Abstract
Alkylative damage to DNA can be induced by environmental chemicals, endogenous metabolites and some commonly prescribed chemotherapeutic agents. The regioisomeric N3-, O2- and O4-ethylthymidine (N3-, O2- and O4-EtdT, respectively) represent an important class of ethylated DNA lesions. Using nonreplicative double-stranded vectors containing an N3-EtdT, O2-EtdT or O4-EtdT at a defined site in the template strand, herein we examined the effects of these lesions on DNA transcription mediated by single-subunit T7 RNA polymerase or multisubunit human RNA polymerase II in vitro and in human cells. We found that O4-EtdT is highly mutagenic and exclusively induces the misincorporation of guanine opposite the lesion, whereas N3-EtdT and O2-EtdT display promiscuous miscoding properties during transcription. In addition, N3-EtdT and O2-EtdT were found to inhibit strongly DNA transcription in vitro and in certain human cells. Moreover, N3-EtdT, but not O2-EtdT or O4-EtdT, is an efficient substrate for transcription-coupled nucleotide excision repair. These findings provide new important insights into how these alkylated DNA lesions compromise the flow of genetic information, which may help to understand the risk of these lesions in living cells.
INTRODUCTION
Environmental and endogenous genotoxic factors can result in a variety of alkylated DNA lesions, which represent a major class of DNA lesions (1). These agents include exogenous compounds like tobacco-specific nitrosamines, N-ethyl-N-nitrosourea and polycyclic aromatic hydrocarbons, as well as endogenous biochemical agents such as S-adenosyl-L-methionine, nitrosated peptides and polyamines (1–6). Hence, alkylative damage to DNA is an unavoidable consequence of endogenous metabolism and environmental exposure (1). Major alkylation sites in DNA include phosphate groups, ring nitrogens and exocyclic oxygen atoms of DNA bases, such as N3 of adenine, O6 and N7 of guanine, and N3, O2 and O4 of thymine (7–13).
Many alkylated DNA lesions have been readily detected in various human tissues and urine samples (8–10,13,14). In particular, the levels of certain ethylated DNA adducts, including N3-, O2- and O4-ethylthymidine (N3-EtdT, O2-EtdT and O4-EtdT) (Figure 1a), were shown to be significantly higher in leukocyte and saliva DNA of smokers than nonsmokers (9,14). In this vein, it was reported that the levels of N3-EtdT, O2-EtdT and O4-EtdT in smokers’ leukocyte DNA were ∼41.1, ∼44.8 and ∼48.3 lesions per 108 nucleosides, respectively, whereas those in nonsmokers were ∼4.1, ∼0.2 and ∼1.0 lesions per 108 nucleosides, respectively (9). In addition, N3-EtdT, O2-EtdT and O4-EtdT were found to be present in smokers’ saliva DNA at frequencies of ∼4.5, ∼5.3 and ∼4.2 lesions per 108 nucleosides, respectively, but none of these lesions were detectable in saliva DNA of nonsmokers (14). Therefore, these three EtdT lesions have been proposed as potential useful biomarkers for exposure to ethylating agents and for cancer risk assessment (9,14).
Figure 1.
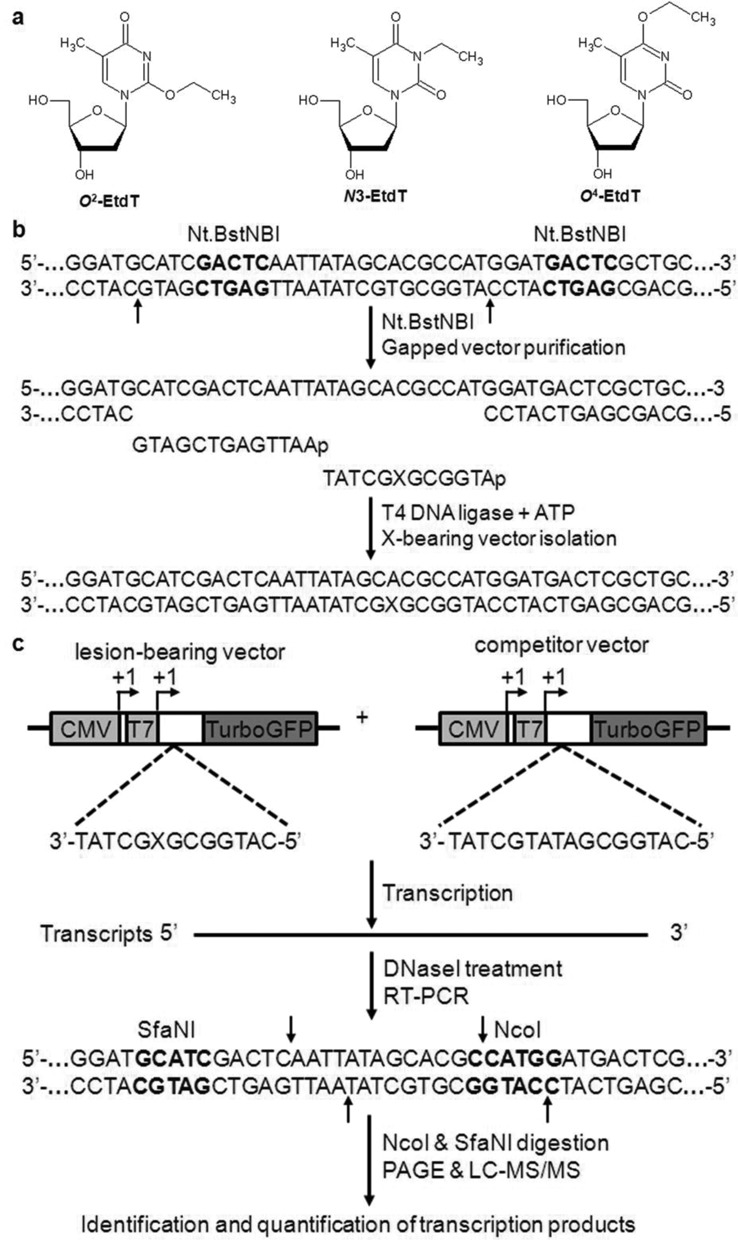
Experimental outline. (a) Chemical structures of N3-EtdT, O2-EtdT and O4-EtdT. (b) Schematic diagrams showing the procedures for the construction of the plasmids harboring a site-specifically incorporated N3-EtdT, O2-EtdT or O4-EtdT. (c) Strategy for assessing the impact of the EtdT lesions on DNA transcription. ‘X’ indicates N3-EtdT, O2-EtdT or O4-EtdT, which was located on the transcribed strand of TurboGFP gene downstream of the CMV and T7 promoters. The +1 transcription start sites are indicated by arrowheads. Lesion-bearing or lesion-free control plasmids were mixed with the competitor vector as DNA templates for in vitro or in vivo transcription assay. Although truncated RNA may be produced when transcription arrests at or near a lesion site, only run-off RNA is shown and used for RT-PCR. Among the RT-PCR products, only the wild-type sequence arising from the lesion-containing vector is shown. The arrows indicate the cleavage sites of Nt.BstNBI, NcoI and SfaNI. The last two enzymes are used to digest the RT-PCR products for subsequent PAGE and LC-MS/MS analyses.
