Abstract
During meiosis programmed DNA double-strand breaks (DSBs) are repaired by homologous recombination using the sister chromatid or the homologous chromosome (homolog) as a template. This repair results in crossover (CO) and non-crossover (NCO) recombinants. Only CO formation between homologs provides the physical linkages guiding correct chromosome segregation, which are essential to produce healthy gametes. The factors that determine the CO/NCO decision are still poorly understood. Using Schizosaccharomyces pombe as a model we show that the Rad51/Dmc1-paralog complexes Rad55-Rad57 and Rdl1-Rlp1-Sws1 together with Swi5-Sfr1 play a major role in antagonizing both the FANCM-family DNA helicase/translocase Fml1 and the RecQ-type DNA helicase Rqh1 to limit hybrid DNA formation and promote Mus81-Eme1-dependent COs. A common attribute of these protein complexes is an ability to stabilize the Rad51/Dmc1 nucleoprotein filament, and we propose that it is this property that imposes constraints on which enzymes gain access to the recombination intermediate, thereby controlling the manner in which it is processed and resolved.
INTRODUCTION
In order to enable correct chromosome segregation during meiosis, the homologous chromosomes have to pair up and get physically linked. This is achieved via the repair of programmed DNA double-strand breaks (DSBs) by homologous recombination. Specifically, repair has to occur using the homolog rather than the sister chromatid as a template (interhomolog recombination) and critically at least some of the interhomolog recombination intermediates have to be processed into crossing overs (1,2).
The crossover (CO)/non-crossover (NCO) decision is a key point for regulation of meiotic recombination, and one that is inextricably linked to the strand exchange reaction driven by Rad51 and/or Dmc1. These proteins load onto single-stranded (ss) DNA, which is formed by nucleolytic resection of the DSB (3). Multiple protomers are loaded creating proteinaceous filaments that encase the DNA. Within these nucleoprotein filaments intact homologous DNA partners are located, paired with the broken DNA and then invaded. Strand invasion forms a displacement (D) loop at which DNA synthesis is primed leading to extension of the invading strand (4,5). The recombination reaction can then take one of the several different paths: the D-loop can be unwound allowing the extended DNA strand to anneal to its complementary strand at the other end of the break in a process known as synthesis-dependent strand annealing (SDSA); the D-loop can be cleaved; or the ssDNA tail at the other end of the DSB can anneal to the displaced strand of the D-loop in a process termed second end capture. SDSA results in an NCO, whereas D-loop cleavage gives rise to a CO. Second end capture leads to the formation of a double Holliday junction (HJ) (or in fission yeast a single HJ seemingly due to nicking of the displaced strand prior to second end capture), which can branch migrate extending the amount of hybrid DNA that is formed (6–9). HJs are resolved by structure-specific endonucleases generating either COs or NCOs depending on which DNA strands are cleaved (10,11). Key factors involved in these processes include the DNA helicase/translocase FANCM (Fml1 in Schizosaccharomyces pombe), RecQ-type helicases and the Mus81-Eme1 endonuclease (12–20).
In vitro FANCM can unwind D-loops and branch migrate HJs, and in plants and fission yeast appears to utilize these activities during meiosis to direct repair via SDSA with the help of its histone-fold co-factors MHF1 and MHF2 (16–18,21–24). RecQ-type helicases, which can also branch migrate HJs, perform a wide range of regulatory roles in homologous recombination including the promotion of NCO formation during meiosis, at least in Saccharomyces cerevisiae and Tetrahymena thermophila (19,20,25). Mus81-Eme1 can cleave a variety of branched DNA molecules in vitro, including D-loops, nicked HJs and fully ligated HJs, albeit its ability to cleave the latter junction remains contentious (12,26–30). In vivo it can promote both CO and NCO formation, and at least in fission yeast is thought to bias resolution toward COs by cleaving D-loops (12–14,16,20,28,31–34). The factors that govern which of these enzymes are used to resolve meiotic recombination intermediates remain largely unknown.
In fission yeast we recently identified the Swi5–Sfr1 complex as a determinant of the CO/NCO decision, seemingly functioning to counteract Fml1-Mhf1-Mhf2 and thereby assigning recombination intermediates to be processed by Mus81-Eme1 (7,12,16). Swi5–Sfr1 mediates Dmc1 loading on to RPA (replication protein A)-coated ssDNA and enhances the stability of both Rad51- and Dmc1-nucleoprotein filaments (35). This latter activity in particular could control the accessibility of the D-loop to Fml1, Mus81-Eme1 and potentially other proteins. However, Swi5–Sfr1 is not the only protein complex that mediates the formation of the Rad51/Dmc1-nucleoprotein filament or enhances its stability. In budding yeast the Rad51-paralog complexes, Rad55-Rad57 and Psy3-Csm2, perform similar functions (36,37).
In this study we employ genetic assays to determine whether the Rad51/Dmc1 paralogs and mediators are determinants of the CO/NCO decision in fission yeast. We find that, along with Swi5-Sfr1, both Rad55-Rad57 and Rdl1-Rlp1 (the fission yeast homologs of Psy3-Csm2) together with Sws1 promote CO formation by antagonizing both Fml1 and the RecQ-type helicase Rqh1. Based on these findings we propose that Rad51/Dmc1 nucleoprotein filament stability is a major determinant of the CO/NCO decision by imposing constraints on which junction processing enzymes can gain access to the underlying DNA.
MATERIALS AND METHODS
Yeast strains and plasmid construction
S. pombe strains used for this study are listed in Supplementary Table S1. Yeast cells were cultured on yeast extract (YE), on pombe minimal glutamate (PMG) and on yeast nitrogen base glutamate (YNG) agar plates containing the required supplements (concentration 250 μg/ml on YE, 75 μg/ml on PMG and YNG). Crosses were performed on malt extract (ME) agar with the required amino acids (concentration 50 μg/ml). Determination of spore viability by random spore analysis and the meiotic recombination assay have been previously described in detail (12,16,38,39).
To make a sfr1 deletion strain marked with a hygromycin B resistance, the hphMX4 cassette from pAG32 (40) was subcloned as a SacI-BamHI fragment into a plasmid carrying up- and downstream flanking sequences of the sfr1 open reading frame (ORF) (16). The deletion cassette was released from the plasmid by digesting with HindIII and transformed into FO652 using a standard Li-acetate protocol (41).
The original sws1Δ strain VM3723 is his3+ (42). Due to the proximity of the his3 and sws1 genes (they are only separated by ∼5 kb on chromosome II) it was unfeasible to create a sws1Δ::kanMX6 his3-D1 strain by crossing. Using oligos located several 100 bp up- and downstream of sws1 (oMW1540 5′-GATCAAATAACTCTCGAAGC-3′ and oMW1541 5′-CTTGATCAAACCTAGACG-3′) a sws1-deletion cassette was PCR-amplified using Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific Inc., Waltham, MA, USA) and genomic DNA from VM3723 as a template. The polymerase chain reaction (PCR) product was then transformed into MCW1221 using a standard Li-acetate protocol (41).
To produce rdl1Δ and rlp1Δ strains marked with a nourseothricin resistance and a sws1Δ strain marked with a hygromycin B resistance we used a one-step marker swap protocol. This makes use of the homology provided by the TEF-promoter and -terminator sequences present in all the MX constructs. Cassettes released by endonuclease digest from plasmids pCR2.1-nat (EcoRI) and pAG32 (BamHI-EcoRV) were used to swap markers in rdl1Δ::kanMX6, rlp1Δ::kanMX6 and sws1Δ::kanMX6 strains (40,43).
All restriction endonucleases were obtained from New England BioLabs (NEB), Inc. (Ipswich, MA, USA).
Construction of a meiotic recombination reporter system flanking ade7
Similar to the meiotic recombination assay at ade6, where we utilized different point mutations (both hotspots and non-hotspots) (12,16,44), we integrated his3+ and ura4+ markers up- and downstream of ade7. The point mutations in ade7-50 and ade7-152 were verified by DNA sequencing, and a discrepancy to the previously reported sequence for ade7-50 was noted; T779del/G780C rather than G780C (45).
DNA sequences ∼6.8 kb upstream (oMW1511 5′-aattaaggatccaacctcattcctctccc-3′ and oMW1512 5′-aattaaactagtggggacgacgagtcg-3′) and ∼12.4 kb downstream (oMW1513 5′-aattaaggatcctcaatttcttgcagttcc-3′ and oMW1514 5′- aattaaactagtctcaagcctcaaacaacc-3′) of the ade7 ORF were PCR-amplified from ALP1596 using Phusion High-Fidelity DNA polymerase, cut with SpeI, and cloned into pAG25 (40) cut with SpeI and PvuII. The his3+-marker was PCR-amplified from pFOX2 (46) (oMW1515 5′-gcttggctgcaggaattc-3′ and oMW1516 5′- aattaactgcagtatcgataagcttgatggc-3′) creating a his3 ORF flanked by two PstI sites, this construct was then cloned into the PstI site of the plasmid carrying the ade7-upstream flanking fragment (giving pALo109). The ura4+-marker was subcloned as a HindIII-fragment from pDUP12 (47) into the HindIII site of the plasmid carrying the ade7-downstream flanking region (giving pALo111). All relevant plasmid sections were verified by DNA sequencing. The cassettes for transformation were released from the plasmids by restriction digest. The ∼3.2-kb EagI-SpeI fragment from pALo109 and the ∼2.5-kb BamHI-SpeI fragment from pALo111 were then introduced into ALP1596 and ALP1593, respectively, by a standard Li-acetate transformation protocol (41).
