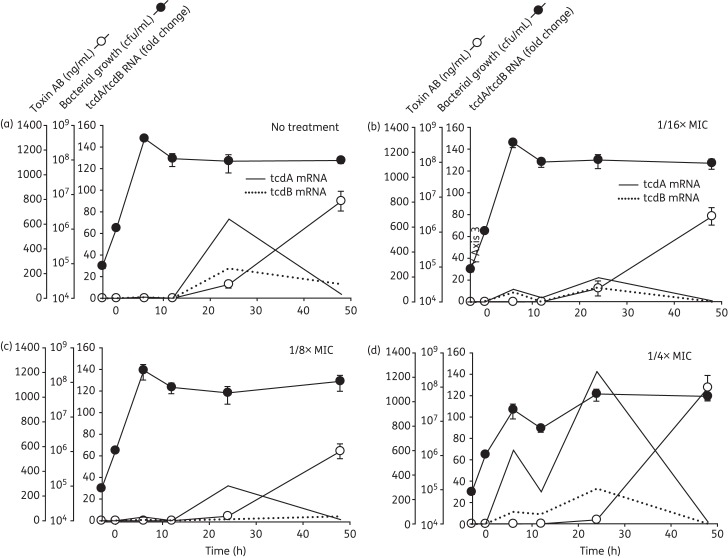
Figure 1.
Effects of tigecycline on the growth, toxin gene expression and soluble toxin production of the historical C. difficile 9689 strain. (a) Antibiotic-free cultures; (b) 1/16× MIC (1.5 mg/L); (c) 1/8× MIC (3 mg/L); and (d) 1/4× MIC (6 mg/L). Tigecycline at the final concentrations indicated was added during early log phase growth (designated Time 0). Samples were collected in duplicate over 48 h for quantification of viable C. difficile, and for measurement of toxin gene expression by real-time RT–PCR and TcdA/B (Toxin AB) production by ELISA.The fold change in mRNA expression for each strain was calculated relative to that of its respective ‘no treatment’ control at 6 h (which was virtually zero).
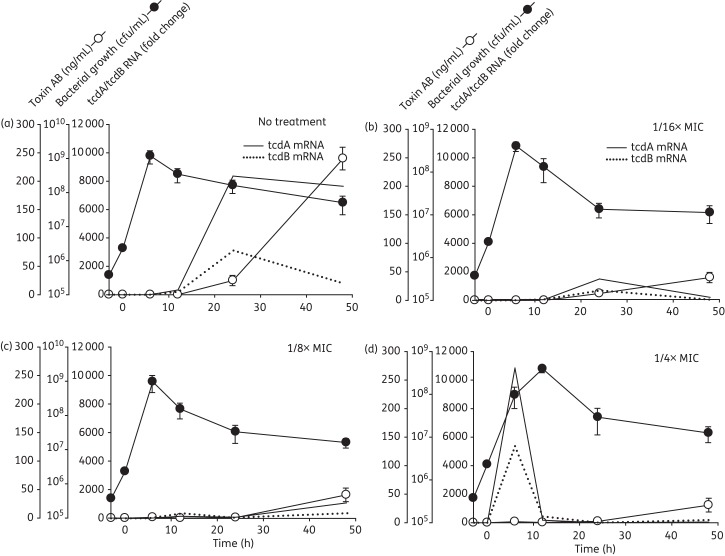
Figure 2.
Effects of tigecycline on the growth, toxin gene expression and soluble toxin production of the hypervirulent C. difficile 5325 strain. (a) Antibiotic-free cultures; (b) 1/16× MIC (3 mg/L); (c) 1/8× MIC (6 mg/L); and (d) 1/4× MIC (12 mg/L). Tigecycline at the final concentrations indicated was added during early log phase growth (designated Time 0). Samples were collected in duplicate over 48 h for quantification of viable C. difficile, and for measurement of toxin gene expression by real-time RT–PCR and TcdA/B (Toxin AB) production by ELISA. The fold change in mRNA expression for each strain was calculated relative to that of its respective ‘no treatment’ control at 6 h (which was virtually zero).