Understanding how DNA lesions perturb the flow of genetic information during DNA replication and transcription may help to understand the risk of these lesions in living cells (15–18). In this context, it has been shown that O4-EtdT is highly mutagenic during DNA replication and it is associated with cancer development in animal studies (11,19–22). In addition, both N3-EtdT and O2-EtdT are able to strongly block DNA replication and substantially induce mutations in vitro and in Escherichia coli cells (20,21,23,24). To date, no studies have been carried out to investigate the effects of the three regioisomeric EtdT lesions on DNA transcription. Herein, we constructed nonreplicative double-stranded vectors containing an N3-EtdT, O2-EtdT or O4-EtdT at a defined site and assessed how these EtdT lesions compromise the efficiency and fidelity of DNA transcription in vitro and in human cells.
MATERIALS AND METHODS
Materials
Unmodified oligodeoxyribonucleotides (ODNs), [γ-32P]ATP, enzymes and chemicals unless otherwise specified were purchased from Integrated DNA Technologies, Perkin-Elmer, New England BioLabs and Sigma-Aldrich, respectively. ON-TARGETplus SMARTpool siRNA against human CSB (L-004888) or XPC (L-016040), and Non-Targeting control siRNA (D-001210) were from Thermo Scientific Dharmacon. The 293T human embryonic kidney epithelial cells were obtained from American Type Culture Collection (ATCC). XPA-complemented (GM15876A) and XPA-deficient (XP12RO) human fibroblast cell lines were kindly provided by Professor Karlene A Cimprich (25). Cells were cultured in Dulbecco's Modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen), 100 μg/ml streptomycin (ATCC) and 100 U/ml penicillin, and incubated at 37°C in 5% CO2 atmosphere.
Transcription template preparation
We prepared the DNA templates for transcription assays as described elsewhere (26–28). To construct the unmodified control vector, a 50-mer ODN with the sequence of 5′-CTAGCGGAT-GCATCGACTCAATTATAGCACGCCATGGTCGACTCATCGCG-3′ was annealed with its complementary strand and ligated to an EcoRI-NheI restriction fragment from the pTGFP-T7-Hha10 plasmid (27). Using the similar method, we also constructed a competitor vector that contains three more nucleotides than the control plasmid near the relevant site of interest (Figure 1c). We next employed Nt.BstNBI to nick the unmodified control vector and generated a gapped plasmid by removing a 25-mer single-stranded ODN (Figure 1b), followed by filling the gap with a 13-mer lesion-free ODN (5′-AATTGAGTCGATG-3′) and a 12-mer lesion-containing ODN (5′-ATGGCGXGCTAT-3′, X = N3-EtdT, O2-EtdT or O4-EtdT) (20). We subsequently incubated the ligation products with ethidium bromide and purified the supercoiled lesion-bearing plasmids by agarose gel electrophoresis, as described previously (26,28). Purified lesion-bearing or lesion-free control plasmids were premixed with the competitor vector at specific molar ratios and used as transcription templates, where the molar ratios of dT, N3-, O2- and O4-EtdT-bearing plasmids over the competitor vector were 3:1, 12:1, 12:1 and 3:1, respectively.
