Abstract
In a fast-growing Escherichia coli cell, most RNA polymerase (RNAP) is allocated to rRNA synthesis forming transcription foci at clusters of rrn operons or bacterial nucleolus, and each of the several nascent nucleoids contains multiple pairs of replication forks. The composition of transcription foci has not been determined. In addition, how the transcription machinery is three-dimensionally organized to promote cell growth in concord with replication machinery in the nucleoid remains essentially unknown. Here, we determine the spatial and functional landscapes of transcription and replication machineries in fast-growing E. coli cells using super-resolution-structured illumination microscopy. Co-images of RNAP and DNA reveal spatial compartmentation and duplication of the transcription foci at the surface of the bacterial chromosome, encompassing multiple nascent nucleoids. Transcription foci cluster with NusA and NusB, which are the rrn anti-termination system and are associated with nascent rRNAs. However, transcription foci tend to separate from SeqA and SSB foci, which track DNA replication forks and/or the replisomes, demonstrating that transcription machinery and replisome are mostly located in different chromosomal territories to maintain harmony between the two major cellular functions in fast-growing cells. Our study suggests that bacterial chromosomes are spatially and functionally organized, analogous to eukaryotes.
INTRODUCTION
Unlike a eukaryotic cell that has defined phases in the cell cycle (S, G2, M and G1), a rapidly growing bacterial cell such as Escherichia coli has no distinct stages in the cell cycle; therefore, all processes, such as transcription, replication and chromosome segregation, are intimately entangled. E. coli is capable of rapid growth, with a growth rate as fast as ∼20 min (Lennox Broth (LB) at 37°C), which is far shorter than the time needed for the completion of one round of chromosome (nucleoid) replication and segregation (>74 min) (1). Consequently, maximum expression of growth-promoting genes and multiple genome replications are concurrently achieved in a fast-growing cell. We have only begun to understand how a fast growth rate influences the distribution of RNA polymerase (RNAP) (2,3); however, how the transcription machinery is spatially organized and, particularly, how transcription and replication machineries maintain harmony in a fast-growing cell, remains unknown.
To maintain a fast growth rate in E. coli, the majority of RNAP molecules form foci and are believed to engage in transcription of growth-promoting genes, most of which are ribosomal RNA operons (rrn) (3,4). There are seven almost identical rrn operons in the origin of chromosome replication (oriC) half of the genome, four of which are located near the oriC. An E. coli cell growing in rich media, such as LB, contains several nascent nucleoids with multiple replication forks (5); thus, copies of the rrn operons are favorably amplified and can be present in up to ∼50 copies due to their locations in the genome (6). Because the number of RNAP foci or transcription foci (these terms are hereafter used interchangeably) revealed by wide-field fluorescent microscopy is significantly smaller than the calculated number of rrn copies in a fast-growing cell, it is inferred that transcription foci are located at clusters of rrn or bacterial nucleolus-like structures (4). Recently, super-resolution microscopy (such as photoactivated localization microscopy, or PALM) was used to examine the distribution of RNAP in fast-growing cells and identified clusters of RNAP, their sizes ranging from 70 to 800 RNAP molecules, which are likely to be transcription foci at single or clustered rrn operons (7). In addition, transcription of rrn is regulated by an antitermination mechanism during elongation (8–10). Genetically, NusA and NusB factors and the boxA sequence in the nascent rRNA are critical for the rrn antitermination system in E. coli, analogous to the lambda N-mediated antitermination system (11–13). NusA is an RNAP-associated protein during elongation (14). NusB together with ribosomal protein S10 binds the boxA RNA (15–17), but it does not associate with RNAP in vitro. The distributions of NusA and NusB in Bacillus subtilis have been reported (18,19); however, whether NusA and NusB are associated with transcription foci in fast-growing E. coli cells remains unknown. The role of RNAP and transcription in the organization of bacterial nucleoid has been established (3,20,21); however, whether transcription machinery is spatially and functionally organized in E. coli chromosome has not been determined.
In a fast-growing E. coli cell, the genome is continuously replicated, with up to five genome equivalent (6) and multiple pairs of replication forks to ensure passage of at least one intact bacterial chromosome into each of the two daughter cells (5). Replication machinery, also called the replisome (22), which consists of the DNA polymerase III holoenzyme (23) and single-stranded DNA-binding (SSB) proteins (24,25), is located at each replication fork. Another protein, SeqA (26), polymerizes with the nascent hemimethylated DNA at or near DNA replication forks (27–31). Genome conformation capture analysis demonstrates that the E. coli chromosome is organized by DNA replication (through SeqA-mediated interactions) and transcription (32); however, the mechanisms underlying nucleoid organization remain to be determined. A longstanding interest in the field has been to determine how the two major cellular functions, transcription and replication, maintain harmony to avoid conflicts between DNA replication and transcription (33), particularly in fast-growing cells.
In this study, we determined the spatial organization and composition of prominent transcription foci which are engaged in active rRNA synthesis, as well as the spatial relationship between transcription and replication machineries in fast-growing E. coli cells using super-resolution-structured illumination microscopy (SR-SIM) (hereafter referred to as SIM) (34,35). This method surpasses the diffraction limit of conventional wide-field fluorescence microscopy. Our results from co-imaging of proteins and DNA by SIM reveal novel features of functional chromosomal territories in fast-growing bacterial cells.
MATERIALS AND METHODS
Bacterial strains, bacterial growth and techniques
The strains used in the study are derivatives of CC72 (MG1655 with a chromosomal rpoC-venus gene fusion) (36). The strains containing mCherry fusions to the carboxy termini of the NusA, NusB, SeqA and SSB proteins were generated by standard phage lambda Red-mediated recombination system (37) using a cassette that contains the mCherry gene linked to a selectable KanR marker and flanked by sequences in the interested genes. All constructed strains were checked by polymerase chain reaction and/or sequencing. All of the fluorescent proteins are regulated and expressed from their original chromosomal positions. The growth rates for all of the strains were similar to that of the parental strain, MG1655. The bacterial media and techniques were as previously described (38). Cells were grown in LB (tryptone 10 g/l, yeast extract 5 g/l, NaCl 5 g/l) overnight at 37ºC before being diluted to an optical density (OD600) of 0.02 in fresh LB and harvested at early log (or exponential) phase.