All restriction endonucleases were obtained from New England BioLabs (NEB), Inc. (Ipswich, MA).
Data presentation and statistics
Data presented as box-and-whisker plots were created in R (version i386, 3.0.1) (http://www.r-project.org/) using the boxplot() function with its standard settings. The lower and upper ‘hinges’ of the box represent the first and the third quartile, respectively, and the black bar within the box indicates the median ( = second quartile). The ‘whiskers’ represent the minimum and maximum of the range, unless they differ more than 1.5 times the interquartile distance from the median. In the latter case, the borders of the 1.5 times interquartile distance around the median are indicated by the ‘whiskers’ and values outside this range (‘outliers’) are shown as open circles. Raw data and R scripts used for the boxplot() function are available online as supporting material (48).
Several data sets did not conform to a normal distribution according to a Shapiro–Wilk test (http://sdittami.altervista.org/shapirotest/ShapiroTest.html). Therefore, all comparisons were done using a two-tailed Mann–Whitney U test, which is non-parametric and does not depend on data sets being normally distributed. Statistical analysis of the genetic recombination data was performed in G*Power 3.1.7 (49,50). P values were calculated with a given statistical power of 1 – β = 0.8.
RESULTS
Assay for monitoring meiotic recombination
We have previously described a genetic recombination assay featuring intragenic markers (ade6-3083 and ade6-469) and flanking intergenic markers (his3+-aim and ura4+-aim2) to identify and characterize determinants of template choice and the CO/NCO decision during meiotic recombination (12,16,44) (Figure 1A). Reductions in gene conversion (GC) and CO frequencies observed in this assay indicate either a lower frequency of DSB formation at the ade6-3083 hotspot allele or reduced interhomolog recombination, whereas the genetic readout for the CO/NCO decision is the percentage of COs associated with a GC event (CO-GC). The linkage of the flanking intergenic markers to the GC event can also provide information about the site of recombination intermediate resolution relative to the position of the intragenic markers (Supplementary Figure S1). In wild-type crosses ∼60% of all ade6+ GC events are also a CO between the flanking his3+ and ura4+ markers, and almost all of these COs are His− Ura− (Figure 1B and C). The remaining ade6+ NCO recombinants are mostly His− Ura+ (Figure 1B and C). This bias toward COs can be explained by the resolution of D-loops and/or unligated HJs by Mus81-Eme1 (Supplementary Figure S1—scenarios 2 and 7), whereas the NCOs likely stem from a combination of SDSA driven by Fml1 (Supplementary Figure S1—scenario 1) and HJ resolution by Mus81-Eme1, which gives equal numbers of COs and NCOs (Supplementary Figure S1—scenarios 3 and 8, and 4 and 9) (12,16,28,44).
Figure 1.
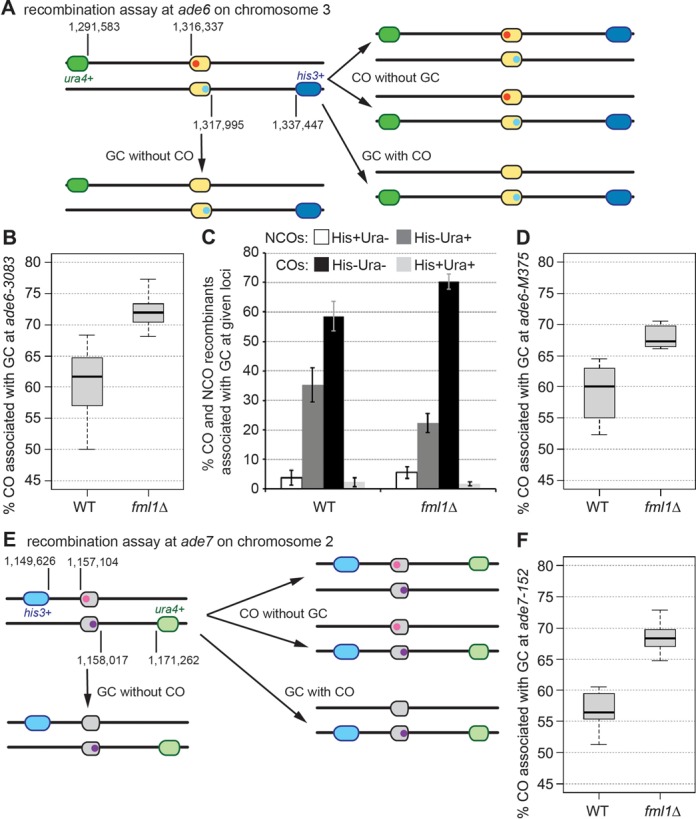
Fml1 is necessary for wild-type levels of non-crossovers associated with gene conversion events at recombination hotspots and non-hotspots. (A) Schematic showing the meiotic recombination assay at ade6 (yellow) and its common outcomes. The positions of ade6 and the artificially introduced markers his3+-aim (blue) and ura4+-aim2 (green) on chromosome 3 are indicated (in bp). Point mutations ade6-M375 and -3083 are shown in red, and ade6-469 is labeled in light blue. (B) Frequency of crossovers and (C) frequencies of different crossover and non-crossover classes associated with a gene conversion event in wild type and fml1Δ at ade6-3083×ade6-469; ALP733×ALP731 (WT, n = 41), ALP1133×FO2608 (fml1Δ, n = 15). (D) Frequency of crossovers associated with a gene conversion event in wild type and fml1Δ at ade6-M375×ade6-469; ALP1541×ALP731 (WT, n = 6), ALP1542×FO2608 (fml1Δ, n = 6). (E) Schematic of the meiotic recombination assay at ade7 (gray) and its common outcomes. The location of ade7 and the artificially introduced markers his3+-aim3 (teal) and ura4+-aim6 (light green) on chromosome 2 are shown (in bp). Point mutations ade7-152 and -50 are labeled in pink and purple, respectively. (F) Frequency of crossovers associated with a gene conversion event in wild type and fml1Δ at ade7-152×ade7-50; ALP1638×ALP1636 (WT, n = 12), ALP1670×ALP1669 (fml1Δ, n = 12). n indicates the number of independent crosses (see also Supplementary Table S2).
The contribution of Fml1 in directing recombination intermediates toward an NCO outcome is indicated by the increase in CO-GCs in fml1Δ crosses (Figure 1B and C), which is observed with both the ade6-3083 hotspot allele and the ade6-M375 non-hotspot allele indicating that it is independent of the level of GC (compare Figure 1B and D; Supplementary Table S2) (16). To corroborate this at an independent locus on a different chromosome, we constructed an equivalent recombination assay at ade7 (Figure 1E). Similar to what is seen at ade6 CO formation associated with GCs at ade7 is significantly increased in the absence of fml1 (compare Figure 1B, D and F).
Destabilization of the strand exchange process allows Mus81-independent repair of meiotic DSBs
In fission yeast Mus81-Eme1 is the structure-selective endonuclease solely responsible for meiotic CO formation. In line with this essential role the deletion of mus81 causes meiotic catastrophe, which results in a dramatic reduction in spore viability, and an almost complete lack of CO recombinants among the few survivors (Figure 2) (12,51). Deletion of the Rad51/Dmc1 mediators Swi5-Sfr1 and Rad55-Rad57 in a mus81Δ background partially rescues spore viability (Figure 2A) (16,52,53). However, the formation of CO-GCs is not restored in sfr1Δ mus81Δ double mutants (P = 0.689) (Supplementary Table S3) (16). We have shown that in this situation Fml1 is capable of processing recombination intermediates into NCOs indicating that Swi5-Sfr1 normally blocks Fml1 from acting on joint molecules thereby biasing the recombination outcome toward CO formation by Mus81-Eme1 processing (16). This ability to block NCO formation is seemingly not restricted to Swi5-Sfr1 as deleting any of the Rad51 paralogs, including Dmc1, also rescues the poor spore viability of a mus81 mutant to a similar degree and without increasing CO formation significantly (Figure 2 and Supplementary Table S3). This suggests that any destabilization of the strand exchange process alleviates the block against DNA helicases processing recombination intermediates into NCOs.
Figure 2.