In vitro transcription assay
Transcription assay using T7 RNA polymerase (T7 RNAP) or human RNA polymerase II (hRNAPII) was performed as described previously (27,29). Briefly, T7 RNAP-mediated reaction contained 50 ng of NotI-linearized DNA templates, 10 U of RNase inhibitor, 20 U of T7 RNAP, and 0.5 mM each of ATP, CTP, GTP and UTP in a 20-μl mixture and was incubated at 37°C for 1 h. The hRNAPII-mediated reaction contained 50 ng of NotI-linearized DNA templates, 10 U of RNase inhibitor, 8 U of HeLa nuclear extract and 0.4 mM each of the four ribonucleotides in a 25-μl mixture and incubated at 30°C for 1 h.
In vivo transcription assay
XPA-deficient (XP12RO) and XPA-complemented (GM15876A) cells in a 24-well plate at ∼70% confluence were transfected with 50 ng DNA templates and 450 ng carrier plasmid (self-ligated pGEM-T; Promega) using Lipofectamine 2000 (Invitrogen), following the manufacturer's protocol. For siRNA experiments, 293T cells in a 24-well plate at 40–60% confluence were transfected with ∼25 pmol siRNAs for each gene. After a 48-h incubation, 50 ng DNA templates, 450 ng carrier plasmid and another aliquot of siRNAs were co-transfected into the cells using Lipofectamine 2000 (Invitrogen). All the cells were harvested for RNA extraction 24 h after transfection with the DNA templates.
RNA extraction and reverse transcription-polymerase chain reaction
The RNA products were extracted using Total RNA Kit I (Omega), and were treated twice with the DNA-free kit (Ambion) to eliminate DNA contamination. cDNA synthesis was performed with M-MLV reverse transcriptase (Promega) and a mixture of oligo(dT)16 and a gene-specific primer (5′-TCGGTGTTGCTGTGAT-3′). Reverse transcription-polymerase chain reaction (RT-PCR) amplification was then performed by using a pair of primers spanning the lesion site and Phusion high-fidelity DNA polymerase as described previously (27). For evaluating the efficiency of siRNA knockdown, real-time RT-PCR was performed with the iQ SYBR Green Supermix kit (Bio-Rad) and gene-specific primers for CSB, XPC or the control gene GAPDH as described elsewhere (27).
Polyacrylamide gel electrophoresis analysis
We performed NcoI- and SfaNI-mediated restriction digestion/postlabeling assay to resolve 13-mer 32P-labeled nonmutagenic fragment d(CATGGCGAGCTAT) from the corresponding products carrying an A→T or A→C mutation, i.e. d(CATGGCGTGCTAT) and d(CATGGCGCGCTAT), respectively (Figures 1c, 2a and b). To this end, a portion of the above RT-PCR products was incubated in a 10-μl mixture containing 5 U NcoI, 1 U shrimp alkaline phosphatase, and 1× NEB buffer 3 at 37°C for 1 h and subsequently at 70°C for 20 min. The resulting dephosphorylated DNA was then incubated in a 15-μl solution containing 5 U T4 polynucleotide kinase, 1× NEB buffer 3 and ATP (50 pmol cold, premixed with 1.66 pmol [γ-32P]ATP). The mixture was incubated at 37°C for 30 min and then at 70°C for 20 min, after which 2 U SfaNI was added and incubated at 37°C for 2 h. The resulting 32P-labeled restriction fragments were resolved by using 30% native polyacrylamide gel (acrylamide:bis-acrylamide = 19:1) and quantified by phosphorimager analysis (27,28). Using a similar restriction digestion/postlabeling assay with MluCI and Cac8I, we were able to completely resolve the 10-mer 32P-labeled fragment d(AATTATAGCG), which corresponds to the product arising from A→G mutation opposite the lesion site, from the nonmutagenic fragment d(AATTATAGCA) and the fragments with an A→T or A→C mutation (Figure 2c and d). The relative bypass efficiency (RBE) was calculated using the following formula, RBE = (lesion signal/competitor signal)/(unmodified control signal/competitor signal) (27,30).
Figure 2.
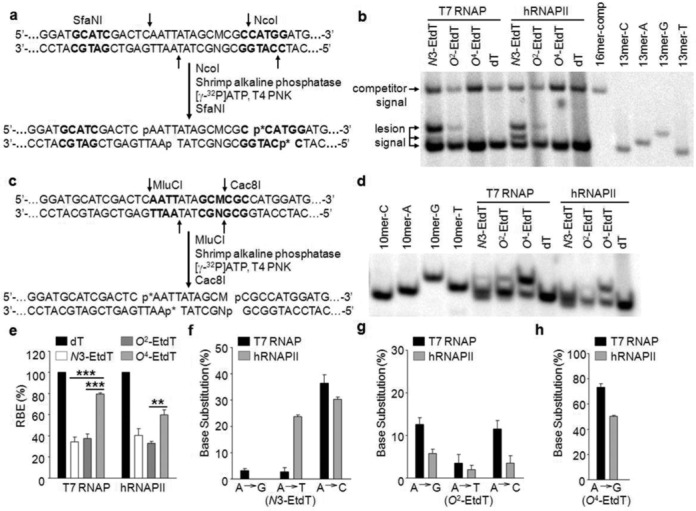
The effects of N3-EtdT, O2-EtdT and O4-EtdT on DNA transcription in vitro. (a) The effects of EtdT lesions on DNA transcription in vitro. (a) Sample processing for NcoI- and SfaNI-mediated restriction digestion/postlabeling assay (p* indicates 32P-labeled phosphate group). (b) Representative gel images showing the NcoI- and SfaNI-treated restriction fragments of interest. ‘16mer-Comp’ represents the standard ODN d(CATGGCGATATGCTAT), which corresponds to the restriction fragment arising from the competitor vector; ‘13mer-C’, ‘13mer-A’, ‘13mer-G’ and ‘13mer-T’ represent the standard ODN d(CATGGCGNGCTAT), where ‘N’ is C, A, G, T, respectively. (c) Sample processing for MluCI- and Cac8I-mediated restriction digestion/postlabeling assay. (d) Representative gel images showing the MluCI- and Cac8I-treated restriction fragments of interest. ‘10mer-C’, ‘10mer-A’, ‘10mer-G’ and ‘10mer-T’ represent the standard ODN d(AATTATAGCM), where ‘M’ is C, A, G, T, respectively. (e) The RBE values of N3-, O2- and O4-EtdT in in vitro transcription systems using T7 RNAP and HeLa nuclear extract (hRNAPII). (f–h) Mutagenic properties of N3- (f), O2- (g) and O4-EtdT (h) in in vitro transcription systems. The data represent the mean and standard error of results from three independent experiments. ‘**’, P < 0.01; ‘***’, P < 0.001. The P-values were calculated by using unpaired two-tailed Student's t-test.
