Abstract
Type 2 diabetes is a genetically heterogeneous disease, with several relatively rare monogenic forms and a number of more common forms resulting from a complex interaction of genetic and environmental factors. Previous studies using a candidate gene approach, family linkage studies, and gene expression profiling uncovered a number of type 2 genes, but the genetic basis of common type 2 diabetes remained unknown. Recently, a new window has opened on defining potential type 2 diabetes genes through genome-wide SNP association studies of very large populations of individuals with diabetes. This review explores the pathway leading to discovery of these genetic effects, the impact of these genetic loci on diabetes risk, the potential mechanisms of action of the genes to alter glucose homeostasis, and the limitations of these studies in defining the role of genetics in this important disease.
We are in the midst of a worldwide epidemic of type 2 diabetes, obesity, and metabolic syndrome—each created by complex interactions between genes and environment. For decades, investigators have worked to unravel the role of genetics in type 2 diabetes through epidemiological studies, studies of candidate genes, and genetic linkage in families. While this has provided important insights into some rare monogenic forms of diabetes, understanding the genetics of common type 2 diabetes remains a major challenge. Over the past year, a number of exciting articles have been published based on high-throughput genome-wide association (GWA) studies. These have not only uncovered a number of new genetic loci associated with diabetes and provided new targets for mechanistic investigation, but are also forcing us to reconsider the degree of genetic heterogeneity and possibly even the role of genetics itself in the pathogenesis of type 2 diabetes. In this Review, we reconstruct the events that led to this major paradigm shift, review the findings emerging from these studies, and discuss the potential implications and limitations of these discoveries for understanding the basic pathophysiology, as well as prediction, prevention, and treatment of type 2 diabetes.
Type 2 Diabetes: A Major Public Health Problem Arising from Environmental and Genetic Exposures
More than 20 million Americans and over 170 million individuals worldwide suffer from diabetes mellitus (Hogan et al., 2003). Despite advances in treatment, diabetes is the leading cause of chronic renal failure, adult blindness, and limb amputation, and a major risk factor for heart disease, stroke, and birth defects (Krolewski and Warram, 2005). As a result, the cost of diabetes care in the U.S. currently exceeds $200 billion annually, representing over 12% of all health-care dollars. These staggering human and economic costs will increase further as the prevalence of diabetes is expected to double worldwide by 2025 (Cowie et al., 2006).
By far, the largest proportion of this public health problem derives from type 2 diabetes, which accounts for more than 90% of diabetes in the US and worldwide (Skyler and Oddo, 2002). Unlike type 1 diabetes, which is caused by insulin deficiency due to autoimmune destruction of pancreatic β cells, type 2 diabetes arises from an impairment in the ability of muscle, fat, and liver to respond to insulin, i.e., insulin resistance, combined with an inability of the β cell to respond normally to glucose by appropriately increasing insulin secretion (Kahn, 1994). While the relative contribution of these two defects to diabetes pathogenesis continues to be debated, longitudinal studies in high-risk individuals suggest that insulin resistance is an early phenomenon, occurring years before any evidence of glucose intolerance, whereas the β cell failure develops later in the pathogenesis of disease (Martin et al., 1992). On the other hand, other studies have shown that the disposition index, reflecting both insulin sensitivity and insulin secretion, is an early marker of and predictor of type 2 diabetes (Lyssenko et al., 2005; Bergman, 2007).
Both insulin resistance and β cell failure are thought to result from the complex interplay of many different pathways under the combined control of environmental and genetic factors (Figure 1). The role of genetics in type 2 diabetes (T2D) is indicated by the familial clustering of insulin sensitivity and insulin secretion, the higher concordance rate of T2D in monozygotic versus dizygotic twins, and the high prevalence of type 2 diabetes in certain ethnic groups (e.g., Pima Indians or Mexican Americans) (Weijnen et al., 2002; Flegal et al., 1991). It has been estimated that 30%–70% of T2D risk can be attributed to genetics (Poulsen et al., 1999). Patterns of inheritance suggest that type 2 diabetes is both polygenic and heterogeneous—i.e., multiple genes are involved and different combinations of genes play a role in different subsets of individuals. Exactly how many genes and what their relative contributions are, however, remains uncertain, with some investigators postulating that type 2 diabetes consists of two or three major types with some minor types (Rich, 1990; Kahn et al., 1996).
Figure 1. Complex Pathogenesis of Type 2 Diabetes.