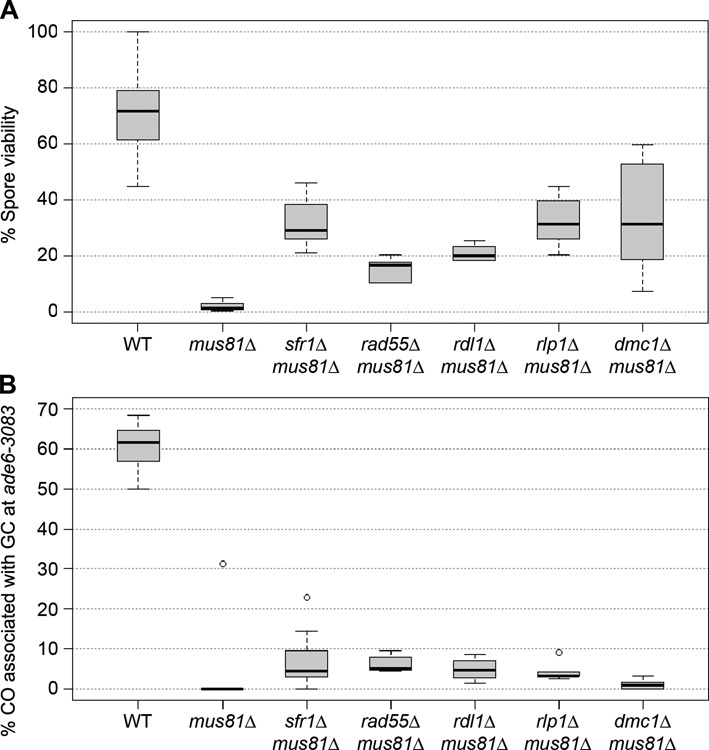
Deletion of strand exchange mediator genes and dmc1 rescue the poor spore viability of a mus81Δ without the formation of crossovers. (A) Viability of progeny from wild-type and mutant crosses; ALP733×ALP731 (WT, n = 20), ALP812×ALP813 (mus81Δ, n = 10), ALP820×ALP814 (sfr1Δ mus81Δ, n = 10), ALP1674×ALP1673 (rad55Δ mus81Δ, n = 6), ALP1676×ALP1675 (rdl1Δ mus81Δ, n = 4), ALP1680×ALP1679 (rlp1Δ mus81Δ, n = 6), ALP1672×ALP1671 (dmc1Δ mus81Δ, n = 12). (B) Frequency of crossovers associated with a gene conversion event in wild-type and mutant crosses (ade6-3083×ade6-469); ALP733×ALP731 (WT, n = 41), ALP802×ALP822 (mus81Δ, n = 10), ALP824×ALP823 (sfr1Δ mus81Δ, n = 19), ALP1674×ALP1673 (rad55Δ mus81Δ, n = 6), ALP1676×ALP1675 (rdl1Δ mus81Δ, n = 4), ALP1680×ALP1679 (rlp1Δ mus81Δ, n = 6), ALP1672×ALP1671 (dmc1Δ mus81Δ, n = 12). n indicates the number of independent crosses (see also Supplementary Table S3).
Rad51 paralogs and Swi5-Sfr1 promote full activation of meiotic recombination independent of each other
In order to assess the contribution of each Rad51 paralog or mediator to meiotic recombination, and determine whether there are any overlaps in function, we performed epistasis analyses. First we measured meiotic recombinant frequencies in rlp1Δ, rdl1Δ and sws1Δ single mutants and compared these to those of a rlp1Δ rdl1Δ double mutant and rlp1Δ rdl1Δ sws1Δ triple mutant (Figure 3A–C). In each case similar modest reductions in GC (2.0- to 3.6-fold), CO (1.6- to 2.1-fold) and CO-GC percentage were observed compared to wild type, which were not associated with any decline in spore viability (Figure 3D). All these differences in recombination frequencies were statistically significant, except the CO-GC of sws1Δ (Supplementary Table S4). These data are consistent with Rlp1, Rdl1 and Sws1 functioning as a complex to promote meiotic recombination similar to their homologs in budding yeast (37,54).
Figure 3.
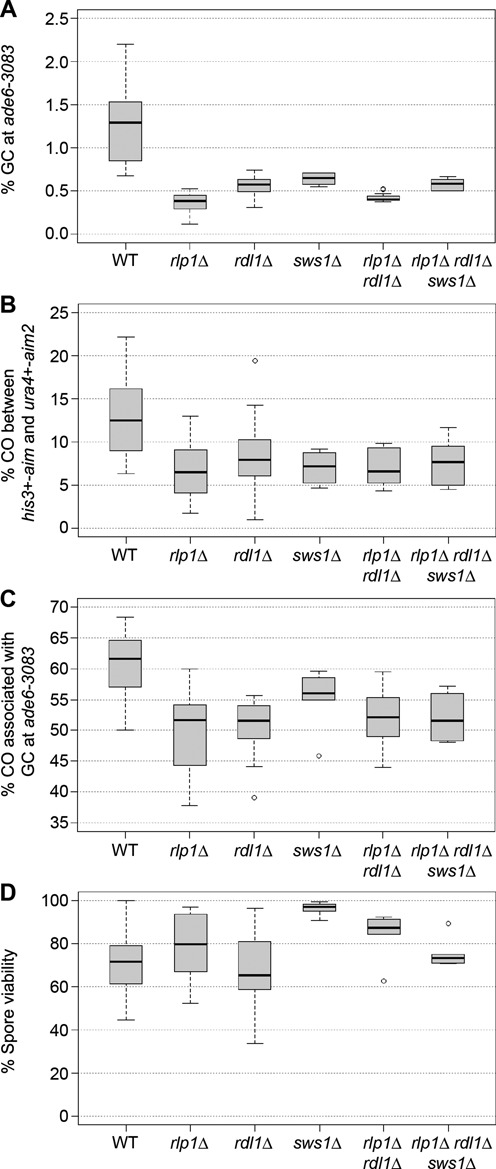
rlp1, rdl1 and sws1 are epistatic for meiotic recombination. (A) Percentage of gene conversions in wild-type and mutant crosses (ade6-3083×ade6-469); ALP733×ALP731 (WT, n = 41), ALP1623×ALP1620 (rlp1Δ, n = 18), ALP1621×ALP1611 (rdl1Δ, n = 18), ALP1708×ALP1707 (sws1Δ, n = 6), ALP1690×ALP1689 (rlp1Δ rdl1Δ, n = 12), ALP1733×ALP1732 (rlp1Δ rdl1Δ sws1Δ, n = 6). (B) Frequency of crossovers between his3+-aim and ura4+-aim2 in wild-type and mutant meioses; crosses as in (A). (C) Frequency of crossovers associated with a gene conversion event in wild-type and mutant crosses (ade6-3083×ade6-469); crosses as in (A). (D) Viability of progeny from wild-type and mutant crosses; ALP733×ALP731 (WT, n = 20), ALP1623×ALP1620 (rlp1Δ, n = 17), ALP1621×ALP1611 (rdl1Δ, n = 18), ALP1708×ALP1707 (sws1Δ, n = 5), ALP1690×ALP1689 (rlp1Δ rdl1Δ, n = 6), ALP1733×ALP1732 (rlp1Δ rdl1Δ sws1Δ, n = 6). n indicates the number of independent crosses (see also Supplementary Table S4).
Next, we compared sfr1Δ, rad55Δ and rlp1Δ single mutants (Figure 4). As previously noted sfr1Δ exhibits a strong reduction in GC (10-fold, P < 3.3 × 10−16) and overall CO (6-fold, P = 3.84 × 10−13) frequency, which has been attributed to a reduction in interhomolog recombination (53). In comparison, deleting rad55 has only a moderate, but highly significant, effect on the general meiotic recombination rate similar to rlp1Δ (Figure 4A–C and Supplementary Table S5). None of these decreases in recombination frequency causes major problems in spore viability, indicating that DSBs are repaired with reasonable efficiency, and each of the three homolog pairs receives sufficient COs to support proper chromosome segregation (Figure 4D). Analysis of the double mutant combinations reveals an epistatic relationship between rad55Δ and rlp1Δ, while both sfr1Δ rad55Δ and sfr1Δ rlp1Δ double mutants exhibit synergistic reductions in GC (in excess of 39-fold compared to wild type), CO (more than 22-fold compared to wild type) and spore viability (at least 7.5-fold lower than in wild type) (Figure 4 and Supplementary Table S5). The sfr1Δ rlp1Δ double mutant also displays a large (5.4-fold) decrease in CO-GC (Figure 4C and Supplementary Table S5). Due to low mating efficiency and strongly reduced spore viability only very few Ade+ recombinants could be recovered from crosses of sfr1Δ rad55Δ double mutants, which makes it unfeasible to determine the CO-GC rate. Together these data indicate that Rad55-Rad57 and Swi5-Sfr1 heterodimers can promote CO-GC events independently of each other, suggesting separate contributions to CO-GC recombination events in wild-type cells. This is consistent with previous findings (52,53,55). The Rlp1–Rdl1–Sws1 complex appears to function together with Rad55-Rad57, which suggests that they contribute to a shared function.