Liquid chromatography-tandem mass spectrometry analysis
The RT-PCR products were treated with 20 U SfaNI and 20 U shrimp alkaline phosphatase in 120 μl NEB buffer 3 at 37°C for 2 h, and then at 70°C for 20 min. To the mixture, 50 U NcoI was added, and the mixture was incubated at 37°C for 2 h. The resulting solution was extracted with phenol/chloroform/isoamyl alcohol (25:24:1, v/v) and the aqueous portion was dried with Speed-vac, desalted with high performance liquid chromatography and dissolved in water. The resultant ODN mixture was subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis following previously described conditions (27,28). Briefly, a 0.50 × 150 mm Zorbax SB-C18 column (Agilent Technologies) was used. The flow rate was 8.0 μl/min, and a 5-min linear gradient of 5–20% methanol followed by a 25 min of 20–60% methanol in 400 mM 1,1,1,3,3,3-hexafluoro-2-propanol buffer (pH was adjusted to 7.0 by the addition of triethylamine) was employed for the separation. The LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific) was set up for monitoring the fragmentation of the [M-3H]3− ions of the complementary 13-mer ODNs, i.e. d(AATTATAGCMCGC), where ‘M’ designates A, T, C or G.
RESULTS
Using our recently developed competitive transcription and adduct bypass assay (27), we investigated how the regioisomeric N3-, O2- and O4-EtdT lesions perturb DNA transcription in vitro and in human cells. To this end, we employed a well-established protocol (26,28) to incorporate N3-, O2- and O4-EtdT at a defined site in the transcribed strand of a nonreplicative double-stranded plasmid, placing the lesions downstream from the cytomegalovirus (CMV) and T7 promoters as described in ‘Materials and Methods’ (Figure 1b and c). The lesion-containing or unmodified control plasmid was premixed with a competitor construct at given molar ratios and used as DNA templates for the in vitro or in vivo transcription assays. We subsequently amplified the run-off transcripts of interest with RT-PCR, digested the resulting RT-PCR products with suitable restriction enzymes and subjected the restriction digestion mixture to polyacrylamide gel electrophoresis (PAGE) and LC-MS/MS analyses (Figure 1c).
We performed the in vitro transcription assays using human RNA polymerase II (hRNAPII) in HeLa cell nuclear extract or purified T7 RNA polymerase (T7 RNAP). The latter is a single-subunit enzyme that is structurally homologous to eukaryotic mitochondrial RNA polymerases (31). The RT-PCR products were digested with NcoI and SfaNI, and the digestion products were subjected to PAGE analysis. In this context, the 32P-labeled wild-type fragment d(p*CATGGCGTGCTAT) could be resolved from the corresponding products harboring an A→T or A→C mutation opposite the lesion site, i.e. d(p*CATGGCGAGCTAT) and d(p*CATGGCGGGCTAT), but could not be resolved from the corresponding product carrying an A→G mutation, i.e. d(p*CATGGCGCGCTAT) (Figure 2a and b). However, by digesting the RT-PCR products with MluCI and Cac8I, we were able to resolve the DNA fragment emanating from A→G mutation opposite the lesion site, i.e. d(p*AATTATAGCG), from the wild-type fragment and the products with an A→T or A→C mutation (Figure 2c and d).
The quantification data from PAGE analysis showed that O4-EtdT did not considerably inhibit T7 RNAP-mediated transcription elongation, whereas N3-EtdT and O2-EtdT impeded substantially DNA transcription mediated by T7 RNAP, with RBE values being ∼34 and ∼38%, respectively (Figure 2e). In addition, we found that the RBE value for O4-EtdT was ∼60% during hRNAPII-mediated transcription reaction, whereas N3-EtdT and O2-EtdT conferred strong inhibitory effects on the in vitro transcription mediated by hRNAPII (Figure 2e).