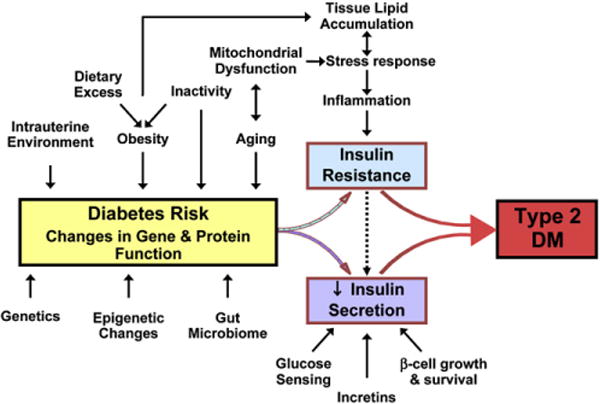
Genetic and environemntal factors may influence the risk of diabetes through the pathways ilustrated in the figure or through as-yet-unidentifed mechanisms affecting insulin sensitivity and/or insulin secretion.
The Search for Type 2 Diabetes Genes in the Pre-Genome-wide Association (GWA) Era
Over the past two decades four approaches have been used to unravel the genetics of type 2 diabetes, each with some success (Table 1). The first approach was to focus on forms of type 2 diabetes transmitted with a Mendelian dominant pattern of inheritance and/or other specific clinical features. This led to the discovery of genes involved in maturity onset diabetes of the young (MODY); the genes involved in several syndromes of severe insulin resistance (the type A syndrome, leprechaunism, Rabson-Mendenhall syndrome, and lipoatrophic diabetes); the genes involved in neonatal diabetes; and the genes involved in mitochondrial diabetes and other rare genetic syndromes (Table 2). Together, these monogenic forms of type 2 diabetes account for less than 2%–5% of this disease. Defining these relatively rare diabetes genes, however, has provided novel insights into pathways involved in the regulation of glucose homeostasis. For instance, elucidating the family of MODY genes has highlighted a complex hierarchical network of transcription factors modulating β cell development and function (Duncan et al., 1998). Similarly, the discovery of mutations in the insulin receptor, Akt2, and PPARγ in syndromes of severe insulin resistance confirms the important role of these signaling proteins in vivo in humans (O’Rahilly, 2007). From a therapeutic perspective, the identification of mutations in the ATP-sensitive potassium channel (Kir6.2/KCNJ11) in infants with neonatal diabetes has been very important, allowing these children to be treated with oral sulfonylureas rather than lifelong insulin injections (Gloyn et al., 2004; Pearson et al., 2006). More interesting from a mechanistic perspective has been the discovery of mutations in genes not previously linked to diabetes, such as the genes for the nuclear membrane protein lamin A or the protein seipin (BSCL2) in certain forms of lipodystrophy and insulin resistance (Shackleton et al., 2000; Magre et al., 2001). However, exactly how mutations in these genes result in either lipodystrophy or severe insulin resistance remains unclear.
Table 1.
Identifying the Genes for Type 2 Diabetes
| Studies of Monogenic Forms of Type 2 Diabetes |
| Maturity Onset Diabetes of the Young (MODY) |
| Rare forms of severe insulin resistance |
| Neonatal diabetes |
| Mitochondrial syndromes of diabetes |
| Candidate Gene Approach |
| Functional |
| Genes involved in insulin action and insulin secretion |
| Genes associated with diseases associated with diabetes (obesity, genetic syndromes, etc) |
| Genes identified in animal models of diabetes |
| Positional |
| Genes in linked intervals identified through family studies |
| Useful for monogenic forms of diabetes (e.g., MODY) |
| Challenging for common, multifactorial Type 2 diabetes |
| Finding Genes with Altered Levels of Expression |
| Subtraction cloning and differential display |
| Microarray analysis (gene expression arrays) |
| Genome-wide Association Studies |
| Microarray-based typing of ≥ 100K SNPs spanning the entire genome |
| Large populations of diabetic cases and non-diabetic controls |
| Populations with intermediate phenotypes – obesity, insulin resistance, polycystic ovarian disease |
Table 2.