Figure 4.
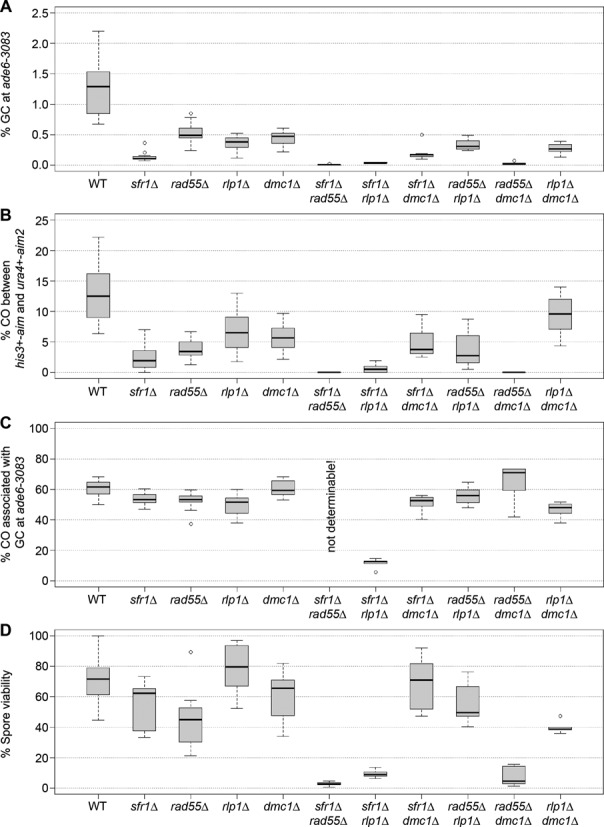
Strand exchange mediators are needed for wild-type levels of meiotic recombination. (A) Percentage of gene conversions in wild-type and mutant crosses (ade6-3083×ade6-469); ALP733×ALP731 (WT, n = 41), ALP800×ALP782 (sfr1Δ, n = 22), ALP1649×ALP1648 (rad55Δ, n = 12), ALP1623×ALP1620 (rlp1Δ, n = 18), ALP1545×ALP1544 (dmc1Δ, n = 12), ALP1735×ALP1734 (sfr1Δ rad55Δ, n = 6), ALP1700×ALP1699 (sfr1Δ rlp1Δ, n = 5), ALP1588×ALP1587 (sfr1Δ dmc1Δ, n = 12), ALP1704×ALP1703 (rad55Δ rlp1Δ, n = 12), ALP1696×ALP1695 (rad55Δ dmc1Δ, n = 6), ALP1694×ALP1693 (rlp1Δ dmc1Δ, n = 12). (B) Frequency of crossovers between his3+-aim and ura4+-aim2 in wild-type and mutant meioses; crosses as in (A). (C) Frequency of crossovers associated with a gene conversion event in wild-type and mutant crosses (ade6-3083×ade6-469); crosses as in (A). (D) Viability of progeny from wild-type and mutant crosses; ALP733×ALP731 (WT, n = 20), ALP800×ALP782 (sfr1Δ, n = 10), ALP1649×ALP1648 (rad55Δ, n = 11), ALP1623×ALP1620 (rlp1Δ, n = 17), ALP1545×ALP1544 (dmc1Δ, n = 10), ALP1735×ALP1734 (sfr1Δ rad55Δ, n = 6), ALP1700×ALP1699 (sfr1Δ rlp1Δ, n = 5), ALP1588×ALP1587 (sfr1Δ dmc1Δ, n = 12), ALP1704×ALP1703 (rad55Δ rlp1Δ, n = 12), ALP1696×ALP1695 (rad55Δ dmc1Δ, n = 6), ALP1694×ALP1693 (rlp1Δ dmc1Δ, n = 5). n indicates the number of independent crosses (see also Supplementary Table S5).
To further define the relationship between these factors, we analyzed their genetic interaction with dmc1 (Figure 4). Loss of dmc1 causes a modest ∼2-fold reduction in GC (P = 2.88 × 10−8) and CO (P = 1.14 × 10−5), while retaining a wild-type level of spore viability (Figure 4A, B and D and Supplementary Table S5), however, unlike sfr1Δ, rad55Δ and rlp1Δ, the CO-GC percentage is not reduced (P = 0.795) (Figure 4C and Supplementary Table S5). The combination of sfr1Δ and dmc1Δ mutations results in an epistatic interaction displaying a sfr1Δ phenotype (Figure 4A and B). It has previously been noted that the levels of interhomolog recombination intermediate in a dmc1Δ swi5Δ double mutant lie between those of dmc1Δ and swi5Δ single mutants (52). Accordingly we observe a slight increase of GC and CO percentage in the dmc1Δ sfr1Δ double mutant compared with the sfr1Δ single mutant, however, the change in GC frequency is not statistically significant (P = 0.354), and the decrease in overall CO percentage is only weakly so (P = 0.065). In contrast a rad55Δ dmc1Δ double mutant exhibits similar low levels of GC, CO and spore viability as a sfr1Δ rad55Δ double mutant. These data indicate that Swi5-Sfr1 and Dmc1 function together to promote meiotic recombination, while Rad55-Rad57 contributes by distinct means.
Intriguingly, rlp1Δ and rdl1Δ exhibit a quite different genetic interaction with dmc1Δ than either sfr1Δ or rad55Δ, with epistasis for GC and CO frequency but a slightly reduced percentage of CO-GCs compared to either single mutant (P = 0.487 for dmc1Δ rlp1Δ versus rlp1Δ, and P = 0.045 for dmc1Δ rdl1Δ versus rdl1Δ) (Figure 4A–C , Supplementary Figure S2 and Supplementary Table S5). Moreover, unlike a rad55Δ dmc1Δ double mutant, which shows a 10-fold reduction in spore viability, rlp1Δ dmc1Δ and rdl1Δ dmc1Δ double mutants display only modest decreases in spore viability compared to wild type (∼2-fold) (Figure 4D and Supplementary Table S5), indicating that DSB repair and meiotic chromosome segregation remain fairly efficient. Altogether these data reveal a more complex interplay between Rad55-Rad57, Rlp1-Rdl1-Sws1 and Swi5-Sfr1 than was suggested by the epistatic relationship between rad55Δ and rlp1Δ.
Absence of Rad51 paralogs/mediators allows both Rqh1 and Fml1 to promote NCO recombinant formation
In sfr1Δ, rad55Δ, rlp1Δ and rdl1Δ single mutants CO-GC events are 50–52%, this is significantly lower than in wild type (Figures 3C and 4C; for P values see Supplementary Table S5). In the case of sfr1Δ we have previously shown that this reduction is reversed by deletion of fml1 (16) (Figure 5A). To determine whether Fml1 is also responsible for suppressing CO-GC in the absence of the Rad51 paralogs we measured the percentage of CO-GC in fml1Δ rad55Δ, fml1Δ rlp1Δ and fml1Δ rdl1Δ double mutants. In each case the percentage of CO-GC increases significantly from the Rad51 paralog mutant level (for P values see Supplementary Table S6), however only in a fml1Δ rad55Δ double mutant does it increase to the ∼70% level observed in a fml1Δ single mutant, in the fml1Δ rlp1Δ and fml1Δ rdl1Δ double mutants the level increases to ∼60%, which is the same as in wild type (Figure 5B and C and Supplementary Figure S3). These data suggest that both Swi5-Sfr1 and Rad55-Rad57 are needed to similar extents to constrain Fml1's NCO-promoting activity and thereby promote CO-GC, whereas Rlp1 and Rdl1 are required to a lesser extent.
Figure 5.