PAGE analysis also allowed us to determine the effects of N3-, O2- and O4-EtdT on the fidelity of transcription mediated by T7 RNAP or hRNAPII in vitro. The quantification data revealed that O4-EtdT induced predominantly one type of mutant transcript that contains a guanine misincorporation opposite the lesion (A→G) during transcription by T7 RNAP and hRNAPII, with base substitution frequencies being ∼73 and ∼50%, respectively (Figure 2f). Unlike O4-EtdT, N3-EtdT and O2-EtdT displayed promiscuous miscoding properties during T7 RNAP-mediated transcription: A→G, A→T and A→C mutations were observed for N3-EtdT at frequencies of ∼3, ∼3 and ∼36%, respectively, and for O2-EtdT at frequencies of ∼13, ∼4 and ∼12%, respectively (Figure 2f–h). During hRNAPII-mediated transcription, N3-EtdT induced A→T and A→C mutations at frequencies of ∼24 and ∼30%, respectively, whereas O2-EtdT induced A→G, A→T and A→C mutations at frequencies of ∼6, ∼2 and ∼4%, respectively (Figure 2f and g). We also confirmed the identities of the above mutant products by LC-MS/MS analysis (Supplementary Figures S1 and S2).
We next asked how N3-, O2- and O4-EtdT compromise DNA transcription in human cells. For this purpose, we premixed either lesion-bearing or unmodified control plasmids with the competitor vector and co-transfected them into XPA-deficient (XP12RO) and XPA-complemented (GM15876A) cells. XPA is a core component of nucleotide excision repair (NER) that is one of the most versatile DNA damage removal pathways (32). After a 24-h incubation, RNA products were extracted from the cells and amplified with RT-PCR, followed by PAGE and LC-MS/MS analyses of the restriction digestion mixture of RT-PCR products as described above.
Our results showed that N3-EtdT, O2-EtdT and O4-EtdT considerably inhibited DNA transcription in GM15876A cells, with the RBE values being ∼37, ∼42 and ∼43%, respectively (Figure 3a and Supplementary Figure S3a). Moreover, we found that the RBE values for N3-EtdT, O2-EtdT and O4-EtdT were significantly lower in XPA-deficient (XP12RO) cells (∼28, ∼26 and 32%, respectively, Figure 3a and Supplementary Figure S3a). We also found that transcriptional bypass of N3-EtdT could induce A→T and A→C mutations at significantly higher frequencies (∼34 and ∼18%, respectively) in XPA-deficient cells than in XPA-complemented cells (∼20 and ∼11%, respectively, Figure 3b and Supplementary Figure S3a and b). On the other hand, transcriptional bypass of O4-EtdT induced A→G mutation at a frequency of ∼34%, and O2-EtdT induced three types of mutant transcripts (A→G, A→T and A→C) at frequencies of ∼2–5% in GM15876A cells; however, depletion of XPA did not considerably alter the mutagenic properties of O2-EtdT or O4-EtdT in human cells (Figure 3c and d and Supplementary Figure S3a and b).
Figure 3.
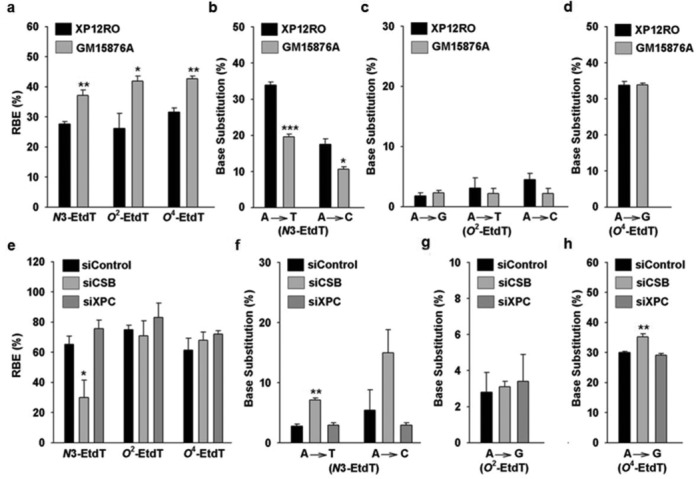
The effects of N3-EtdT, O2-EtdT and O4-EtdT on DNA transcription in human cells. (a) The RBE values of N3-, O2- and O4-EtdT in XPA-deficient (XP12RO) and XPA-complemented (GM15876A) cells. (b–d) Mutagenic properties of N3- (b), O2- (c) and O4-EtdT (d) in XP12RO and GM15876A cells. (e) The RBE values of N3-, O2- and O4-EtdT in 293T cells treated with CSB or XPC siRNAs. (f–h) Mutagenic properties of N3- (f), O2- (g) and O4-EtdT (h) in 293T cells treated with CSB or XPC siRNAs. The data represent the mean and standard error of results from three independent experiments. ‘*’, P < 0.05; ‘**’, P < 0.01; ‘***’, P < 0.001. The P-values were calculated by using unpaired two-tailed Student's t-test.