Microscopy and image analysis
The microcopy was performed as previously described (20). Cells were fixed using formaldehyde (3.7% v/v final) before being mounted on a microscope slide for imaging (4). An inverted microscope (Zeiss Axio Observer) with a Plan-Apochromat 63x/1.46 Oil DIC objective and an ELYRA S.1 SIM module was used for SIM microscopy. Images with a high signal-to-noise ratio were captured with a high-resolution EMCCD camera. We performed three grid rotations per image. For each channel, we set the exposure such that all fluorescence proteins in the cell were imaged (no more signal was detectable at the end of the acquisition process). The high-resolution SIM images were reconstituted using Zeiss proprietary software (ZEN, black edition). Prior to image acquisition, the microscope was calibrated using fluorescence beads, and the calibration profile was used to realign each channel after image acquisition to correct for chromatic aberration. Each condition consists of three independent biological replicates. The imaged fields were chosen randomly to better represent the entire population of cells.
Images were processed and analyzed using MATLAB. The heat map images were determined from the log2 of the normalized intensities of the signals from the two channels involved. The segmentations of nucleoids, foci of RNAP-Venus and other proteins (mCherry fusions) in different channels were performed by applying the Laplacian of Gaussian edge detection method (39) with optimal parameters. To refine the detection of mCherry fusion and RNAP-Venus, a watershed segmentation method was used (40) to separate adjacent foci. Because E. coli cells are rod shaped (cylinder-like) the placement of a population of cells on microscope slides will be random and reflecting different rotations of cells on the short axis. Thus, co-localization analyses were based on two-dimensional (2D) images as best estimates. The foci-clustering frequencies of a particular protein-mCherry fusion and RNAP-Venus in each cell were measured using a subcellular co-localization analysis as described (41). Specifically, the positions of the weighted centroids of the foci in the two channels were determined and the distance between each mCherry-weighted centroid and its closest RNAP-Venus-weighted centroid was recorded in the 2D image analysis. Two foci were scored as co-localized if the distance of the two centroids was below the theoretical microscope resolution for the mCherry (140 nm). The co-localization frequency is represented as the fraction of mCherry foci which co-localized with at least one of the transcription foci (RNAP-Venus) in a cell and the central tendency of the distribution (average) in a population of cells is represented by the median. Co-localization of mCherry foci and RNAP-Venus foci was also represented as a cumulative distribution of pairwise distance as described (42). To generate the random data set, the regions of interest from different channels were merged. Within the merged region, the position of each mCherry centroid was randomly shuffled and similar analyses were performed. Only the distances ranging from 0 to 1000 nm were included in the plots to avoid scaling issue and increase the readability. The cells illustrated are representatives of the majority of the observed cells. Pictures were processed uniformly and false-colored with Adobe Photoshop.
RESULTS
Spatial compartmentalization of transcription foci in the nucleoids of fast-growing cells
We wished to know where transcription foci are spatially located in the nucleoids of fast-growing cells. To determine the spatial organization of the transcription machinery in the bacterial nucleoid, we performed SIM co-imaging of RNAP and DNA in fast-growing cells (LB, 37°C, doubling time 20 min). Given its resolution (∼140-nm lateral and ∼300-nm axial for a typical Venus fluorophore), SIM would be as effective as PALM in detecting small transcription foci in fast-growing cells, because analysis of PALM images reveals that even small clusters (foci) of 70 RNAPs form a sphere of ∼160 nm in diameter (7), which is larger than the SIM resolution. The enhanced optical resolution by SIM would also reveal more details of bacterial chromosome, thus enabling the determination of the spatial relationship between transcription foci and the nucleoids in a fast-growing cell.
Compared to previous reports (4,20,36,43) our SIM images of RNAP and DNA in a representative fast-growing cell were superior in quality (Figure 1A), with rich textural details, largely due to an increased signal-to-noise ratio and improved resolution. As with the RNAP picture, the DNA images revealed a striking heterogeneity in the nucleoid structure, containing areas of high and low DNA density, as well as apparent voids, very similar to what was recently described in super-resolution microscopy (44) and unlike the blob morphology typically described in the literature (45). There were, on average, four apparent nascent nucleoids per cell, although the degree of entanglement between nascent nucleoids varies from cell to cell. Transcription foci were present in each of the four apparent nascent nucleoids (overlay of RNAP/DNA). This feature indicates that the transcription machinery was duplicated along with newly replicated and segregated chromosomes in the fast-growing cells. The median number of transcription foci revealed by SIM was eight per cell in the fast-growing cell populations (Figure 1B), compared with the median number of four per cell detected by wide-field fluorescence microscopy under the same growth conditions (3). The extra transcription foci identified by SIM are likely attributed to small clusters of 70 RNAPs revealed by PALM (7). Thus, on average there are about two transcription foci, per nascent nucleoid in a fast-growing E. coli cell.
Figure 1.
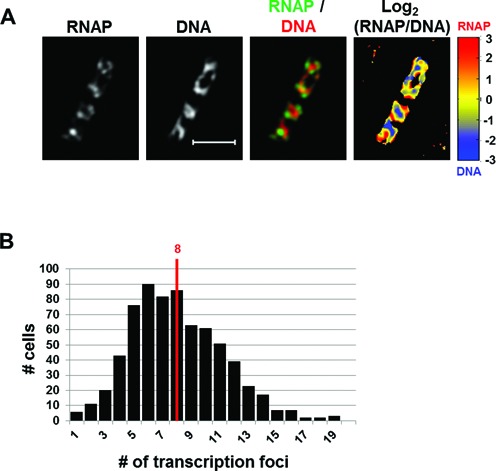
SIM co-imaging of RNAP and DNA reveals spatial compartmentalization of transcription foci in fast-growing cells. (A) Images of RNAP, DNA (nucleoid), and an overlay of RNAP and DNA from a representative fast-growing E. coli cell (LB, 37°C). On the overlay image, the RNAP is false-colored in green and the nucleoid in red. The scale bar represents 2 μm. The log2 (RNAP/DNA) plot (heat map) is a quantitative representation of the relationship between RNAP and DNA, which is represented by a color scale bar with values ranging from −3 to 3. Note that regions enriched up to 8-fold in RNAP over DNA are at the periphery of the nucleoid (red foci) and regions enriched up to 8-fold in DNA over RNAP in the center of the nucleoid (blue regions). (B) The histogram showing the distribution of apparent RNAP-Venus foci in fast-growing cells. The red line indicates the median number of transcription foci in the population of cells.