Monogenic Causes of Type 2 Diabetes
| Gene Name | Common/Other Name | Cellular Function | Chromosome Location | OMIM Number | Diabetes Phenotype |
|---|---|---|---|---|---|
| Monogenic Causes of β Cell Dysfunction | |||||
| MODY Diabetes | |||||
| HNF4A | HNF4alpha | Transcription factor | 20q12 | 600281 | MODY1; β-cell dysfunction |
| GCK | Glucokinase | Glucose phosphorylation | 7p15 | 138079 | MODY2; mild life-long fasting hyperglycemia |
| TCF1 | HNF1alpha | Transcription factor | 12q24 | 142410 | MODY3; progressive β-cell dysfunction |
| PDX1 | Insulin promoter factor 1 (IPF1) | Homeodomain transcription factor | 13q12 | 600733 | MODY4: Heterozygote: similar to HNF1A (rare); homozygote: pancreatic agenesis and neonatal diabetes |
| TCF2 | HNF1beta | Transcription factor | 17q21 | 189907 | MODY5; Renal cysts and diabetes, pancreatic atrophy, neonatal diabetes |
| NEUROD1 | Beta2 | bHLH Transcription factor | 2q32 | 601724 | MODY6; Similar to HNF1A (rare) |
| KLF11 | Kruppel-like factor 11 | TGF-beta inducible Transcription Factor | 2p25 | 610508 | MODY7; impairs activation of insulin promoter |
| CEL | Carboxyl-ester lipase | Lipid metabolism | 9q34.3 | 114840 | MODY8; Exocrine and β-cell dysfunction (rare) |
| Neonatal Diabetes | |||||
| KCNJ11 | Kir6.2 | Potassium channel | 11p15.1 | 600937 | Permanent and transient neonatal diabetes |
| ABCC8 | Sur1 | Sulfonylurea receptor | 11p15.1 | 600509 | Permanent and transient neonatal diabetes |
| EIF2AK3 | PERK | Pancreatic eIF2-alpha kinase | 2p12 | 604032 | Wolcott-Rallison Syndrome |
| PLAGL1 | Pleomorhpic adenoma | Plagl1 – Nuclear zinc finger | 6p24 | 606546 | Imprinted region, exact gene |
| HYMA1 | gene 1; hydatidiform mole transcript | protein | 603044 | unclear; transient neonatal diabetes type 1 | |
| PTF1A | Pancreas transcription factor 1 | Alpha subunit of PTF1 | 10p12 | 607194 | Permanent neonatal diabetes with cerebellar agenesis |
| INS | Insulin | Hormone | 11p15.5 | 176730 | Mutation in insulin, proinsulin, and proinsulin processing |
| Mitochondrial Diabetes | |||||
| Mitochondrial genome | MIDD | tRNA for leucine | Mutation at 3243 mtDNA | 590050 | Maternally inherited diabetes and deafness; other mitochondrial mutation also observed |
| Mitochondrial genome | Mitochondrial myopathy, lipid type | tRNA for glutamic acid | Mutation at 14709 mtDNA | 500002 | Mitochondrial myopathy with diabetes |
| Other | |||||
| WFS1 | Wolframin | 10 transmembrane domain protein, function unknown | 4p16.1 | 2223000 | Diabetes insipidus and mellitus with optic atrophy and deafness; DIDMOAD; Wolfram Syndrome |
| ZCD2 | ERIS | Zinc finger protein ZCD2 | 4q22–q24 | 604928 | Wolfram Syndrome 2 |
| Monogenic Causes of Insulin Resistance | |||||
| Severe Insulin Resistance | |||||
| INSR | Insulin receptor | Receptor tyrosine kinase | 19p13 | 147670 | Insulin-resistant diabetes with various phenotypes: leprechaunism, Rabson-Mendenhall or type A syndrome |
| AKT2 | PKB-beta | Serine-threonine kinase | 19q1 | 164731 | Severe insulin resistance |
| Lipoatrophic Forms of Diabetes | |||||
| LMNA | Lamin A/C | Inner nuclear membrane protein | 1q21 | 150330 | Face-sparing partial lipodystrophy with peripheral fat loss; mutations also associated with cardiomyopathy; muscular dystrophy; and Hutchinson-Gilford Progeria |
| LMNB2 | Lamin B2 | Inner nuclear membrane protein | 19p13 | 150341 | Partial lipodystrophy sparing legs (Barraquer-Simon Syndrome) |
| AGPAT2 | 1-acylglycerol-3-phosphophate O-acyltransferase 2 | Enzyme of phospholipid metabolism | 9q34 | 603100 | Congenital generalized lipodystrophy with skeletal lytic lesions (Bernardinelli-Seip Syndrome) |
| BSCL2 | Seipin | 398 amino acid protein of unknown function | 11q13 | 606158 | Congenital generalized lipodystrophy, learning disabilities |
| PPARG | Peroxisome proliferator activated receptor γ | Nuclear receptor for prostaglands and thiazolidine-diones | 3p25 | 601487 | Rare variants in ligand binding domain associated with insulin resistance, hypertension, buttock lipodystrophy |
The second approach to identify diabetes genes has been to search for genetic variants in candidate genes that might be associated, i.e., more frequent, in individuals with common type 2 diabetes (Hansen and Pedersen, 2005). In general, these studies have focused on functional candidate genes—i.e., genes whose products are known to play a role in glucose homeostasis, or positional candidate genes—i.e., genes located in chromosomal regions that had been identified in linkage studies. By focusing on genes already implicated in glucose homeostasis, the functional candidate gene approach is geared toward confirming the role of genes in diabetes rather than discovering as-yet-unknown disease pathways. The positional candidate gene strategy has more potential for discovery of new genes; however, this approach is limited by the low sensitivity of linkage studies for multifactorial disorders and the large size of linked genetic intervals.