We further examined the potential roles of XPC and CSB, which are key players in global-genome NER (GG-NER) and transcription-coupled NER (TC-NER), respectively (32), in transcriptional alternations induced by N3-EtdT, O2-EtdT and O4-EtdT in human cells. Our results showed that, compared to nontargeting control siRNA treatment, siRNA-mediated downregulation of CSB led to a significant decrease in transcriptional bypass efficiency of N3-EtdT, along with a marked elevation in transcriptional mutations induced by this lesion in 293T cells (Figure 3e and f and Supplementary Figures S4 and S5a). We also found that siRNA knockdown of CSB caused a slight, yet statistically significant increase in A→G mutation induced by O4-EtdT; however, depletion of CSB did not considerably change the transcriptional bypass efficiency of O4-EtdT in 293T cells (Figure 3e and h and Supplementary Figure S5a and b). In addition, depletion of CSB exerted no significant effect on the transcriptional bypass efficiency or mutation frequency of O2-EtdT in 293T cells (Figure 3e and g and Supplementary Figure S5a and b). On the other hand, siRNA knockdown of XPC did not significantly change the effects of all three EtdT lesions on the efficiency and fidelity of DNA transcription in 293T cells (Figure 3e–h and Supplementary Figures S4 and S5a and b).
DISCUSSION
Alkylative damage to DNA is generally unavoidable because of the abundant presence of alkylating agents in the environment and within cells (1). Thymine is known to be alkylated at the N3, O2 and O4 positions (8,9,11–13,24). In this context, the regioisomeric N3-EtdT, O2-EtdT and O4-EtdT lesions have been detected at significantly higher levels in smokers than in nonsmokers, suggesting their potential roles as useful biomarkers for exposure to ethylating agents and possibly for cancer risk assessment (9,14). Although little is known about the repair of N3-EtdT, O2-EtdT and O4-EtdT are known to be poorly repaired and thus accumulate as highly persistent DNA lesions in mammalian tissues (12,19,33).
Accurate transmission of genetic information is essential for normal physiological processes of a living organism (15,16,34). Emerging evidence indicates that, aside from errors in DNA replication, transient transcriptional mutagenesis may contribute to genomic instability and stable phenotypic changes, which may ultimately result in carcinogenesis and the development of other diseases (15,16,34–39). It has been suggested that transient errors in the mRNA encoding a transcription factor involved in a bistable switch can promote heritable change in cellular phenotype (36,37). In addition, a recent study showed that mutagenic transcriptional bypass of a site-specifically inserted 8-oxoguanine lesion in the transcribed strand of the Ras oncogene could lead to a constitutively active mutant Ras protein and activation of downstream oncogenic signaling (35). Whereas the effects of N3-EtdT, O2-EtdT and O4-EtdT on DNA replication have been well studied (20–24), it is desirable to determine how the three regioisomeric EtdT lesions compromise the efficiency and fidelity of DNA transcription.
In the present study, we showed that N3-EtdT and O2-EtdT, but not O4-EtdT, strongly block DNA transcription by single-subunit T7 RNA polymerase or multisubunit human RNA polymerase II in vitro. In addition, we found that transcriptional bypass of O4-EtdT is highly mutagenic and exclusively induces the misincorporation of guanine opposite the lesion (i.e. A→G); N3-EtdT and O2-EtdT are also miscoding lesions, but display promiscuous mutagenic properties during transcription. These findings are consistent with results from previous replication studies of the corresponding alkylated DNA adducts (20–24), suggesting the similar mechanisms underlying base misincorporation opposite these lesions during DNA replication and transcription. The distinct miscoding properties of three regioisomeric EtdT lesions may be attributed to their unique chemical properties. In this respect, it has been shown that alkylation of thymine at the O4 position promotes its favorable pairing with guanine (40). The addition of an alkyl group to the N3 position, however, can block the Watson–Crick hydrogen bonding face of thymine and thus abolish its base-pairing capability; alkylation of thymine at the O2 position confers the inability of the nucleobase to pair preferentially with any of the canonical nucleobases (20,41). Thus, relative to O4-EtdT, nucleotide incorporation opposite N3-EtdT and O2-EtdT is much less selective.
We further demonstrated that all three regioisomeric EtdT lesions exhibit strong inhibitory and mutagenic effects on DNA transcription in certain human cells. Moreover, we found that XPA and CSB, but not XPC, are involved in the repair of N3-EtdT when located on the template DNA strand. Thus, these results suggest that the nonbulky N3-EtdT lesion is an efficient substrate for TC-NER, which is a subpathway of NER that preferentially repairs DNA lesions in the transcribed strand of active genes (32). Along this line, TC-NER has also been shown to efficiently repair several other nonbulky lesions including abasic sites in the transcribed strand in cells (27,29,34,42). On the other hand, although we found that depletion of XPA reduces the transcriptional bypass efficiencies of O2-EtdT and O4-EtdT, such depletion does not appreciably affect the mutagenic properties of the two lesions in human cells. In addition, siRNA-mediated knockdown of CSB has no or little impact on the transcriptional alternations induced by O2-EtdT and O4-EtdT in human cells. Thus, it is very likely that TC-NER has negligible roles in the removal of O2-EtdT and O4-EtdT from the template strand of actively transcribed genes in human cells.
In conclusion, we have examined, for the first time, how the regioisomeric N3-EtdT, O2-EtdT and O4-EtdT lesions perturb the flow of genetic information during transcription mediated by single-subunit T7 RNA polymerase or multisubunit human RNA polymerase II in vitro and in human cells. We have also investigated the potential roles of NER proteins, including XPA, CSB and XPC, in the repair of these EtdT lesions in human cells. Our results demonstrated that TC-NER has an important role in the removal of N3-EtdT, but not O2-EtdT or O4-EtdT, in the DNA template strand in human cells. Together, our findings provide novel and important insights into the biological consequences of these ethylated DNA lesions in living cells.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
Acknowledgments
We thank Professor Timothy R. O'Connor for providing the initial pTGFP-Hha10 vector, and Professor Karlene A Cimprich for kindly providing XPA-deficient (XP12RO) and XPA-complemented (GM15876A) cell lines.