An RNAP/DNA overlay indicated that high intensities signals for RNAP and DNA were located at different locations in the nucleoids of a fast-growing cell (Figure 1A, RNAP/DNA overlay). The inverse relationship between the signal intensities of RNAP and DNA was also clearly visualized with the normalized log2 (RNAP/DNA) density plot or heat map, in which the ratios of intensities between pairs of pixels from the two channels were determined [Figure 1A, log2 (RNAP/DNA)]. In this heat map, warmer colors (yellow to red) represent the areas being enriched with RNAP over DNA; conversely, colder colors (yellow to dark blue) indicate the areas enriched with DNA over RNAP. There is a clear clustering of pixels indicating regions enriched in RNAP over DNA (red color) and vice versa (blue color). As expected, the transcription foci show an enrichment of RNAP over DNA approaching 10-fold, whereas the high-density DNA regions show an enrichment of DNA over RNAP, also close to 10-fold, in the cell. Together, our results indicate that the RNAP and DNA signals are not homogenously distributed in the nucleoid, but rather in large, mutually exclusive organizations and that the transcription machinery is located in compartments at the periphery of the nucleoid in a fast-growing cell.
The rrn antitermination system (NusA and NusB) co-localizes with transcription foci
RNAP forms prominent transcription foci for active rRNA synthesis in fast-growing E. coli cells (3,4). To determine the composition of transcription machinery in fast-growing cells we chose two Nus factors. NusA and NusB are components of the rrn antitermination system and are likely to locate at positions where active rRNA synthesis occurs. We constructed NusA-mCherry and NusB-mCherry fusions at the original chromosomal loci in the CC72 strain that has the chromosomal rpoC-venus fusion and examined their relationship with RNAP. Strains with these fusions have wild-type growth phenotypes. These constructs enabled us to simultaneously image NusA or NusB along with RNAP and the nucleoid in fast-growing cells.
Figure 2A shows the images of NusA, nucleoid and RNAP in a typical fast-growing cell. The distribution of NusA is very similar to that of RNAP:NusA forms foci and it locates at the nucleoid and its surroundings (DNA/NusA overlay). The co-localization of NusA-mCherry foci and transcription (RNAP-Venus) foci is apparent from the RNAP/NusA overlay. The normalized log2 (RNAP/NusA) heat map from the signal intensities of RNAP and NusA also clearly showed a similar distribution pattern between RNAP and NusA (Figure 2A), as indicated by an overall yellow color of the heat map, in contrast to the log2 (RNAP/DNA) heat map described above (Figure 1A). There were seven NusA foci per cell on average (Figure 2B), a value that is similar to the RNAP foci in fast-growing cells (Figure 1). To quantify the co-localization of the NusA foci with the RNAP foci in fast-growing cells, we performed foci co-localization analysis in a population of cells (>100 cells) by measuring the distance between their weighted centroids (see the Materials and Methods section). Our measurements showed that on average, 86% of NusA foci are co-localized with at least one RNAP focus in the cells (Figure 2C), whereas a random data set showed an average of 0%. Additionally, the cumulative distribution of the distance between NusA foci and RNAP foci showed an unequivocal co-localization of both proteins with 82.3% of them being within 140 nm of each other (Figure 2D), while the distribution of the random data set showed essentially no co-localization (1.9%) between the two proteins.
Figure 2.
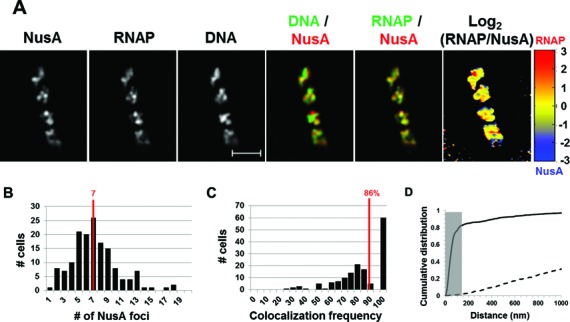
NusA involved in rRNA transcription antitermination forms foci and co-localizes with transcription foci in fast-growing cells. (A) Images of NusA, RNAP, DNA (nucleoid), overlays of NusA (red) and DNA (green), RNAP (green) and NusA (red) and log2 (RNAP/NusA) (heat map) from a typical fast-growing E. coli cell as described in the legend to Figure 1. Note that NusA foci are at the periphery of the nucleoid and that NusA signals perfectly co-localize with RNAP signals (overall yellow color on the RNAP/NusA overlay and on the heat map). (B) The histogram depicting the distribution of apparent NusA-mCherry foci in fast-growing cells. The red line indicates the median number of NusA foci in the population of cells. Note that the median number of NusA foci is close to that of transcription foci in fast-growing cells. (C) Co-localization frequency between the NusA foci and transcription foci in fast-growing cells. The red line indicates the median co-localization frequency of NusA foci with transcription foci in the population of cells as described in the Materials and Methods section. Most (86%) of the NusA-mCherry foci co-localize with at least one of the transcription foci in the cells. The average co-localization frequency of NusA foci and RNAP foci from a random data set is 0%. (D) Cumulative distribution of the distances between NusA foci and their closest RNAP foci in the population of cells as described in the Materials and Methods section. (──) NusA-mCherry RNAP-Venus and (- - -) NusA-mCherry RNAP-Venus random. The gray rectangle represents the co-localization area (≤140 nm). 82.3% of the NusA foci are within 140 nm of the closest RNAP foci. Only the distances ranging from 0 to 1000 nm were included in the plot to avoid scaling issue and increase the readability.