Tigecycline at 1/16× and 1/8× MIC had little effect on these dynamics (Figure 1b and c and Figure 2b and c). At 1/4× MIC, tigecycline reduced the viability of both the historical 9689 and NAP1 5325 strains by nearly 1 log at 6 h (Figures 1d and 2d), although the numbers of viable C. difficile 9689 ultimately recovered by 48 h were comparable to the levels observed in the untreated control culture.
Effects of tigecycline on cytotoxin gene expression and protein production
In non-antibiotic-treated cultures of the historical 9689 C. difficile strain, measureable toxin gene expression began at ∼12 h, peaked in the stationary phase at 24 h and gradually returned to baseline over the next 24 h period (Figure 1a; continuous and broken lines). TcdA/B protein production was detectable by 12 h and peaked at ∼800 ng/mL by 48 h (Figure 1a; open circles).
In this strain, tigecycline induced a biphasic toxin gene expression response (Figure 1b–d) in which the first peaks of tcdA and tcdB expression occurred during the log phase of growth (6 h). The resultant mRNA species were short lived, having waned by 12 h, and were followed by a second peak of gene expression at 24 h. Although this latter peak occurred as expected during the stationary phase, the magnitude of the response was decidedly different than in untreated control cultures. Specifically, the 1/4× MIC increased peak tcdA expression 2.3-fold, whereas lower tigecycline doses suppressed toxin gene expression below control values. Despite the varied tigecycline-induced changes in the dynamics and magnitude of the responses, toxin protein production remained largely unchanged (Figure 1a–d).
The effects of tigecycline on the NAP1 5325 strain were in part comparable to those of the historical 9689 strain. Similar to the 9689 strain, tigecycline at 1/4× MIC induced pronounced early peaks of tcdA and tcdB gene expression during the log phase of growth (Figure 2d). However, unlike the 9689 strain, all antibiotic doses halted virtually all toxin gene expression in the stationary phase (Figure 2b–d) and little to no toxin production was observed in any of the tigecycline-treated 5325 cultures (Figure 2b–d).
Similarly, increased TcdA/B production was also observed by the 9689 strain following exposure to subinhibitory concentrations of both vancomycin and metronidazole. In contrast, only vancomycin at 1/4× MIC induced protein toxin production by the hypervirulent 5325 strain (Figure 3).
Figure 3.
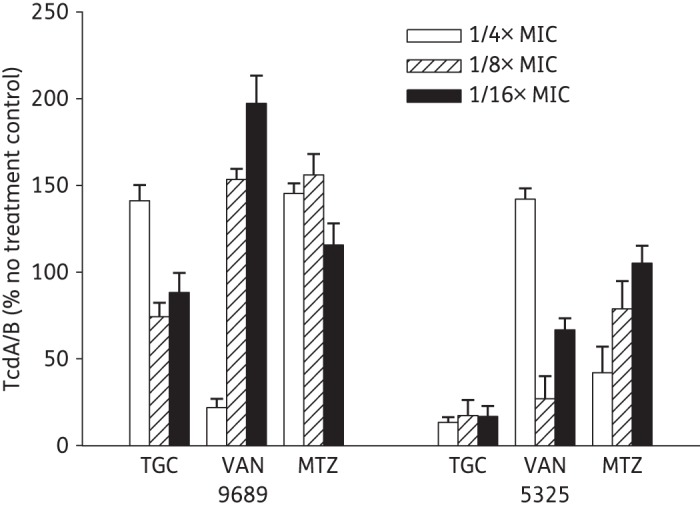
Comparison of soluble TcdA/TcdB production at 48 h following exposure to subinhibitory concentrations of antibiotics. Supernatant samples were collected at 48 h from historical 9689 and hypervirulent 5325 C. difficile cultures containing nothing, tigecycline (TGC), vancomycin (VAN) or metronidazole (MTZ) at the final concentrations indicated. The collected samples were screened to detect TcdA/TcdB using commercial ELISA and the results are given as relative values compared with the drug-free culture. Data shown are the means of three replicates from two experiments.