FUNDING
National Institutes of Health (NIH) [ES025121 to Y.W.]. Funding for open access charge: NIH [R01 ES025121].
Conflict of interest statement. None declared.
REFERENCES
- 1.Fu D., Calvo J.A., Samson L.D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beranek D.T. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 3.Rydberg B., Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. EMBO J. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sedgwick B. Nitrosated peptides and polyamines as endogenous mutagens in O6-alkylguanine-DNA alkyltransferase deficient cells. Carcinogenesis. 1997;18:1561–1567. doi: 10.1093/carcin/18.8.1561. [DOI] [PubMed] [Google Scholar]
- 5.Hecht S.S. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat. Res. 1999;424:127–142. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 6.Thompson C.L., McCoy Z., Lambert J.M., Andries M.J., Lucier G.W. Relationships among benzo[a]pyrene metabolism, benzo[a]pyrene-diol-epoxide:DNA adduct formation, and sister chromatid exchanges in human lymphocytes from smokers and nonsmokers. Cancer Res. 1989;49:6503–6511. [PubMed] [Google Scholar]
- 7.Chen L., Wang M., Villalta P.W., Hecht S.S. Liquid chromatography-electrospray ionization tandem mass spectrometry analysis of 7-ethylguanine in human liver DNA. Chem. Res. Toxicol. 2007;20:1498–1502. doi: 10.1021/tx700147f. [DOI] [PubMed] [Google Scholar]
- 8.Anna L., Kovacs K., Gyorffy E., Schoket B., Nair J. Smoking-related O4-ethylthymidine formation in human lung tissue and comparisons with bulky DNA adducts. Mutagenesis. 2011;26:523–527. doi: 10.1093/mutage/ger011. [DOI] [PubMed] [Google Scholar]
- 9.Chen H.J., Wang Y.C., Lin W.P. Analysis of ethylated thymidine adducts in human leukocyte DNA by stable isotope dilution nanoflow liquid chromatography-nanospray ionization tandem mass spectrometry. Anal Chem. 2012;84:2521–2527. doi: 10.1021/ac203405y. [DOI] [PubMed] [Google Scholar]
- 10.Chao M.R., Wang C.J., Chang L.W., Hu C.W. Quantitative determination of urinary N7-ethylguanine in smokers and non-smokers using an isotope dilution liquid chromatography/tandem mass spectrometry with on-line analyte enrichment. Carcinogenesis. 2006;27:146–151. doi: 10.1093/carcin/bgi177. [DOI] [PubMed] [Google Scholar]
- 11.Swenberg J.A., Dyroff M.C., Bedell M.A., Popp J.A., Huh N., Kirstein U., Rajewsky M.F. O4-ethyldeoxythymidine, but not O6-ethyldeoxyguanosine, accumulates in hepatocyte DNA of rats exposed continuously to diethylnitrosamine. Proc. Natl Acad. Sci. U.S.A. 1984;81:1692–1695. doi: 10.1073/pnas.81.6.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Den Engelse L., De Graaf A., De Brij R.J., Menkveld G.J. O2- and O4-ethylthymine and the ethylphosphotriester dTp(Et)dT are highly persistent DNA modifications in slowly dividing tissues of the ethylnitrosourea-treated rat. Carcinogenesis. 1987;8:751–757. doi: 10.1093/carcin/8.6.751. [DOI] [PubMed] [Google Scholar]
- 13.Godschalk R., Nair J., van Schooten F.J., Risch A., Drings P., Kayser K., Dienemann H., Bartsch H. Comparison of multiple DNA adduct types in tumor adjacent human lung tissue: effect of cigarette smoking. Carcinogenesis. 2002;23:2081–2086. doi: 10.1093/carcin/23.12.2081. [DOI] [PubMed] [Google Scholar]
- 14.Chen H.J., Lee C.R. Detection and simultaneous quantification of three smoking-related ethylthymidine adducts in human salivary DNA by liquid chromatography tandem mass spectrometry. Toxicol. Lett. 2014;224:101–107. doi: 10.1016/j.toxlet.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Bregeon D., Doetsch P.W. Transcriptional mutagenesis: causes and involvement in tumour development. Nat. Rev. Cancer. 2011;11:218–227. doi: 10.1038/nrc3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morreall J.F., Petrova L., Doetsch P.W. Transcriptional mutagenesis and its potential roles in the etiology of cancer and bacterial antibiotic resistance. J. Cell Physiol. 2013;228:2257–2261. doi: 10.1002/jcp.24400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sale J.E., Lehmann A.R., Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange S.S., Takata K., Wood R.D. DNA polymerases and cancer. Nat. Rev. Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scherer E., Timmer A.P., Emmelot P. Formation by diethylnitrosamine and persistence of O4-ethylthymidine in rat liver DNA in vivo. Cancer Lett. 1980;10:1–6. doi: 10.1016/0304-3835(80)90058-0. [DOI] [PubMed] [Google Scholar]