Despite these shortcomings, these approaches have had some success. Candidate gene studies led to the identification of a common amino acid substitution in the nuclear receptor and adipogenic transcription factor PPARγ (Pro12Ala, rs1801282), which has a modest, yet extensively replicated effect on the risk of type 2 diabetes (Beamer et al., 1998). Similarly, the diabetes-associated Glu23Lys variant was identified in the KCNJ11 gene, which encodes Kir6.2, one of the two components of the KATP channel essential for normal glucose-stimulated insulin secretion (Barroso et al., 2003). Functionally significant polymorphisms have also been identified in proteins involved in insulin action, including a “gain-of-function” polymorphism (K121Q) in the insulin action inhibitor ENPP1 (Pizzuti et al., 1999), the insulin receptor substrates IRS1 and IRS2 and phosphatidylinositol 3-kinase (Almind et al., 1996; Almind et al., 2002). Some, but not all, of these associations with T2 DM have been confirmed by meta-analysis or studies in large, diverse populations (McAteer et al., 2008; Florez et al., 2004a). Positional candidate efforts, on the other hand, have been mostly inconclusive, despite identification of multiple genomic regions linked to diabetes (e.g., 1q, 20q) (Stern, 2002). One notable exception is the identification of TCF7L2 as a type 2 diabetes gene (discussed below) (Grant et al., 2006).
A number of recent efforts have used microarray gene expression analysis in attempt to define genetic alterations in type 2 diabetes. These have uncovered two important findings. One is a defect in skeletal muscle of humans with T2D characterized by a coordinated decrease in the expression of nuclear-encoded genes involved in mitochondrial oxidative phosphorylation (Patti et al., 2003; Mootha et al., 2003). This appears to be secondary to reduced expression of the transcriptional coactivators PGC-1α and PGC-1β. Similar changes in expression have been observed in some cohorts of first-degree relatives of diabetic individuals, suggesting that these may be heritable traits (Patti et al., 2003), but thus far no primary genetic defect has been linked to these changes. Although polymorphisms in PGC-1 and epigenetic modification of complex subunits may contribute to age-associated reductions in expression (Ling et al., 2004, 2007, Ronn et al., 2008), other studies have suggested that this expression phenotype may be secondary, at least to some extent, to poor physical fitness, obesity, intramyocellular lipid accumulation, or insulin resistance itself (Crunkhorn et al., 2007). Furthermore, the role of these changes in diabetes pathogenesis is uncertain, since an animal model with primary reduced mitochondrial oxidative phosphorylation in muscle shows increased, rather than decreased, insulin sensitivity (Pospisilik et al., 2007).
A second potential type 2 gene was discovered via microarray analysis is the transcription factor ARNT/Hif1β. This was identified using islets isolated from humans with type 2 diabetes and was associated with altered expression of many other genes in the β cell involved in glucose sensing, insulin signaling, and transcriptional control (Gunton et al., 2005). Again, however, it is not clear which, if any, of these changes represent primary defects or are secondary to trans-acting genes or metabolic/environmental factors.