Similarly, the distribution of NusB mimicked that of RNAP in a fast-growing cell (Figure 3A). The overlay of RNAP/NusB revealed an almost complete co-localization of the NusB-mCherry foci and transcription foci (RNAP-Venus) in the cell. The normalized log2 (RNAP/NusB) heat map also exhibited homogenous distribution between the signals of RNAP and NusB, similar to the relationship between RNAP and NusA. There were six NusB foci per cell on average (Figure 3B), a value that is close to that of RNAP foci and NusA foci. Foci co-localization analysis of a population of cells showed that on average, 100% of NusB foci are co-localized with at least one RNAP focus under this optimal growth condition (Figure 3C), and a random data set showed an average of 0%. Similar to NusA, the cumulative distribution of NusB foci and RNAP foci also exhibited almost perfect co-localization with 87.1% of the foci of NusB and RNAP being co-localized (Figure 3D). The random data set showed very poor co-localization (4.6%). Note that the transcription foci or the co-localized foci of RNAP/NusA/NusB have different sizes: while most are large, some are smaller, and few are punctate spots. We postulate that the apparent heterogeneity of the foci, which are fortuitously captured in the images, reflects the dynamic nature of transcription machinery in fast-growing cells where DNA replication is active.
Figure 3.
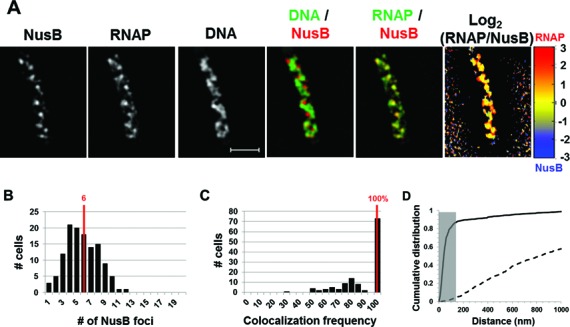
Nascent rRNA-binding protein NusB forms foci and co-localizes with transcription foci in fast-growing cells. (A) Images of NusB, RNAP, DNA (nucleoid), overlays of NusB (red) and DNA (green), RNAP (green) and NusB (red) and log2 (RNAP/NusB) (heat map) from a representative fast-growing E. coli cell as described in the legend to Figure 1. Similar to NusA, NusB foci are at the periphery of the nucleoid and that NusB signals perfectly co-localize with RNAP signals (overall yellow color on the RNAP/NusB overlay and on the heat map). (B) The distribution of apparent NusB-mCherry foci in fast-growing cells. The red line in the histogram indicates the median number of NusB foci in the population of cells. Note that the median number of NusB foci is close to that of NusA or transcription foci in fast-growing cells. (C) Co-localization frequency between the NusB foci and transcription foci in fast-growing cells. The red line indicates the median co-localization frequency of NusB foci with transcription foci in the population of cells as described in the Materials and Methods section. Almost all of the NusB-mCherry foci co-localize with at least one transcription foci in the cells. The average co-localization frequency of NusB foci and RNAP foci from a random data set is 0%. (D) Cumulative distribution of the distances between NusB foci and their closest RNAP foci in the population of cells as described in the Materials and Methods section. (──) NusB-mCherry RNAP-Venus and (- - -) NusB-mCherry RNAP-Venus random. The gray rectangle represents the co-localization area (≤140 nm). 87.1% of the NusB foci are within 140 nm of the closest transcription foci.
Together, our results show significant foci co-localization of RNAP with both Nus factors involved in the rrn antitermination system, thus demonstrating that the two Nus factors are integral components of transcription machinery engaged in active rRNA synthesis in fast-growing cells. Importantly, these foci numbers are far fewer than the ∼ 50 copy numbers of rrn operons per cell calculated from the growth rate (6), further supporting the notion that these larger foci of transcription machinery are located at clusters of rrn operons or nucleolus-like structures (3,4).
Spatial segregation of transcription and replication machineries in fast-growing cells
Having found spatial compartmentalization of transcription machinery in the nucleoid, we wished to determine the spatial relationship between transcription and replication machineries in fast-growing cells using SIM. We hypothesized that one effective way to maintain the harmony between replication and transcription machineries is to have spatial segregation of these two important cellular functions in a fast-growing cell. To test this hypothesis, we made two new constructs in the parental strain with the RNAP-Venus fusion background. One resulting strain contained an additional chromosomal SeqA-mCherry fusion and the other an SSB-mCherry fusion. SeqA foci or SSB foci serve as a proxy for replisome localization in the cell. While SeqA, with potentially several hundred molecules, binds nascent hemimethylated DNA behind each replication fork (26,28,31,46), SSB coats the single-stranded DNA at each replication fork and interacts with the DNA polymerase (24,25).
Images from SIM co-imaging of RNAP, SeqA and DNA in a typical fast-growing cell (LB, 37°C) are shown in Figure 4A. The overlay of SeqA and DNA showed two to three SeqA foci in each of the four nascent nucleoids in the cell, consistent with previous reports stating that each nascent nucleoid contains multiple pairs of replication forks in fast-growing cells in LB (5,47). The RNAP/SeqA overlay also revealed that the locations of SeqA foci tend to separate from those of transcription foci. This feature is also manifested in the normalized log2 (RNAP/SeqA) heat map (Figure 4A), in which RNAP foci (warm color) appear to be separated from SeqA foci (cold color). Although in this study we focused on the visualization of the spatial organization of transcription and replication machineries in fast-growing cells, the overlay of DNA and SeqA (Figure 4A) revealed that SeqA foci tend to separate from high intensities of DNA signals, indicating that SeqA foci are also mainly segregated from the mass of DNA in the nucleoids in the cell.
Figure 4.