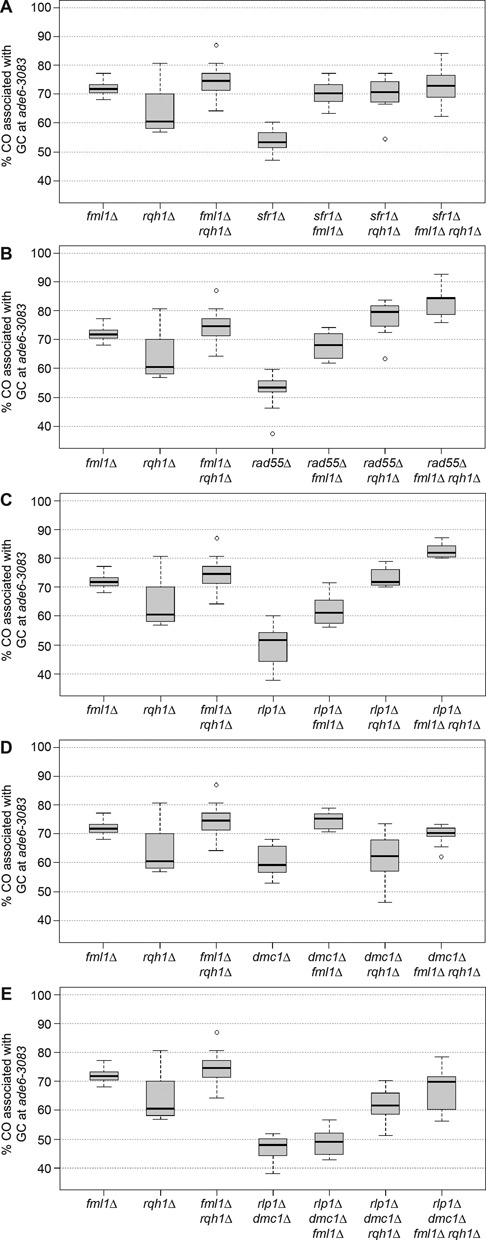
DNA helicases, strand exchange mediators and Dmc1 shape the crossover/non-crossover decision. (A–E) Frequency of crossovers associated with a gene conversion event in mutant crosses (ade6-3083×ade6-469); ALP1133×FO2608 (fml1Δ, n = 15), ALP781×ALP780 (rqh1Δ, n = 10), ALP945×ALP944 (fml1Δ rqh1Δ, n = 22). (A) ALP800×ALP782 (sfr1Δ, n = 22), ALP1134×FO2609 (sfr1Δ fml1Δ , n = 12), ALP801×ALP821 (sfr1Δ rqh1Δ, n = 10), ALP1363×ALP1362 (sfr1Δ fml1Δ rqh1Δ, n = 12). (B) ALP1649×ALP1648 (rad55Δ, n = 11), ALP1658×ALP1657 (rad55Δ fml1Δ , n = 14), UoA328×UoA327 (rad55Δ rqh1Δ, n = 12), UoA330×UoA329 (rad55Δ fml1Δ rqh1Δ, n = 9). (C) ALP1623×ALP1620 (rlp1Δ, n = 18), ALP1664×ALP1663 (rlp1Δ fml1Δ, n = 12), FO3158×FO3159 (rlp1Δ rqh1Δ, n = 10), FO3142×FO3143 (rlp1Δ fml1Δ rqh1Δ, n = 10). (D) ALP1545×ALP1544 (dmc1Δ, n = 10), ALP1590×ALP1589 (dmc1Δ fml1Δ, n = 12), UoA295×UoA294 (dmc1Δ rqh1Δ, n = 8), UoA297×UoA296 (dmc1Δ fml1Δ rqh1Δ, n = 11). (E) ALP1694×ALP1693 (rlp1Δ dmc1Δ, n = 12), FO3146×FO3147 (rlp1Δ dmc1Δ fml1Δ, n = 10), UoA322×UoA321 (rlp1Δ dmc1Δ rqh1Δ, n = 12), UoA299×UoA298 (rlp1Δ dmc1Δ fml1Δ rqh1Δ, n = 6). n indicates the number of independent crosses (see also Supplementary Table S6).
Similar to rad55Δ and rlp1Δ mutants, a dmc1Δ mutant exhibits a moderate decrease in GC and CO frequency (Figure 4A and B and Supplementary Table S5), but unlike them its CO-GC percentage is not reduced (Figure 4C and Supplementary Table S5). This suggests that Dmc1 is not required for constraining Fml1 from promoting NCO formation. Accordingly, the CO-GC percentages in a dmc1Δ fml1Δ double mutant and fml1Δ single mutant are similar (P = 0.145) (Figure 5D). However, a different pattern emerges in a dmc1Δ rlp1Δ double mutant background (Figure 5E). This double mutant exhibits a similar, maybe slightly lower, CO-GC percentage than a rlp1Δ mutant, but, unlike in either single mutant, deletion of fml1 does not increase this percentage to wild-type or higher levels (P = 0.687). It does, however, improve GC frequency (P = 4.77 × 10−4) and cause a synergistic reduction in spore viability (Figure 6E and Supplementary Table S6). The heightened GC frequency of a dmc1Δ rlp1Δ fml1Δ triple compared to a dmc1Δ rlp1Δ double mutant could indicate that in the absence of both dmc1 and rlp1 Fml1 becomes capable of driving template choice away from the homologous chromosome toward the sister chromatid.
Figure 6.
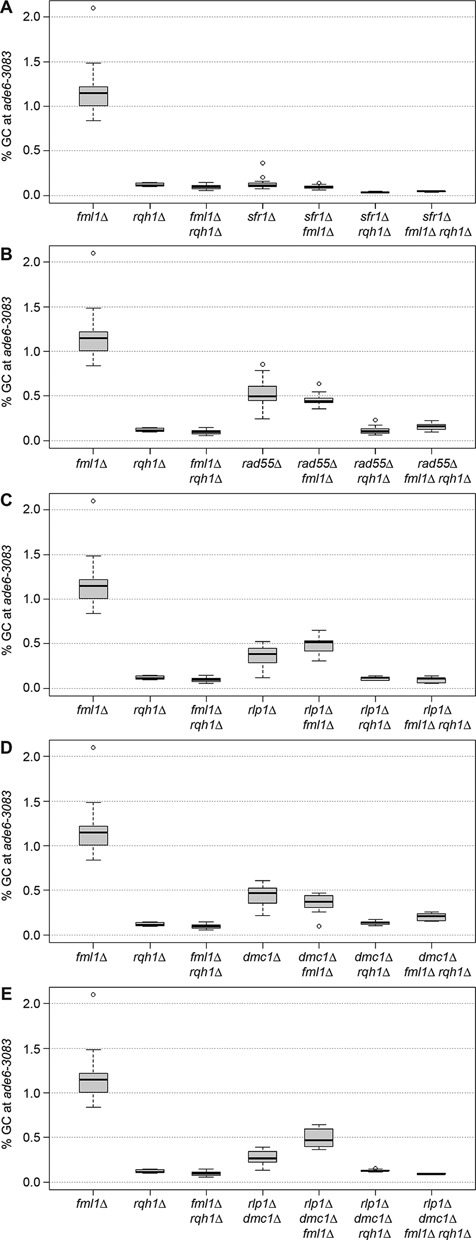
DNA helicases, strand exchange mediators and Dmc1 affect the interhomolog bias. (A–E) Percentage of gene conversion in mutant crosses (ade6-3083×ade6-469), crosses as in Figure 5A–E (see also Supplementary Table S6).
RecQ-type helicases in other organisms have been implicated in promoting NCOs during meiosis (19,20,25). However, deletion of the RecQ-type helicase in S. pombe Rqh1 has no significant effect on the percentage of CO-GC in either a wild type (P = 0.631) or a fml1Δ mutant (P = 0.458) (Figure 5) (16) even though it strongly limits COs in mitotic cells (56). To see whether Rqh1 might be similarly constrained as Fml1 during meiosis, we measured the percentage of CO-GC in double mutants of rqh1Δ with sfr1Δ, rad55Δ and rlp1Δ (Figure 5A–C). In each case CO-GCs increased to 70% or more (for P values see Supplementary Table S6). Unlike fml1, deletion of rqh1 in a dmc1Δ rlp1Δ mutant background also resulted in a significant increase in CO-GC percentage (P = 1.63 × 10−5) (Figure 5E). The deletion of fml1 in rqh1Δ rad55Δ, rqh1Δ rlp1Δ and rqh1Δ dmc1Δ rlp1Δ mutants resulted in a further increase in CO-GC percentage, whereas no additional change was observed in a fml1Δ rqh1Δ sfr1Δ triple mutant (Figure 5). These data indicate that Rqh1 can promote NCO formation during meiosis, but is normally constrained from doing so by Rad51 paralogs/mediators. Moreover, Rqh1 and Fml1 can seemingly promote NCOs by both common and separate means depending on which Rad51 paralogs/mediators are present.
Consistent with previous findings a rqh1Δ mutant exhibits an ∼10-fold reduction in GC (P = 1.32 × 10−11) and ∼2.5-fold reduction in CO (P = 1.62 × 10−4) compared with wild type (Figure 6 and Supplementary Table S6) (57). This loss of recombinogenic activity is not associated with a decline in DSB formation or repair, a change in the ratio of interhomolog versus intersister recombination or a dramatic reduction in spore viability (Supplementary Table S6) (57). Instead it has been proposed that Rqh1 might promote hybrid DNA formation necessary for GC (57). This putative function of Rqh1 would appear to operate alongside Rad55 and Rlp1 to support GC formation as both rqh1Δ rad55Δ (P = 0.80) and rqh1Δ rlp1Δ (P = 0.70) double mutants exhibit similar GC frequencies as a rqh1Δ single mutant (Figure 6B and C). However, a rqh1Δ rad55Δ double mutant exhibits a synergistic reduction in spore viability (Supplementary Table S6) indicating that Rqh1 and Rad55 also have non-overlapping roles in promoting reproductive success. A rqh1Δ sfr1Δ double mutant exhibits a 3- to 4-fold reduction in GC frequency compared to a rqh1Δ (P = 2.38 × 10−8) or a sfr1Δ (P = 1.97 × 10−4) single mutant (Figure 6A). At least in part this can be explained by an additive effect of a weakened interhomolog bias and reduced hybrid DNA formation during interhomolog recombination.