The GWA Revolution
By 2006 it had become clear that identifying the major genes of type 2 diabetes would require a paradigm shift. Such a shift was made possible by several parallel developments. One was the completion of the HapMap project aimed at characterizing the genome-wide pattern of linkage disequilibrium (Frazer et al., 2007). Linkage disequilibrium (LD) is a statistical association between alleles at separate but linked loci, usually resulting from a particular ancestral haplotype being common in the population studied (Strachan and Read, 2003). This phenomenon causes adjacent polymorphisms to be correlated to the point of being strong proxies for each other. With linkage disequilibrium, therefore, it becomes possible to select a set of 300,000 to 1 million single nucleotide polymorphisms (SNPs) that can represent most of the 10 million common SNPs estimated to be present in the human genome. Coupled with improved microarray technology allowing precise typing of a large number of polymorphisms and access to DNA from large cohorts of patients with diabetes, the possibility of large GWA studies of diabetes became a reality.
It is important to keep in mind that the GWA approach is not without its own limitations. First, in its current implementation, it assumes that the genetic variants conferring susceptibility to type 2 diabetes are common, i.e., have a frequency of ≥5% in the population. While theoretical models support this assumption, rare variants have been shown to contribute significantly to the modulation of complex metabolic traits (Brunham et al., 2006). Second, the GWA strategy poses significant challenges in terms of study design and data interpretation. For example, type 2 diabetes is an age-dependent disease, and, thus any population of controls will include many individuals who might later develop diabetes. Third, given the hundreds of thousands of comparisons performed in the analysis, a large number of polymorphisms can be significant at the p < 0.05 or p < 0.001 level by chance alone. Thus, a much more stringent significance threshold must be used (e.g., p < 2 × 10−7 for a 300K array), meaning that a very large sample size is needed in order to preserve power. This, in turn, creates a financial challenge. The final, and perhaps most important, challenge is what to study. Type 2 diabetes is not only polygenic and heterogeneous, but also is closely linked to other metabolic phenotypes and has a progressive pathogenesis, starting with insulin resistance and β cell dysfunction progressing to clinical hyperglycemia (Figure 1). Since there are no precise markers for defining stages in this progress, GWA studies were forced to focus on cases with clinically defined type 2 diabetes, adding elements of complexity to interpretation of results.
The GWA Studies for Type 2 Diabetes Genes
By the late spring of 2007, the results of five independent GWA screens for type 2 diabetes genes had been published (Sladek et al., 2007; Zeggini et al., 2007; Scott et al., 2007; Saxena et al., 2007; Steinthorsdottir et al., 2007) (see Table S1 for characteristics of each study), and these were followed by five smaller GWA studies (Salonen et al., 2007; Rampersaud et al., 2007; Hayes et al., 2007; Florez et al., 2007; Hanson et al., 2007). The five large studies were all conducted using a two-stage strategy consisting of a GWA screen in an initial cohort of unrelated cases and controls followed by replication of the most significant findings in additional sets. The screening and replication sets consisted primarily of European Whites, with the exception of the Decode study, which included groups of Chinese from Hong Kong and West Africans (Table S1). Altogether, the screening sets included more than 7,000 cases and 12,000 controls; the replication sets consisted of about 20,000 cases and 26,000 controls. The screening set of the McGill/Imperial College study stands out from others for the relatively young age at diagnosis (average ~45 years) and the low BMI (average 25.8) of the individuals with diabetes, a focus hoping to increase the probability of detecting genetic effects, while excluding genes related to the etiology of obesity. In the other four large studies, the phenotype of cases was closer to that of common type 2 diabetes, with average age at diagnosis in the sixth decade. In the DGI study, a large proportion of controls were matched to cases by BMI, whereas in the other studies, the BMI was lower in controls than cases by two or three points, similar to what is observed in the corresponding source populations. The GWA screens were based on the Illumina 300K or Affymetrix 500K genotyping array, both capturing about 80% of common variants present in the human genome of Caucasian individuals.
While each GWA screen identified dozens of potential candidates, at least 15 loci emerged as being most consistently associated with risk of type 2 diabetes across multiple studies (Table 3). Three of these correspond to genes that had already been implicated in type 2 diabetes (TCF7L2, KCNJ11, PPARG); the other 12 represent new potential type 2 diabetes genes. Given the large number of cases and controls, these associations are highly significant. However, these large p values should not be confused with the magnitude of the genetic effect as quantified by its odds ratio (OR), which in all cases is relatively small (Table 3). Two other striking features of these studies were that most of the genes identified would not be considered typical candidate genes for type 2 diabetes, and in most cases the variants associated with type 2 diabetes were in noncoding regions of the gene, suggesting alterations in regulatory elements and gene expression rather than amino acid sequence. The following is a brief summary of the evidence linking these 15 loci to type 2 diabetes and some thoughts about possible mechanisms involved in their effects.