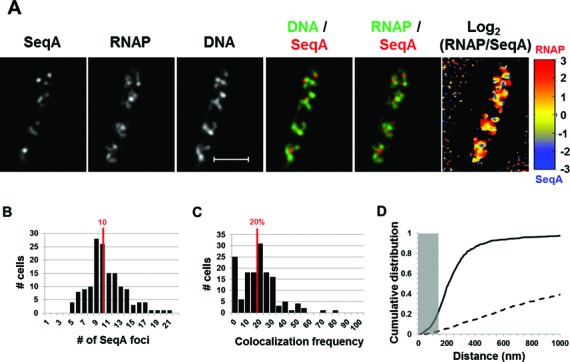
Spatial segregation of transcription foci and replication forks tracked by SeqA in fast-growing cells. (A) Images of SeqA, RNAP, DNA (nucleoid), overlays of SeqA (red) and DNA (green), RNAP (green) and SeqA (red) and log2 (RNAP/SeqA) (heat map) from a representative fast-growing E. coli cell as described in the legend to Figure 1. SeqA foci are largely located at different positions from transcription foci (red and green colors on the overlay of SeqA and RNAP and red and blue color on the heat map). Note also that SeqA foci appear mainly to be separated from high intensities of DNA signals in the nucleoids (red and green colors on the overlay of SeqA and DNA). (B) The distribution of apparent SeqA-mCherry foci in a population of fast-growing cells. The red line in the histogram indicates the median number of SeqA foci in those cells. (C) Co-localization frequency between the SeqA foci and transcription foci in fast-growing cells. The red line indicates the median co-localization frequency of SeqA foci with transcription foci in the population of cells as described in the Materials and Methods section. Majority of SeqA foci are separated from transcription foci in the fast-growing cells. The average co-localization frequency of SeqA foci and RNAP foci from a random data set is 0%. (D) Cumulative distribution of the distances between SeqA foci and their closest RNAP foci in the population of cells as described in the Materials and Methods section. (──) SeqA-mCherry RNAP-Venus and (- - -) SeqA-mCherry RNAP-Venus random. The gray rectangle represents the co-localization area (≤140 nm). Only 21.5% of the SeqA foci are within 140 nm of the closest transcription foci.
Images analysis of a population of fast-growing cells showed that on average, each cell contains 10 SeqA foci (Figure 4B). Considering that the SIM has a higher resolution, this value is consistent with the previous reported numbers of SeqA foci in fast-growing cells by wide-field fluorescence microscopy (29,48). The co-localization efficiency of SeqA foci with transcription foci in the population of cells was determined and found that the clustering frequency of SeqA and RNAP foci is low, with an average of 20% (Figure 4C), confirming that foci of SeqA and RNAP are largely segregated in fast-growing cells. The same analysis performed on the random data sets showed no co-localization (0%). Additionally, the cumulative distribution of the distance between SeqA foci and RNAP foci (Figure 4D) revealed that despite the low co-localization frequency (21.5%) the spatial relationship between the foci of two proteins is not random (3% of co-localization). Interestingly, most of the SeqA foci (≥90%) were constrained within 470 nm of the closest transcription foci. To resolve whether the observed spatial segregation of transcription and replication machineries is correlated with cell cycle parameters, we also performed an analysis to determine whether there is a relationship between the number of SeqA foci per cell and the co-localization frequency of SeqA and RNAP foci, but found no such relationship (r = 0.02).
SSB behaved similarly as SeqA in a typical fast-growing cell (Figure 5A). The spatial segregation of RNAP foci and SSB foci was manifested in the overlay of RNAP and SSB and the quantitative log2 (RNAP/SeqA) heat map. SSB foci also appeared to be located mainly at nucleoid regions containing low intensities of DNA signals (DNA/SSB overlay). Also similar to the distribution of SeqA foci in a population of fast-growing cells, on average there were 11 SSB foci per cell, a value that corresponds to two or three SSB foci in each of the four nascent nucleoids in a cell (Figure 5B). In addition, on average only 16% of SSB foci are co-localized with RNAP foci in the population of cells (Figure 5C), which is above the random (0%). Similar to SeqA, the cumulative distribution of distance between foci of SSB and RNAP (Figure 5D) showed a poor non-random co-localization with only 22.3% being co-localized (4.3% for random). Note also that the vast majority (≥90%) of SSB foci are constrained within 670 nm of the closest RNAP foci. Additionally, no relationship between the number of SSB foci per cell and the co-localization frequency of SSB and RNAP foci was found (r = 0.04). Thus, SSB foci and transcription foci are also mostly spatially segregated in fast-growing cells. Together, our results show that in contrast to the NusA/B foci that are co-localized with RNAP foci, there is an apparent spatial segregation of transcription and replication machineries in fast-growing cells.
Figure 5.
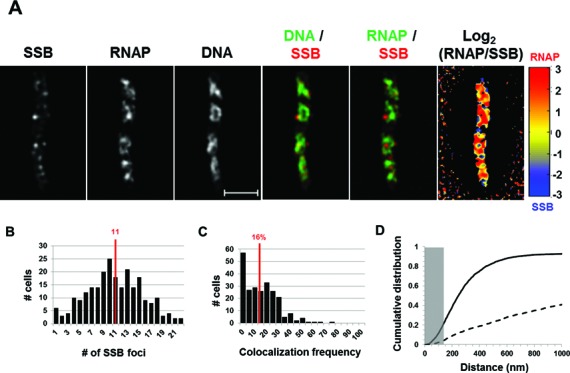
Spatial segregation of transcription foci and replisomes tracked by SSB. (A) Images of SSB, RNAP, DNA (nucleoid), overlays of SSB (red) and DNA (green), RNAP (green) and SSB (red) and log2 (RNAP/SSB) (heat map) from a typical fast-growing E. coli cell as described in the legend to Figure 1. SSB foci tend to separate from transcription foci (red and green colors on the overlay of SSB and RNAP and red and blue color on the heat map). Note also that SSB foci seem largely to be separated from high intensities of DNA signals in the nucleoids (red and green colors on the overlay of SSB and DNA). (B) The distribution of apparent SSB-mCherry foci in fast-growing cells. The red line in the histogram indicates the median number of SSB foci in the population of cells. Note that the median number of SSB foci is close to that of SeqA foci in fast-growing cells. (C) Co-localization frequency between the SSB foci and transcription foci in fast-growing cells. The red line indicates the median co-localization frequency of SSB foci with transcription foci in the population of cells as described in the Materials and Methods section. Most of the SSB foci are segregated from transcription foci in fast-growing cells. The average co-localization frequency of SSB foci and RNAP foci from a random data set is 0%. (D) Cumulative distribution of the distances between SSB foci and their closest RNAP foci in the population of cells as described in the Materials and Methods section. (──) SSB-mCherry RNAP-Venus and (- - -) SSB-mCherry RNAP-Venus random. The gray rectangle represents the co-localization area (≤140 nm). Only 22.3% of the SSB foci are within 140 nm of the closest RNAP foci.