The Rlp1–Rdl1–Sws1 complex, Dmc1, and Fml1 limit hybrid DNA formation driven in part by Rqh1
Not only are the overall levels of CO-GC events strongly decreased in dmc1Δ rlp1Δ double mutants (Figure 5E), but the distribution of NCO recombinants among GCs is also starkly different from that in wild-type or either single mutant (Figure 7A). In wild type, the vast majority of NCO-Ade+ recombinants are linked to the flanking marker of the hotspot allele (i.e. ura4+-aim2). This can be explained by the formation of hybrid DNA not extending as far as the cold allele (i.e. ade6-469), as envisaged in Supplementary Figure S1 (scenarios 1–4 and 6–9). However, in dmc1Δ rlp1Δ and dmc1Δ rdl1Δ double mutants, and to some extent in fml1Δ rlp1Δ and fml1Δ rdl1Δ double mutants, a high proportion of NCO-GC recombinants are linked to the flanking marker of the cold allele (Figure 7A and Supplementary Figure S4), this skew is even more pronounced in a dmc1Δ rlp1Δ fml1Δ triple mutant (Figure 7A). One likely explanation for this is extensive branch migration beyond the position of the cold allele (Supplementary Figure S1, scenarios 5 and 10). These data suggest that the Rlp1–Rdl1(–Sws1) complex and Dmc1 perform redundant roles in limiting hybrid DNA formation, with an additional contribution by Fml1. Deletion of rqh1 in dmc1Δ rlp1Δ and dmc1Δ rlp1Δ fml1Δ backgrounds, while significantly reducing the overall frequency of NCO-GCs, does not restore their wild-type ratio (Figure 7A). These data are consistent with Rqh1's proposed role in promoting hybrid DNA formation, but also suggest that extensive hybrid DNA can be formed without it when Dmc1 and Rlp1 are absent.
Figure 7.
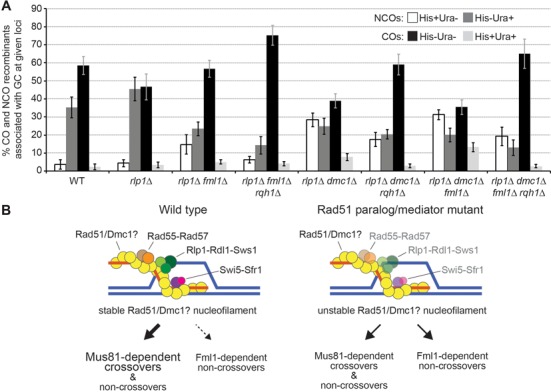
Double and triple mutant combinations of rlp1, dmc1 and fml1 show a skew in non-crossover recombination classes. (A) Frequencies of different crossover and non-crossover classes associated with a gene conversion event in wild-type and mutant crosses (ade6-3083×ade6-469); ALP733×ALP731 (WT, n = 41), ALP1623×ALP1620 (rlp1Δ, n = 18), ALP1664×ALP1663 (fml1Δ rlp1Δ, n = 12), FO3142×FO3143 (fml1Δ rlp1Δ rqh1Δ, n = 10), ALP1694×ALP1693 (dmc1Δ rlp1Δ, n = 12), UoA322×UoA321 (dmc1Δ rlp1Δ rqh1Δ, n = 12), FO3146×FO3147 (dmc1Δ fml1Δ rlp1Δ, n = 10), UoA299×UoA298 (dmc1Δ fml1Δ rlp1Δ rqh1Δ, n = 6). n indicates the number of independent crosses (see also Supplementary Table S7). (B) Model depicting possible outcomes of processing a D-loop in the presence or absence of Rad51 paralogs/mediators. Whether strand exchange by both Rad51- and Dmc1-nucleofilaments is supported by Rad55-57 and/or Rlp1-Rdl1-Sws1 has not been characterized biochemically yet. Thickness and style of arrows indicates the prevalence of Mus81-dependent cleavage or Fml1-dependent non-crossover formation via synthesis-dependent strand annealing (SDSA). DNA strands are shown as blue and orange lines.
DISCUSSION
The Rad51 paralogs/mediators in meiotic recombination
In agreement with previous studies (52,53) we have shown that Rad55-Rad57 and Swi5-Sfr1 have both distinct and overlapping/compensatory functions for promoting meiotic recombination. Both protein complexes are capable of mediating the nucleation of Rad51 onto RPA-coated ssDNA and enhancing nucleoprotein filament stability, albeit Swi5-Sfr1 is a much better mediator for Dmc1 than it is for Rad51 (35,36,58). Accordingly, sfr1 and dmc1 exhibit an epistatic interaction for meiotic recombination and spore viability, as opposed to the synergistic interaction observed between rad55 and dmc1. Epistasis analysis also indicates that the Rlp1–Rdl1–Sws1 complex (42) functions together with Rad55-Rad57, which accords with recent data on the homologous complex in budding yeast (comprised of Shu1, Shu2, Psy3 and Csm2), which was shown to physically interact with Rad55-Rad57 and function epistatically with it for mitotic recombination (59). Similar to Rad55-Rad57, Psy3-Csm2 enhance Rad51 nucleoprotein filament stability in vitro and plays an important role in promoting meiotic recombination, which becomes essential in the absence of dmc1 (37,54). However, unlike its budding yeast equivalent and in contrast to a rad55Δ dmc1Δ double mutant, a rlp1Δ dmc1Δ double mutant in fission yeast still exhibits moderate levels of meiotic recombination and spore viability. One possible explanation is that in fission yeast the Rlp1–Rdl1–Sws1 complex imposes some kind of constraint over Swi5-Sfr1 assisting Rad51 nucleoprotein filament formation and function. It is also possible that the absence of Dmc1 allows Swi5-Sfr1 to support Rad51-mediated recombination more often. Swi5-Sfr1 loads and stabilizes Dmc1 filaments, whereas it only stabilizes, but does not load, Rad51 filaments (35), and consequently Swi5-Sfr1 might tend to promote Dmc1- rather than Rad51-driven recombination because of its earlier association with Dmc1. If these constraints are removed Swi5-Sfr1 might be available to promote Rad51 activity (either together with or in parallel to Rad55-Rad57) sufficiently well to partially compensate for the lack of Rlp1 and Dmc1. This potential interplay between Rad51 paralogs and mediators, influencing the contributions that Rad51 and Dmc1 make to meiotic recombination, contrasts with the situation in S. cerevisiae where Rad51 itself functions as a mediator of Dmc1-driven strand exchange (60), and its own strand exchange activity is massively curbed to avoid competition between it and Dmc1. This is achieved by a specialized and Saccharomyces-specific factor, called Hed1, which downregulates Rad51's activity (61–63).
Based on our data deletion of any of the Rad51/Dmc1 paralogs causes a significant reduction in intragenic recombination. This contradicts a previous study, which found no effect of deleting rad55/rad57 or rlp1 on GC frequency (64). We suspect that this difference is due to analyzing GC at a hotspot allele (our study) versus a non-hotspot allele (64), and could relate to the phenomenon of CO invariance (53). According to CO invariance, recombination at non-hotspots, but not at hotspots, strongly depends on Dmc1 (53). Together with the observation that Rad55-Rad57 and Rlp1 play a less important role for GC formation at non-hotspots (64), one might surmise that there is a division of labor between Dmc1 and Rad55-Rad57 plus Rdl1-Rlp1-Sws1 at sites with different levels of meiotic recombination competence. However, our data indicate that these suggested relationships do not hold true at the ade6-3083 hotspot. In wild type a similar level of GC frequency is observed at the ade6-3083 hotspot compared with the ade6-3049 hotspot (∼1.3% versus ∼1.4%), whereas in a dmc1Δ mutant GC rate is reduced 3-fold (P = 2.88 × 10−8) at ade6-3083 (Figure 4A and Supplementary Table S5) in contrast to ade6-3049 where it is reduced by only 1.2-fold (53). Intriguingly, the Smith lab reported a 3-fold reduction in GC frequency for the coldest ade6 allele combination (3057 × M375) they tested in a wild-type versus dmc1Δ comparison (53), similar to what we see at the ade6-3083 hotspot.