Tigecycline decreases sporulation by C. difficile
Little to no expression of the sporulation-related transcription factor spo0A or formation of viable spores was observed prior to 24 h of growth under any culture condition with either strain (data not shown); however, both spo0A gene transcription and sporulation were prominent by 48 h in both non-antibiotic-treated cultures, albeit at decidedly different levels (data not shown). The production of a markedly greater number of spores by the 5325 strain was expected since NAP1 strains are commonly high spore producers.17
The effects of tigecycline on sporulation were strain-specific and paralleled those observed for exotoxin production. Like exotoxin gene expression, tigecycline at 1/4× MIC significantly increased spo0A transcription in the historical 9689 strain (data not shown), but this tigecycline-induced increase in spo0A mRNA did not result in a commensurate increase in spore production in this strain (Figure 4a). In fact, viable spore formation was significantly reduced by tigecycline at all concentrations tested (Figure 4a). In the hypervirulent NAP1 5325 strain, tigecycline at 1/4× MIC did not significantly alter the high level of spo0A transcription at 48 h, but it markedly suppressed spore formation (Figure 4b). Compared with the number of spores estimated by visual inspection using light microscopic analysis (data not shown), ∼0.1% of spores germinated and grew as cfu on BHI agar plates. These values are in agreement with germination rates and outgrowth efficiencies reported by Lawley et al.15
Figure 4.
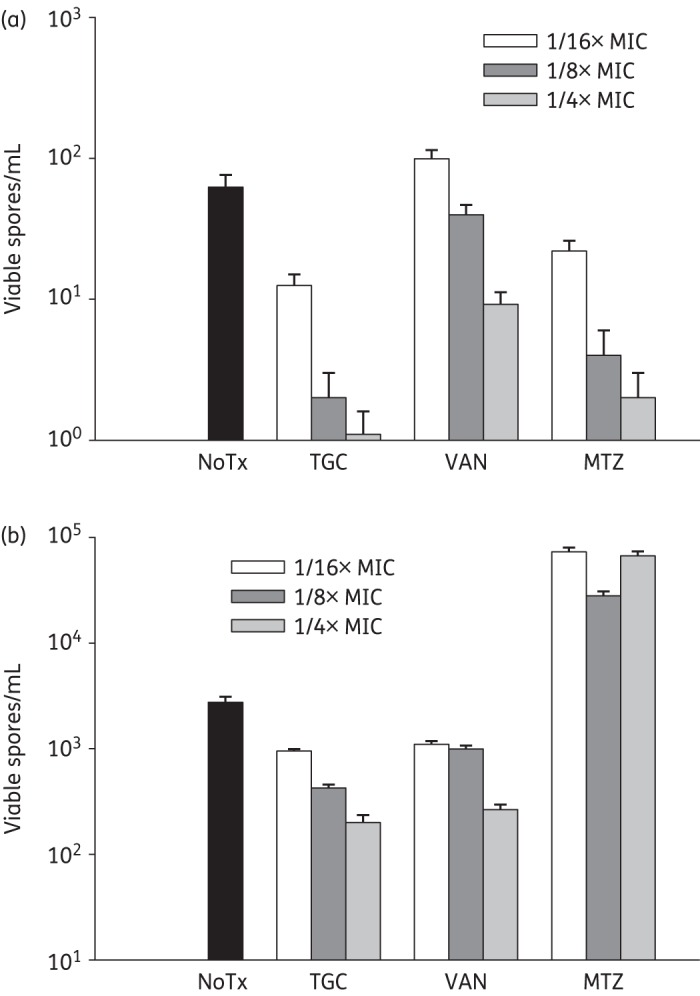
Comparison of spore production at 48 h following exposure to subinhibitory concentrations of antibiotics. (a) Historical 9689 and (b) hypervirulent 5325 strains of C. difficile were exposed to nothing (NoTx), tigecycline (TGC), vancomycin (VAN) or metronidazole (MTZ) at the final concentrations indicated. At 48 h, samples were collected and spores isolated by mixing an equal volume of culture supernatant with 100% ethanol, followed by centrifugation. Harvested spores were enumerated by serially diluting in PBS and plating onto BHI agar plates. The plates were incubated anaerobically at 37°C for 72 h.