- 20.Andersen N., Wang P., Wang Y. Replication across regioisomeric ethylated thymidine lesions by purified DNA polymerases. Chem. Res. Toxicol. 2013;26:1730–1738. doi: 10.1021/tx4002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhai Q., Wang P., Wang Y. Cytotoxic and mutagenic properties of regioisomeric O2-, N3- and O4-ethylthymidines in bacterial cells. Carcinogenesis. 2014;35:2002–2006. doi: 10.1093/carcin/bgu085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein J.C., Bleeker M.J., Lutgerink J.T., van Dijk W.J., Brugghe H.F., van den Elst H., van der Marel G.A., van Boom J.H., Westra J.G., Berns A.J., et al. Use of shuttle vectors to study the molecular processing of defined carcinogen-induced DNA damage: mutagenicity of single O4-ethylthymine adducts in HeLa cells. Nucleic Acids Res. 1990;18:4131–4137. doi: 10.1093/nar/18.14.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhai Q., Wang P., Cai Q., Wang Y. Syntheses and characterizations of the in vivo replicative bypass and mutagenic properties of the minor-groove O2-alkylthymidine lesions. Nucleic Acids Res. 2014;42:10529–10537. doi: 10.1093/nar/gku748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grevatt P.C., Donahue J.M., Bhanot O.S. The role of N3-ethyldeoxythymidine in mutagenesis and cytotoxicity by ethylating agents. J. Biol. Chem. 1991;266:1269–1275. [PubMed] [Google Scholar]
- 25.Bomgarden R.D., Lupardus P.J., Soni D.V., Yee M.C., Ford J.M., Cimprich K.A. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. EMBO J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker D.J., Wuenschell G., Xia L., Termini J., Bates S.E., Riggs A.D., O'Connor T.R. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J. Biol. Chem. 2007;282:22592–22604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- 27.You C., Dai X., Yuan B., Wang J., Brooks P.J., Niedernhofer L.J., Wang Y. A quantitative assay for assessing the effects of DNA lesions on transcription. Nat. Chem. Biol. 2012;8:817–822. doi: 10.1038/nchembio.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan B., You C., Andersen N., Jiang Y., Moriya M., O'Connor T.R., Wang Y. The roles of DNA polymerases kappa and iota in the error-free bypass of N2-carboxyalkyl-2′-deoxyguanosine lesions in mammalian cells. J. Biol. Chem. 2011;286:17503–17511. doi: 10.1074/jbc.M111.232835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You C., Dai X., Yuan B., Wang Y. Effects of 6-thioguanine and S6-methylthioguanine on transcription in vitro and in human cells. J. Biol. Chem. 2012;287:40915–40923. doi: 10.1074/jbc.M112.418681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaney J.C., Essigmann J.M. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 31.Masters B.S., Stohl L.L., Clayton D.A. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 32.Hanawalt P.C., Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 33.Thomale J., Huh N.H., Nehls P., Eberle G., Rajewsky M.F. Repair of O6-ethylguanine in DNA protects rat 208F cells from tumorigenic conversion by N-ethyl-N-nitrosourea. Proc. Natl Acad. Sci. U.S.A. 1990;87:9883–9887. doi: 10.1073/pnas.87.24.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaillard H., Herrera-Moyano E., Aguilera A. Transcription-associated genome instability. Chem. Rev. 2013;113:8638–8661. doi: 10.1021/cr400017y. [DOI] [PubMed] [Google Scholar]
- 35.Saxowsky T.T., Meadows K.L., Klungland A., Doetsch P.W. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl Acad. Sci. U.S.A. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon A.J., Halliday J.A., Blankschien M.D., Burns P.A., Yatagai F., Herman C. Transcriptional infidelity promotes heritable phenotypic change in a bistable gene network. PLoS Biol. 2009;7:e44. doi: 10.1371/journal.pbio.1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon A.J., Satory D., Halliday J.A., Herman C. Heritable change caused by transient transcription errors. PLoS Genet. 2013;9:e1003595. doi: 10.1371/journal.pgen.1003595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strathern J.N., Jin D.J., Court D.L., Kashlev M. Isolation and characterization of transcription fidelity mutants. Biochim. Biophys. Acta. 2012;1819:694–699. doi: 10.1016/j.bbagrm.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bregeon D., Peignon P.A., Sarasin A. Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 2009;5:e1000577. doi: 10.1371/journal.pgen.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brennan R.G., Pyzalska D., Blonski W.J., Hruska F.E., Sundaralingam M. Crystal structure of the promutagen O4-methylthymidine: importance of the anti conformation of the O4 methoxy group and possible mispairing of O4-methylthymidine with guanine. Biochemistry. 1986;25:1181–1185. doi: 10.1021/bi00353a036. [DOI] [PubMed] [Google Scholar]
- 41.Huff A.C., Topal M.D. DNA damage at thymine N-3 abolishes base-pairing capacity during DNA synthesis. J. Biol. Chem. 1987;262:12843–12850. [PubMed] [Google Scholar]
- 42.Kim N., Jinks-Robertson S. Abasic sites in the transcribed strand of yeast DNA are removed by transcription-coupled nucleotide excision repair. Mol. Cell Biol. 2010;30:3206–3215. doi: 10.1128/MCB.00308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