Swi5-Sfr1 and Rad51 paralogs function together to restrict Fml1 promoting NCO formation
We have previously shown that Fml1 restricts CO-GC percentage in wild-type meiosis most likely by directing repair via SDSA (16). However, its ability to do this is normally restricted by Swi5-Sfr1, and consequently it cannot compensate for any loss of Mus81-Eme1, which results in meiotic catastrophe due to unprocessed recombination intermediates (16,21). The finding that mus81Δ spore viability is dramatically improved following the deletion not only of sfr1 but also rad55, rlp1, rdl1 and dmc1, without resulting in a recovery of CO formation, suggests that Swi5-Sfr1 and the Rad51 paralogs function together to constrain Fml1 and possibly other DNA helicases from processing recombination intermediates (Figure 7B). Our favored model is that they physically exclude Fml1 from accessing D-loop DNA by promoting the formation and stabilization of Rad51- (and Dmc1-) nucleoprotein filaments. Consistent with this model deletion of sfr1 or most of the Rad51-paralog genes reduces CO-GC percentage in a Fml1-dependent manner. One exception is dmc1, which, as already mentioned, is not required for CO-GC formation during meiotic recombination in fission yeast.
Rqh1 might promote NCOs by driving hybrid DNA formation
Unlike Fml1 loss of Rqh1 does not result in a significant increase in CO-GC percentage in a wild-type background. However, in both sfr1Δ and Rad51-paralog mutant backgrounds increases in CO-GC percentage are observed upon removal of rqh1. At first glance this could indicate that Rqh1, like Fml1, drives NCO formation but is normally prevented from doing so by fully stabilized Rad51- and Dmc1-nucleoprotein filaments. However, it has been postulated that Rqh1 promotes hybrid DNA formation (57), and one way this could be achieved is through driving the conversion of D-loops into HJs, which could then branch migrate away from the site of strand invasion. In our assay such an activity if left unfettered could lower the CO-GC percentage by decreasing the number of D-loops that are resolved by Mus81-Eme1 and forcing more HJ resolution, which would generate equal numbers of COs and NCOs (Supplementary Figure S1; less of scenarios 2 and 7 and more of scenarios 3–5 and 8–9). The conversion of D-loops into HJs might normally be controlled through the combined action of Swi5-Sfr1 and the Rad51 paralogs, and an impairment of the D-loop stabilization function of these factors might promote a more rapid conversion rate, providing less opportunity for D-loop cleavage. However, some D-loops may remain accessible to Fml1 enabling it to promote NCO formation via the SDSA pathway. The additive increase in CO-GC percentage observed in both rad55Δ fml1Δ rqh1Δ and rlp1Δ fml1Δ rqh1Δ triple mutants compared to the rad55Δ fml1Δ (P = 1.42 × 10−5) and rlp1Δ fml1Δ (P = 4.45 × 10−6) double mutants, respectively (Figure 5B and C), might therefore be explained by the combined effect of less D-loop unwinding and conversion to HJs, resulting in more D-loops being available for cleavage by Mus81-Eme1 (Supplementary Figure S1—scenarios 2 and 7).
Once formed the branch migration of the HJ would either extend or limit the amount of hybrid DNA that is formed depending on its direction. Rlp1-Rdl1-Sws1 and Dmc1 appear to play overlapping roles in limiting hybrid DNA formation based on the increase in the proportion of His+ Ura− GC recombinants observed in rlp1Δ/rdl1Δ dmc1Δ double mutants (Figure 7A, Supplementary Figure S4 and Supplementary Figure S1—scenarios 5 and 10). The rapid conversion of all D-loops into HJs would prevent Fml1 from driving NCO formation via its D-loop unwinding activity, which could explain why no increase in CO-GC percentage is observed in a rlp1Δ dmc1Δ fml1Δ triple mutant. However, Fml1's inability to promote NCOs in the absence of Rlp1 and Dmc1 is seemingly restored upon deletion of Rqh1 based on the additive increase in CO-GC percentage observed in a rlp1Δ dmc1Δ rqh1Δ fml1Δ quadruple mutant (Figure 5E). Intriguingly, the presence of fml1 is required to maintain the reasonably high levels of spore viability in a rlp1Δ dmc1Δ mutant even though its NCO-promoting activity appears to be inactive (Supplementary Table S6). Possible explanations are that either the efficiency of overall DNA repair is strongly negatively affected in a rlp1Δ dmc1Δ fml1Δ triple mutant leaving unrepaired DSBs or that Fml1 might promote the resolution of HJs by Mus81-Eme1 by catalyzing their branch migration to existing DNA strand discontinuities. Further studies are needed to test the validity of these ideas.
CONCLUSION
The role of the Rad51 paralogs/mediators in promoting homologous recombination is well established; however their role in directing homologous recombination outcome by influencing the processing of recombination intermediates is only recently becoming recognized. A common theme that is emerging is one where Rad51 paralogs/mediators play a role in promoting Rad51-nucleoprotein filament stability, which restricts the ability of DNA helicases/translocases to act. This has been observed in vitro for human RAD51B and RAD51C limiting the antirecombinogenic activity of the RecQ-type helicase BLM on RAD51 (65), and for budding yeast Rad55-Rad57 enabling Rad51 to resist displacement from DNA by the Srs2 DNA helicase (36). Similarly Rad51 paralogs/mediators appear to antagonize the Rad51 nucleoprotein filament disrupting activity of the Fbh1 DNA helicase in fission yeast (66,67). While these studies reveal the important role that Rad51 paralogs play in preventing DNA helicases from aborting homologous recombination, our study highlights how their role in promoting Rad51-nucleoprotein filament stability may also be important for controlling D-loop processing. We propose that the full ensemble of Rad51 paralogs together with Swi5-Sfr1 are required to create and maintain a Rad51(/Dmc1?)-nucleoprotein filament that remains permissive to Mus81-Eme1 nuclease activity but at the same time acts to occlude and/or constrain Fml1 and Rqh1 activity on the underlying D-loop. Modulation of the recruitment of these proteins during Rad51(/Dmc1?)-nucleoprotein filament assembly could therefore provide a mechanism for influencing both hybrid DNA formation and the CO/NCO decision.
AVAILABILITY
Supporting online material for this publication is available at http://dx.doi.org/10.6084/m9.figshare.1197171.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful to Jürg Kohli, Ramsay J. McFarlane, Paul Russell, Gerald R. Smith, Walter W. Steiner and the National BioResource Project (NBRP) Japan for providing strains and to C. Bryer for technical assistance.
FUNDING
Wellcome Trust [090767/Z/09/Z to M.C.W.]; College of Life Sciences and Medicine, University of Aberdeen [to A.L., in part]. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pradillo M., Santos J.L. The template choice decision in meiosis: is the sister important. Chromosoma. 2011;120:447–454. doi: 10.1007/s00412-011-0336-7. [DOI] [PubMed] [Google Scholar]
- 2.Humphryes N., Hochwagen A. A non-sister act: recombination template choice during meiosis. Exp. Cell Res. 2014;329:53–60. doi: 10.1016/j.yexcr.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neale M.J., Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krogh B.O., Symington L.S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 5.Krejci L., Altmannova V., Spirek M., Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwacha A., Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 7.Cromie G.A., Hyppa R.W., Taylor A.F., Zakharyevich K., Hunter N., Smith G.R. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cromie G.A., Smith G.R. Branching out: meiotic recombination and its regulation. Trends Cell Biol. 2007;17:448–455. doi: 10.1016/j.tcb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Youds J.L., Boulton S.J. The choice in meiosis—defining the factors that influence crossover or non-crossover formation. J. Cell Sci. 2011;124:501–513. doi: 10.1242/jcs.074427. [DOI] [PubMed] [Google Scholar]
- 10.Whitby M.C. Making crossovers during meiosis. Biochem. Soc. Trans. 2005;33:1451–1455. doi: 10.1042/BST0331451. [DOI] [PubMed] [Google Scholar]
- 11.Svendsen J.M., Harper J.W. GEN1/Yen1 and the SLX4 complex: solutions to the problem of Holliday junction resolution. Genes Dev. 2010;24:521–536. doi: 10.1101/gad.1903510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osman F., Dixon J., Doe C.L., Whitby M.C. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 13.Berchowitz L.E., Francis K.E., Bey A.L., Copenhaver G.P. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 2007;3:e132. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins J.D., Buckling E.F., Franklin F.C.H., Jones G.H. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 2008;54:152–162. doi: 10.1111/j.1365-313X.2008.03403.x. [DOI] [PubMed] [Google Scholar]
- 15.Ciccia A., McDonald N., West S.C. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 16.Lorenz A., Osman F., Sun W., Nandi S., Steinacher R., Whitby M.C. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science. 2012;336:1585–1588. doi: 10.1126/science.1220111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crismani W., Girard C., Froger N., Pradillo M., Santos J.L., Chelysheva L., Copenhaver G.P., Horlow C., Mercier R. FANCM limits meiotic crossovers. Science. 2012;336:1588–1590. doi: 10.1126/science.1220381. [DOI] [PubMed] [Google Scholar]