Similar responses were also observed for the 9689 and 5325 strains following exposure to vancomycin. As shown in Figure 4(a and b), the addition of vancomycin at 1/4× MIC (1 mg/L) and 1/8× MIC (0.5 mg/L) reduced spore production by both strains at 48 h (by between 10% and 50%), despite slight increases in spo0A expression (data not shown). Vancomycin at 1/16× MIC (0.25 mg/L) also inhibited spore production by the 5325 strain (∼15%) but not the 9689 strain as viable spore counts were comparable to those of the antibiotic-free control (Figure 4a and b). In addition, all subinhibitory concentrations of metronidazole suppressed spore formation by the historical 9689 strain at 48 h, and this was concentration dependent (Figure 4a). This is in contrast to the hypervirulent 5325 strain, for which nearly 2 log increases in spore production were measured for all doses of metronidazole tested (Figure 4b).
Discussion
Tigecycline is a glycylcycline antibiotic that has in vitro activity against numerous Gram-positive and Gram-negative bacteria, including C. difficile.6 Tigecycline was originally approved by the US FDA to treat complicated skin and soft tissue infections, intra-abdominal infections and cases of community-acquired pneumonia.6 However, case studies describing the use of intravenous tigecycline to treat severe cases of CDI have recently been reported. Herpers et al.8 showed that intravenous tigecycline was successfully used to treat four patients with severe CDI, all of whom had no evidence of recurrent infection during a 3 month follow-up period. Similar studies by Lu et al.18 and Cheong et al.19 also described the improvement of two patients with severe CDI following the administration of tigecycline after all other antibiotic treatments had failed. As with the results form Herpers et al.,8 no recurrent CDIs were reported in these patients. However, one case report by Kopterides et al.20 described the failure of tigecycline to treat a fatal case of CDI in a 70-year-old patient. These largely positive findings have been attributed to the in vivo stability of tigecycline, as unmetabolized drug (3–14 mg/kg) is recovered in the stool of patients21,22 at concentrations well above the in vitro susceptibility range for multiple isolates of C. difficile.6
We hypothesize that the mechanism of action of tigecycline may be responsible for its efficacy in treating severe or recurrent CDI. Specifically, data presented here suggest that tigecycline's ability to block protein synthesis is largely responsible for reducing both toxin production and sporulation. Interestingly, a growing body of evidence suggests that some pathogens sense and respond to low-dose antibiotic challenge with a putative SOS response that includes the up-regulation of virulence-associated genes,9–11,23 and our data show that the exposure of C. difficile to tigecycline is no exception. In this study, tigecycline-induced toxin synthesis was observed in the 1/4× MIC culture of the historical 9689 strain, resulting in a ∼1.4-fold increase in TcdA/B production. Interestingly, subinhibitory concentrations of both vancomycin (a cell wall synthesis inhibitor) and metronidazole (a nucleic acid synthesis inhibitor) also stimulated increased TcdA/B production by the 9689 strain. This is in contrast to the hypervirulent 5325 strain, for which only vancomycin at 1/4× MIC induced increased toxin production. These results suggest that fluctuations in the timing, magnitude and onset of antibiotic-induced stress responses vary between C. difficile isolates and antimicrobial stress. However, given that only three antibiotics and two strains of C. difficile were examined in this study, these effects may only reflect strain-dependent responses to these antibiotics and not global SOS reactions by C. difficile. The mechanisms governing this phenomenon of strain and antibiotic variability on inducing toxin production by C. difficile are currently being explored in our laboratory.