DISCUSSION
In this work, we used SIM to focus on the spatial organization of transcription machinery, the composition of transcription machinery and its relationship with the replisome in a fast-growing E. coli cell. There are mainly three findings from this study: (i) NusA and NusB are integral components of transcription machinery engaged in active rRNA synthesis; (ii) transcription machinery is spatially organized into functional compartments, indication of an important functional landscape in bacterial chromosome for maximal rRNA synthesis; and (iii) transcription and replication machineries are spatially segregated, indicating an important mechanism whereby the two cellular functions maintain harmony and avoid conflicts. Our study indicates that super-resolution co-imaging of DNA and components of cellular functions is an effective technique to determine spatial and functional organization of bacterial chromosome.
Composition and spatial organization of transcription machinery in fast-growing cells
Although it has been known that NusA and NusB are important for the rrn antitermination function in E. coli genetically and biochemically, it remained undetermined whether the two Nus factors are associated with transcription foci in fast-growing E. coli cells, in which rRNA synthesis is most active. Our study provides the first cell biology-based evidence that the two Nus factors are integral components of the transcription machinery because their foci co-localize with RNAP foci with a frequency of 86–100% in fast-growing cells. Our results indicate that NusB binds to nascent pre-rRNAs cotranscriptionally and intimately associates with transcription foci in fast-growing cells. Our results thus support the notion that transcription machinery forms foci for rRNA synthesis at clusters of rrn operons or bacterial nucleolus, thus functioning as specialized transcription machinery in fast-growing cells. Identification of other components or factors in the transcription machinery of fast-growing cells is warranted for future studies. Under the optimal growth condition, there are up to four nascent nucleoids per cell, and each nascent nucleoid has, on average, about two foci of the transcription machinery, indicating that transcription machinery is duplicated and distributed during DNA replication and segregation.
During replication of rrn operons, it is conceivable that transcription foci are somehow disassembled allowing replication forks to pass through the region and followed by reassembly of transcription machinery at rrn clusters. This proposition is suggested by the apparent heterogeneity of the foci of RNAP/NusA/NusB and by the poor but above random co-localization of transcription and replication machineries. Although formation of transcription foci does not happen concomitantly with cell division, newly divided daughter cells will ‘inherit’ the functional machinery without the need for de novo synthesis and/or assembly. Because transcription foci at nucleoid-like structures are critical for growth rate regulation in E. coli (2), this feature can explain, in part, why newly divided daughter cells maintain the same fast growth rate as the parental cells.
Co-imaging of RNAP with DNA using SIM enables us to visualize the spatial relationship between transcription machinery and bacterial chromosome. Our results show that transcription foci are located at the nucleoid surface where DNA density is low, possibly in DNA loops containing clusters of rrn operons or bacterial nucleolus. This spatial organization of the transcription machinery has logistical advantages, such as enhancing the coupling of rrn transcription and ribosomal assembly, and possibly channeling nucleoside triphosphates (NTPs) effectively in the regions. It would be interesting to determine experimentally whether different rrn operons located at different parts of genome are clustered with the transcription machinery, and other hyperstructures (49), such as nucleotide biosynthesis machinery, are also located near the transcription machinery in fast-growing cells. In addition, because highly expressed genes are in a highly interactive environment and present in large clusters (32), transcription foci are likely to be networking hubs for the transcription of growth-promoting genes, most of which are rrn operons, in fast-growing cells (3). The spatial compartmentation of transcription foci also suggests that these foci could be used as RNAP pools to effectively interact with DNA loops containing non-rrn growth-promoting genes located a distance away by hopping three-dimensionally (50), rather than by traveling linearly or laterally. Such an organization would reduce the transcription traffic jam and allow for maximum use of limited RNAP in the cell (2). It would be interesting to identify other non-rrn genes and RNAs in these potential transcription hubs.
Segregated functional chromosomal territories for transcription and replication machineries in fast-growing cells
We also studied the spatial relationship between transcription and replication machineries in fast-growing cells by co-imaging RNAP with SSB, which is an integral component of replisomes, or SeqA, which tracks replication forks. Our results show that SeqA and SSB behave similarly; there are multiple SSB or SeqA foci in each of the nascent nucleoids, reflecting the presence of multiple pairs of replication forks in a fast-growing cell. Remarkably, transcription and replication machineries are largely spatially segregated, possibly into functional chromosomal territories; thus, our study reveals an important mechanism for maintaining harmony between transcription and replication in fast-growing cells. Our finding could explain the reported observations based on Chip-chip analysis of cells grown in LB that genome-wide regions with strong RNAP binding signals such as the rRNA operons are not bound by SeqA (51). The spatial segregation of the two machineries is not absolute because SeqA or SSB foci partially (∼16–22%) associate with RNAP foci in fast-growing cells. This partial co-localization between transcription and replication machineries could be attributed to the 2D measurements of 3D cells. In addition, transcription machinery and replisomes are dynamic; locally the two machineries will likely intercept and resolve at some point of the cell cycle. While foci of SeqA and SSB are maintained after inhibition of transcription and other cellular perturbations (24,48), transcription foci disappear under those conditions because they are extremely sensitive to environmental cues which affect growth rate and/or physiology (3,4,21,36). Thus, development of fast super-resolution time-lapse live-cell imaging techniques will be necessary to address the dynamic interaction and segregation of the two active cellular functions in fast-growing cells.
Although it was not the focus of this study, co-imaging of SeqA (or SSB) and DNA revealed that replication machinery appears to be mainly located at regions of nascent nucleoids containing low intensities of DNA in a fast-growing cell, suggesting that replisomes also locate at the surface of the nucleoids. We speculate that replisomes are also spatially organized into compartmentation in bacterial nucleoid, similarly to transcription machinery but located in different locations. Because both replication and transcription machineries are likely associated with DNA loops at the nucleoid peripheries, it follows that their spatial segregation is constrained by the perimeter of a single nascent nucleoid. A fast growing E. coli cell on average is ∼5 μm in length and has 4 nascent nucleoids with their cumulative length being ∼50% of the cell length (20); thus, the size of each nascent nucleoid is estimated to be ∼500–650 nm, a value very close to the distance constraints we observed between the transcription and replication machineries in the cumulative distribution plots. Further research is needed to study spatial and functional organization of replication machinery during the cell cycle in fast-growing E. coli cells, in which cell cycle parameters are much more complicated (1) than in slow-growing cells (doubling time >100 min) (24,52,53).