- 18.Knoll A., Higgins J.D., Seeliger K., Reha S.J., Dangel N.J., Bauknecht M., Schröpfer S., Franklin F.C.H., Puchta H. The Fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in Arabidopsis. Plant Cell. 2012;24:1448–1464. doi: 10.1105/tpc.112.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J., Dayani Y., Lichten M. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell. 2012;46:43–53. doi: 10.1016/j.molcel.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukaszewicz A., Howard-Till R.A., Loidl J. Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex. Nucleic Acids Res. 2013;41:9296–9309. doi: 10.1093/nar/gkt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenz A., Whitby M.C. How not to get cross(ed): a novel role for FANCM orthologs in meiotic recombination. Cell Cycle. 2012;11:3347–3348. doi: 10.4161/cc.21844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharjee S., Osman F., Feeney L., Lorenz A., Bryer C., Whitby M.C. MHF1–2/CENP-S-X performs distinct roles in centromere metabolism and genetic recombination. Open Biol. 2013;3:130102. doi: 10.1098/rsob.130102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Dangel N.J., Knoll A., Puchta H. MHF1 plays Fanconi anaemia complementation group M protein (FANCM)-dependent and FANCM-independent roles in DNA repair and homologous recombination in plants. Plant J. 2014;78:822–833. doi: 10.1111/tpj.12507. [DOI] [PubMed] [Google Scholar]
- 24.Girard C., Crismani W., Froger N., Mazel J., Lemhemdi A., Horlow C., Mercier R. FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 2014;42:9087–9095. doi: 10.1093/nar/gku614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rockmill B., Fung J.C., Branda S.S., Roeder G.S. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 2003;13:1954–1962. doi: 10.1016/j.cub.2003.10.059. [DOI] [PubMed] [Google Scholar]
- 26.Gaillard P.-H.L., Noguchi E., Shanahan P., Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- 27.Ciccia A., Constantinou A., West S.C. Identification and characterization of the human Mus81-Eme1 endonuclease. J. Biol. Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- 28.Gaskell L.J., Osman F., Gilbert R.J.C., Whitby M.C. Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates. EMBO J. 2007;26:1891–1901. doi: 10.1038/sj.emboj.7601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ehmsen K.T., Heyer W.-D. A junction branch point adjacent to a DNA backbone nick directs substrate cleavage by Saccharomyces cerevisiae Mus81-Mms4. Nucleic Acids Res. 2009;37:2026–2036. doi: 10.1093/nar/gkp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz E.K., Wright W.D., Ehmsen K.T., Evans J.E., Stahlberg H., Heyer W.-D. Mus81-Mms4 functions as a single heterodimer to cleave nicked intermediates in recombinational DNA repair. Mol. Cell. Biol. 2012;32:3065–3080. doi: 10.1128/MCB.00547-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De los Santos T., Hunter N., Lee C., Larkin B., Loidl J., Hollingsworth N.M. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agostinho A., Meier B., Sonneville R., Jagut M., Woglar A., Blow J., Jantsch V., Gartner A. Combinatorial regulation of meiotic holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 2013;9:e1003591. doi: 10.1371/journal.pgen.1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Neil N.J., Martin J.S., Youds J.L., Ward J.D., Petalcorin M.I.R., Rose A.M., Boulton S.J. Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during Caenorhabditis elegans meiosis. PLoS Genet. 2013;9:e1003582. doi: 10.1371/journal.pgen.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito T.T., Lui D.Y., Kim H.-M., Meyer K., Colaiácovo M.P. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet. 2013;9:e1003586. doi: 10.1371/journal.pgen.1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murayama Y., Kurokawa Y., Tsutsui Y., Iwasaki H. Dual regulation of Dmc1-driven DNA strand exchange by Swi5-Sfr1 activation and Rad22 inhibition. Genes Dev. 2013;27:2299–2304. doi: 10.1101/gad.218693.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J., Renault L., Veaute X., Fabre F., Stahlberg H., Heyer W.-D. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature. 2011;479:245–248. doi: 10.1038/nature10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasanuma H., Tawaramoto M.S., Lao J.P., Hosaka H., Sanda E., Suzuki M., Yamashita E., Hunter N., Shinohara M., Nakagawa A., et al. A new protein complex promoting the assembly of Rad51 filaments. Nat. Commun. 2013;4:1676. doi: 10.1038/ncomms2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabatinos S.A., Forsburg S.L. Molecular genetics of Schizosaccharomyces pombe. Methods Enzymol. 2010;470:759–795. doi: 10.1016/S0076-6879(10)70032-X. [DOI] [PubMed] [Google Scholar]
- 39.Smith G.R. Genetic analysis of meiotic recombination in Schizosaccharomyces pombe. Methods Mol. Biol. 2009;557:65–76. doi: 10.1007/978-1-59745-527-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 41.Keeney J.B., Boeke J.D. Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martín V., Chahwan C., Gao H., Blais V., Wohlschlegel J., Russell P., Yates J.R., McGowan C.H. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato M., Dhut S., Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- 44.Lorenz A., West S.C., Whitby M.C. The human Holliday junction resolvase GEN1 rescues the meiotic phenotype of a Schizosaccharomyces pombe mus81 mutant. Nucleic Acids Res. 2010;38:1866–1873. doi: 10.1093/nar/gkp1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parvanov E., Kohli J., Ludin K. The mating-type-related bias of gene conversion in Schizosaccharomyces pombe. Genetics. 2008;180:1859–1868. doi: 10.1534/genetics.108.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osman F., Adriance M., McCready S. The genetic control of spontaneous and UV-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Curr. Genet. 2000;38:113–125. doi: 10.1007/s002940000145. [DOI] [PubMed] [Google Scholar]
- 47.Szankasi P., Heyer W.D., Schuchert P., Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe. Wild-type and mutant alleles including the recombination hot spot allele ade6-M26. J. Mol. Biol. 1988;204:917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- 48.Supporting online material. 2014. figshare. 10.6084/m9.figshare.1197171.
- 49.Faul F., Erdfelder E., Lang A.-G., Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 50.Faul F., Erdfelder E., Buchner A., Lang A.-G. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav. Res. Methods. 2009;41:1149–1160. doi: 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
- 51.Boddy M.N., Gaillard P.H., McDonald W.H., Shanahan P., Yates J.R., Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 52.Ellermeier C., Schmidt H., Smith G.R. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics. 2004;168:1891–1898. doi: 10.1534/genetics.104.034280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyppa R.W., Smith G.R. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell. 2010;142:243–255. doi: 10.1016/j.cell.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong S., Sung Y., Yu M., Lee M., Kleckner N., Kim K.P. The logic and mechanism of homologous recombination partner choice. Mol. Cell. 2013;51:440–453. doi: 10.1016/j.molcel.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akamatsu Y., Dziadkowiec D., Ikeguchi M., Shinagawa H., Iwasaki H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl Acad. Sci. U.S.A. 2003;100:15770–15775. doi: 10.1073/pnas.2632890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun W., Nandi S., Osman F., Ahn J.S., Jakovleska J., Lorenz A., Whitby M.C. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cromie G.A., Hyppa R.W., Smith G.R. The fission yeast BLM homolog Rqh1 promotes meiotic recombination. Genetics. 2008;179:1157–1167. doi: 10.1534/genetics.108.088955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 59.Godin S., Wier A., Kabbinavar F., Bratton-Palmer D.S., Ghodke H., Van Houten B., Vandemark A.P., Bernstein K.A. The Shu complex interacts with Rad51 through the Rad51 paralogues Rad55-Rad57 to mediate error-free recombination. Nucleic Acids Res. 2013;41:4525–4534. doi: 10.1093/nar/gkt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cloud V., Chan Y.-L., Grubb J., Budke B., Bishop D.K. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012;337:1222–1225. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsubouchi H., Roeder G.S. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006;20:1766–1775. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lao J.P., Cloud V., Huang C.-C., Grubb J., Thacker D., Lee C.-Y., Dresser M.E., Hunter N., Bishop D.K. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 2013;9:e1003978. doi: 10.1371/journal.pgen.1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y., Gaines W.A., Callender T., Busygina V., Oke A., Sung P., Fung J.C., Hollingsworth N.M. Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand breaks. PLoS Genet. 2014;10:e1004005. doi: 10.1371/journal.pgen.1004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grishchuk A.L., Kohli J. Five RecA-like proteins of Schizosaccharomyces pombe are involved in meiotic recombination. Genetics. 2003;165:1031–1043. doi: 10.1093/genetics/165.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amunugama R., Groden J., Fishel R. The HsRAD51B-HsRAD51C stabilizes the HsRAD51 nucleoprotein filament. DNA Repair. 2013;12:723–732. doi: 10.1016/j.dnarep.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Osman F., Dixon J., Barr A.R., Whitby M.C. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol. Cell. Biol. 2005;25:8084–8096. doi: 10.1128/MCB.25.18.8084-8096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simandlova J., Zagelbaum J., Payne M.J., Chu W.K., Shevelev I., Hanada K., Chatterjee S., Reid D.A., Liu Y., Janscak P., et al. FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J. Biol. Chem. 2013;288:34168–34180. doi: 10.1074/jbc.M113.484493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