The ability of C. difficile to shed highly resistant spores in hospital environments perpetuates the development of new and recurrent CDIs. Our analysis demonstrates that tigecycline reduces sporulation in both the historical 9689 and hypervirulent 5325 strains. These findings agree with those of Garneau et al.24 who also demonstrated that subinhibitory concentrations of tigecycline reduced spore formation in multiple isolates of C. difficile. Our findings suggest that this decreased sporulation is attributable to an inhibition of translation of sporulation-related mRNA species by tigecycline. These data may explain, at least in part, the extremely low recurrence rate of CDI in patients receiving tigecycline therapy.6,8 Interestingly, as both strains displayed similar decreases in sporulation following a challenge with tigecycline and vancomycin, only metronidazole stimulated sporulation (a nearly 2 log increase) by the high-spore-forming hypervirulent 5325 strain. Collectively, these data imply that the impact of antibiotics on C. difficile sporulation can also vary depending on both the strain of C. difficile being studied and the antibiotic (and its specific mode of action) being tested.
In conclusion, there is a pressing need for the development of new treatments for severe and recurrent cases of CDI. Based on the current study demonstrating tigecycline's ability to inhibit both toxin production and sporulation better than other accepted CDI antimicrobial therapies, and on publications of largely positive case studies, tigecycline may offer physicians an alternative treatment for severe or refractory CDI.
Funding
This material is based upon work supported in part by the U.S. Department of Veterans Affairs, Office of Research and Development Biomedical Laboratory Research Program (M. J. A., A. E. B., D. L. S.), the National Institutes of Health (Grant P20RR016454/P20GM103408) and Pfizer Pharmaceuticals, Incorporated (ASPIRE award).
Transparency declarations
None to declare.
References
- 1.Hookman P, Barkin JS. Clostridium difficile associated infection, diarrhea and colitis. World J Gastroenterol. 2009;15:1554–80. doi: 10.3748/wjg.15.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter GP, Douce GR, Govind R, et al. The anti-sigma factor TcdC modulates hypervirulence in an epidemic BI/NAP1/027 clinical isolate of Clostridium difficile. PLoS Pathog. 2011;7:e1002317. doi: 10.1371/journal.ppat.1002317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Connor JR, Johnson S, Gerding DN. Clostridium difficile infection caused by the epidemic BI/NAP1/027 strain. Gastroenterology. 2009;136:1913–24. doi: 10.1053/j.gastro.2009.02.073. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett JG. Antibiotic-associated diarrhea. Clin Infect Dis. 1992;15:573–81. doi: 10.1093/clind/15.4.573. [DOI] [PubMed] [Google Scholar]
- 5.Shah D, Dang MD, Hasbun R, et al. Clostridium difficile infection: update on emerging antibiotic treatment options and antibiotic resistance. Expert Rev Anti Infect Ther. 2010;8:555–64. doi: 10.1586/eri.10.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larson KC, Belliveau PP, Spooner LM. Tigecycline for the treatment of severe Clostridium difficile infection. Ann Pharmacother. 2011;45:1005–10. doi: 10.1345/aph.1Q080. [DOI] [PubMed] [Google Scholar]
- 7.Lao D, Chiang T, Gomez E. Refractory Clostridium difficile infection successfully treated with tigecycline, rifaximin, and vancomycin. Case Rep Med. 2012;2012:702910. doi: 10.1155/2012/702910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herpers BL, Vlaminckx B, Burkhardt O, et al. Intravenous tigecycline as adjunctive or alternative therapy for severe refractory Clostridium difficile infection. Clin Infect Dis. 2009;48:1732–5. doi: 10.1086/599224. [DOI] [PubMed] [Google Scholar]