In summary, our study shows the first SIM images revealing that transcription machinery is spatially organized into functional compartments and that transcription and replication machineries are largely spatially segregated in the nucleoids of fast-growing E. coli cells. These features are biologically significant. Like eukaryotes, in which functional chromosomal territories are important for chromosome biology (54–56), our study demonstrates that spatial and functional compartmentation and segregation are also conserved in bacterial chromosome biology. Thus, E. coli will be an attractive model system to study chromosome biology in cells during growth and stress responses.
Acknowledgments
We thank Thomas Schneider (NCI) for many helpful discussions during the course of the study, and acknowledge the support from the Optical Microscopy and Analysis Laboratory (Leidos Biomedical Research) for the SIM imaging system. We also thank Bo Xiao for his participation in the Matlab scripts and Jerome Izard for comments on the manuscript.
FUNDING
The Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stokke C., Flatten I., Skarstad K. An easy-to-use simulation program demonstrates variations in bacterial cell cycle parameters depending on medium and temperature. PLoS One. 2012;7:e30981. doi: 10.1371/journal.pone.0030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin D.J., Cagliero C., Zhou Y.N. Growth rate regulation in Escherichia coli. FEMS Microbiol. Rev. 2012;36:269–287. doi: 10.1111/j.1574-6976.2011.00279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin D.J., Cagliero C., Zhou Y.N. Role of RNA polymerase and transcription in the organization of the bacterial nucleoid. Chem. Rev. 2013;113:8662–8682. doi: 10.1021/cr4001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabrera J.E., Jin D.J. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol. Microbiol. 2003;50:1493–1505. doi: 10.1046/j.1365-2958.2003.03805.x. [DOI] [PubMed] [Google Scholar]
- 5.Nielsen H.J., Youngren B., Hansen F.G., Austin S. Dynamics of Escherichia coli chromosome segregation during multifork replication. J. Bacteriol. 2007;189:8660–8666. doi: 10.1128/JB.01212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bremer H., Dennis P.P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella. Vol. 2. Washington, DC: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- 7.Endesfelder U., Finan K., Holden S.J., Cook P.R., Kapanidis A.N., Heilemann M. Multiscale spatial organization of RNA polymerase in Escherichia coli. Biophys. J. 2013;105:172–181. doi: 10.1016/j.bpj.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quan S., Zhang N., French S., Squires C.L. Transcriptional polarity in rRNA operons of Escherichia coli nusA and nusB mutant strains. J. Bacteriol. 2005;187:1632–1638. doi: 10.1128/JB.187.5.1632-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Squires C.L., Greenblatt J., Li J., Condon C. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc. Natl Acad. Sci. U.S.A. 1993;90:970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan E.A. Antitermination mechanisms in rRNA operons of Escherichia coli. J. Bacteriol. 1986;168:1–5. doi: 10.1128/jb.168.1.1-5.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Court D.L., Oppenheim A.B., Adhya S.L. A new look at bacteriophage lambda genetic networks. J. Bacteriol. 2007;189:298–304. doi: 10.1128/JB.01215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S.C., Squires C.L., Squires C. Antitermination of E. coli rRNA transcription is caused by a control region segment containing lambda nut-like sequences. Cell. 1984;38:851–860. doi: 10.1016/0092-8674(84)90280-0. [DOI] [PubMed] [Google Scholar]
- 13.Torres M., Balada J.M., Zellars M., Squires C., Squires C.L. In vivo effect of NusB and NusG on rRNA transcription antitermination. J. Bacteriol. 2004;186:1304–1310. doi: 10.1128/JB.186.5.1304-1310.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenblatt J., Li J. Interaction of the sigma factor and the nusA gene protein of E. coli with RNA polymerase in the initiation-termination cycle of transcription. Cell. 1981;24:421–428. doi: 10.1016/0092-8674(81)90332-9. [DOI] [PubMed] [Google Scholar]
- 15.Stagno J.R., Altieri A.S., Bubunenko M., Tarasov S.G., Li J., Court D.L., Byrd R.A., Ji X. Structural basis for RNA recognition by NusB and NusE in the initiation of transcription antitermination. Nucleic Acids Res. 2011;39:7803–7815. doi: 10.1093/nar/gkr418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greive S.J., Lins A.F., von Hippel P.H. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. J. Biol. Chem. 2005;280:36397–36408. doi: 10.1074/jbc.M507146200. [DOI] [PubMed] [Google Scholar]
- 17.Nodwell J.R., Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993;72:261–268. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- 18.Davies K.M., Dedman A.J., van Horck S., Lewis P.J. The NusA:RNA polymerase ratio is increased at sites of rRNA synthesis in Bacillus subtilis. Mol. Microbiol. 2005;57:366–379. doi: 10.1111/j.1365-2958.2005.04469.x. [DOI] [PubMed] [Google Scholar]
- 19.Doherty G.P., Meredith D.H., Lewis P.J. Subcellular partitioning of transcription factors in Bacillus subtilis. J. Bacteriol. 2006;188:4101–4110. doi: 10.1128/JB.01934-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabrera J.E., Cagliero C., Quan S., Squires C.L., Jin D.J. Active transcription of rRNA operons condenses the nucleoid in Escherichia coli: examining the effect of transcription on nucleoid structure in the absence of transertion. J. Bacteriol. 2009;191:4180–4185. doi: 10.1128/JB.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin D.J., Cabrera J.E. Coupling the distribution of RNA polymerase to global gene regulation and the dynamic structure of the bacterial nucleoid in Escherichia coli. J. Struct. Biol. 2006;156:284–291. doi: 10.1016/j.jsb.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Yao N.Y., O'Donnell M. SnapShot: the replisome. Cell. 2010;141:1088. doi: 10.1016/j.cell.2010.05.042. 1088.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Donnell M. Replisome architecture and dynamics in Escherichia coli. J. Biol. Chem. 2006;281:10653–10656. doi: 10.1074/jbc.R500028200. [DOI] [PubMed] [Google Scholar]
- 24.Reyes-Lamothe R., Possoz C., Danilova O., Sherratt D.J. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marceau A.H., Bahng S., Massoni S.C., George N.P., Sandler S.J., Marians K.J., Keck J.L. Structure of the SSB-DNA polymerase III interface and its role in DNA replication. EMBO J. 2011;30:4236–4247. doi: 10.1038/emboj.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu M., Campbell J.L., Boye E., Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 27.Waldminghaus T., Skarstad K. The Escherichia coli SeqA protein. Plasmid. 2009;61:141–150. doi: 10.1016/j.plasmid.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Waldminghaus T., Weigel C., Skarstad K. Replication fork movement and methylation govern SeqA binding to the Escherichia coli chromosome. Nucleic Acids Res. 2012;40:5465–5476. doi: 10.1093/nar/gks187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiraga S., Ichinose C., Niki H., Yamazoe M. Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA-protein complexes in E. coli. Mol. Cell. 1998;1:381–387. doi: 10.1016/s1097-2765(00)80038-6. [DOI] [PubMed] [Google Scholar]
- 30.Yamazoe M., Adachi S., Kanaya S., Ohsumi K., Hiraga S. Sequential binding of SeqA protein to nascent DNA segments at replication forks in synchronized cultures of Escherichia coli. Mol. Microbiol. 2005;55:289–298. doi: 10.1111/j.1365-2958.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 31.Brendler T., Sawitzke J., Sergueev K., Austin S. A case for sliding SeqA tracts at anchored replication forks during Escherichia coli chromosome replication and segregation. EMBO J. 2000;19:6249–6258. doi: 10.1093/emboj/19.22.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cagliero C., Grand R.S., Jones M.B., Jin D.J., O'Sullivan J.M. Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription. Nucleic Acids Res. 2013;41:6058–6071. doi: 10.1093/nar/gkt325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merrikh H., Zhang Y., Grossman A.D., Wang J.D. Replication-transcription conflicts in bacteria. Nat. Rev. Microbiol. 2012;10:449–458. doi: 10.1038/nrmicro2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlton P.M. Three-dimensional structured illumination microscopy and its application to chromosome structure. Chromosome Res. 2008;16:351–365. doi: 10.1007/s10577-008-1231-9. [DOI] [PubMed] [Google Scholar]
- 35.Langhorst M.F., Schaffer J., Goetze B. Structure brings clarity: structured illumination microscopy in cell biology. Biotechnol. J. 2009;4:858–865. doi: 10.1002/biot.200900025. [DOI] [PubMed] [Google Scholar]
- 36.Cagliero C., Jin D.J. Dissociation and re-association of RNA polymerase with DNA during osmotic stress response in Escherichia coli. Nucleic Acids Res. 2013;41:315–326. doi: 10.1093/nar/gks988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller J.H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 39.Canny J. A computational approach to edge-detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986;8:679–698. [PubMed] [Google Scholar]
- 40.Meyer F. Topographic distance and watershed lines. Signal Process. 1994;38:113–125. [Google Scholar]
- 41.Bolte S., Cordelieres F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 42.Nicolas E., Upton A.L., Uphoff S., Henry O., Badrinarayanan A., Sherratt D. The SMC complex MukBEF recruits topoisomerase IV to the origin of replication region in live Escherichia coli. MBio. 2014;5:e01001–e01013. doi: 10.1128/mBio.01001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakshi S., Siryaporn A., Goulian M., Weisshaar J.C. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol. Microbiol. 2012;85:21–38. doi: 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spahn C., Endesfelder U., Heilemann M. Super-resolution imaging of Escherichia coli nucleoids reveals highly structured and asymmetric segregation during fast growth. J. Struct. Biol. 2014;185:243–249. doi: 10.1016/j.jsb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Margolin W. Imaging the Bacterial Nucleoid. In: Dame RT, Dorman CJ, editors. In Bacterial Chromatin. Heidelberg: Springer Dordrecht; 2012. pp. 13–30. [Google Scholar]
- 46.Onogi T., Niki H., Yamazoe M., Hiraga S. The assembly and migration of SeqA-Gfp fusion in living cells of Escherichia coli. Mol. Microbiol. 1999;31:1775–1782. doi: 10.1046/j.1365-2958.1999.01313.x. [DOI] [PubMed] [Google Scholar]
- 47.Morigen, Odsbu I., Skarstad K. Growth rate dependent numbers of SeqA structures organize the multiple replication forks in rapidly growing Escherichia coli. Genes Cells. 2009;14:643–657. doi: 10.1111/j.1365-2443.2009.01298.x. [DOI] [PubMed] [Google Scholar]
- 48.Molina F., Skarstad K. Replication fork and SeqA focus distributions in Escherichia coli suggest a replication hyperstructure dependent on nucleotide metabolism. Mol. Microbiol. 2004;52:1597–1612. doi: 10.1111/j.1365-2958.2004.04097.x. [DOI] [PubMed] [Google Scholar]
- 49.Norris V., den Blaauwen T., Cabin-Flaman A., Doi R.H., Harshey R., Janniere L., Jimenez-Sanchez A., Jin D.J., Levin P.A., Mileykovskaya E., et al. Functional taxonomy of bacterial hyperstructures. Microbiol. Mol. Biol. Rev. 2007;71:230–253. doi: 10.1128/MMBR.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang F., Redding S., Finkelstein I.J., Gorman J., Reichman D.R., Greene E.C. The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion. Nat. Struct. Mol. Biol. 2013;20:174–181. doi: 10.1038/nsmb.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez-Romero M.A., Busby S.J., Dyer N.P., Ott S., Millard A.D., Grainger D.C. Dynamic distribution of seqa protein across the chromosome of escherichia coli K-12. MBio. 2010;1 doi: 10.1128/mBio.00012-10. doi:10.1128/mBio.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bates D., Epstein J., Boye E., Fahrner K., Berg H., Kleckner N. The Escherichia coli baby cell column: a novel cell synchronization method provides new insight into the bacterial cell cycle. Mol. Microbiol. 2005;57:380–391. doi: 10.1111/j.1365-2958.2005.04693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen H.J., Li Y., Youngren B., Hansen F.G., Austin S. Progressive segregation of the Escherichia coli chromosome. Mol. Microbiol. 2006;61:383–393. doi: 10.1111/j.1365-2958.2006.05245.x. [DOI] [PubMed] [Google Scholar]
- 54.Meaburn K.J., Misteli T. Cell biology: chromosome territories. Nature. 2007;445:379–781. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- 55.Cremer T., Cremer M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cavalli G., Misteli T. Functional implications of genome topology. Nat. Struct. Mol. Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]