- 9.Drummond LJ, Smith DG, Poxton IR. Effects of sub-MIC concentrations of antibiotics on growth of and toxin production by Clostridium difficile. J Med Microbiol. 2003;52:1033–8. doi: 10.1099/jmm.0.05387-0. [DOI] [PubMed] [Google Scholar]
- 10.Gerber M, Walch C, Loffler B, et al. Effect of sub-MIC concentrations of metronidazole, vancomycin, clindamycin and linezolid on toxin gene transcription and production in Clostridium difficile. J Med Microbiol. 2008;57:776–83. doi: 10.1099/jmm.0.47739-0. [DOI] [PubMed] [Google Scholar]
- 11.Aldape MJ, Packham AE, Nute DW, et al. The effects of ciprofloxacin on the expression and production of exotoxins by Clostridium difficile. J Med Microbiol. 2013;62:741–7. doi: 10.1099/jmm.0.056218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore P, Kyne L, Martin AJ, et al. Germination efficiency of clinical Clostridium difficile spores and correlation with ribotype, disease severity and therapy failure. J Med Microbiol. 2013;62:1405–13. doi: 10.1099/jmm.0.056614-0. [DOI] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria: Seventh Informational Supplement M11-A7. Wayne, PA, USA: CLSI; 2007. [Google Scholar]
- 14.Merrigan M, Venugopal A, Mallozzi M, et al. Human hypervirulent Clostridium difficile strains exhibit increased sporulation as well as robust toxin production. J Bacteriol. 2010;192:4904–11. doi: 10.1128/JB.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawley TD, Croucher NJ, Yu L, et al. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J Bacteriol. 2009;191:5377–86. doi: 10.1128/JB.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noren T, Alriksson I, Akerlund T, et al. In vitro susceptibility to 17 antimicrobials of clinical Clostridium difficile isolates collected in 1993–2007 in Sweden. Clin Microbiol Infect. 2010;16:1104–10. doi: 10.1111/j.1469-0691.2009.03048.x. [DOI] [PubMed] [Google Scholar]
- 17.Akerlund T, Persson I, Unemo M, et al. Increased sporulation rate of epidemic Clostridium difficile type 027/NAP1. J Clin Microbiol. 2008;46:1530–3. doi: 10.1128/JCM.01964-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu CL, Liu CY, Liao CH, et al. Severe and refractory Clostridium difficile infection successfully treated with tigecycline and metronidazole. Int J Antimicrob Agents. 2010;35:311–2. doi: 10.1016/j.ijantimicag.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Cheong EY, Gottlieb T. Intravenous tigecycline in the treatment of severe recurrent Clostridium difficile colitis. Med J Aust. 2011;194:374–5. doi: 10.5694/j.1326-5377.2011.tb03018.x. [DOI] [PubMed] [Google Scholar]
- 20.Kopterides P, Papageorgiou C, Antoniadou A, et al. Failure of tigecycline to treat severe Clostridium difficile infection. Anaesth Intensive Care. 2010;38:755–8. doi: 10.1177/0310057X1003800339. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann M, DeMaio W, Jordan RA, et al. Metabolism, excretion, and pharmacokinetics of [14C]tigecycline, a first-in-class glycylcycline antibiotic, after intravenous infusion to healthy male subjects. Drug Metab Dispos. 2007;35:1543–53. doi: 10.1124/dmd.107.015735. [DOI] [PubMed] [Google Scholar]
- 22.Nord CE, Sillerstrom E, Wahlund E. Effect of tigecycline on normal oropharyngeal and intestinal microflora. Antimicrob Agents Chemother. 2006;50:3375–80. doi: 10.1128/AAC.00373-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens DL, Ma Y, Salmi DB, et al. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus. J Infect Dis. 2007;195:202–11. doi: 10.1086/510396. [DOI] [PubMed] [Google Scholar]
- 24.Garneau JR, Valiquette L, Fortier LC. Prevention of Clostridium difficile spore formation by sub-inhibitory concentrations of tigecycline and piperacillin/tazobactam. BMC Infect Dis. 2014;14:29. doi: 10.1186/1471-2334-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]