Table 3.
Genetic Loci Implicated in “Common Variety” Type 2 Diabetes
| Chromosome Location | Gene Symbol | Common/Other name | Cellular Function | OMIM Number | OR | Frequency of Risk Allele |
|---|---|---|---|---|---|---|
| 1p12 | NOTCH2 | Notch 2 preproprotein | Regulator of cell differentiation | 600275 | 1.13 | 0.11 |
| 2p21 | THADA | Thyroid adenoma-associated gene | Unknown | 611800 | 1.15 | 0.90 |
| 3p14 | ADAMTS9 | Disintegrin-like and metalloproteinase with thrombospondin type 1 motif | Proteolytic enzyme regulating extracellular matrix | 605421 | 1.09 | 0.76 |
| 3p25 | PPARG | Peroxisome proliferator activating receptor gamma (PPARγ) | Transcription factor receptor for TZDs and prostaglandins | 601487 | 1.17 | 0.85 |
| 3q28 | IGF2BP2 | IMP2 | IGF2 mRNA-binding protein 2 | 608289 | 1.14 | 0.29 |
| 6p22.3 | CDKAL1 | CDK5 regulatory subunit associated protein 1-like 1 | Presumed regulator of cyclin kinase | 611259 | 1.0–1.20 | 0.31 |
| 7p15 | JAZF1 | Juxtaposed with another zinc finger gene 1 | Zinc-finger protein of unknown function | 606246 | 1.10 | 0.50 |
| 8q24.11 | SLC30A8 | ZNT8 | Zinc transporter 8 | 611145 | 1.18 | 0.65 |
| 9p21 |
CDKN2A CDKN2B |
p16 (INK4a) p14(ARF)p15 (INK4b) | Cyclin-dependent kinase inhibitor 2A and 2B | 600431 | 1.20 | 0.83 |
| 10p13-p14 | CDC123 | Cell division cycle protein 123 homolog | Required for S phase entry of the cell cycle | – | 1.11 | 0.18 |
| CAMK1D | Calcium/calmodulin-dependent protein kinase i-delta | Mediator of chemokine signal transduction in granulocytes | 607957 | |||
| 10q23-q25 | IDE | Insulin degrading enzyme | Neutral metallopeptidase that can degrade many peptides | 146680 | 1.13 | 0.53 |
| HHEX | Hematopoietically expressed homeobox; PRHX | Homeobox transcription factor | 604420 | |||
| KIF11 | Kinesin family member 11; Homologue of Xenopus EG-5 | Kinesin related motor in microtubule & spindle function | 148760 | |||
| 10q25.3 | TCF7L2 | TCF4 | High mobility group transcription factor | 602228 | 1.31–1.71 | 0.26 |
| 11p15.1 | KCNJ11 | Kir6.2 | Inwardly rectifying potassium channel | 600937 | 1.14 | 0.47 |
| 12q21 | TSPAN8 | Tetraspanin 8 | Cell surface glycoprotein | 600769 | 1.09 | 0.27 |
| LGR5 | Leucine-rich repeat-containing G proteincoupled | Orphan G protein-receptor | 606667 | |||
| 16q12.2 | FTO | Fat Mass- and Obesity-Associated Gene | 58kD protein with nuclear localization signal | 610966 | 1.27 | 0.38 |
OR = Odss ratio of type 2 diabetes per risk allele.
TCF7L2 (10q25)
The strongest association in all of the studies is with a region on chromosome 10q25 located in a 92 kb linkage disequilibrium (LD) block in the TCF7L2 gene. While the identification of this type 2 diabetes locus predates the GWA era (Grant et al., 2006), the association of the TCF7L2 locus with T2D is by far the strongest and most consistent signal across the GWA studies. In a meta-analysis of the WTCCC, Fusion, and DGI studies, the combined odds ratio of type 2 diabetes per copy of the “high-risk” allele is 1.37 (95% confidence interval, 1.31–1.43), with a combined p value of 10−48 (Zeggini et al., 2007; Scott et al., 2007; Saxena et al., 2007). Individuals homozygous for the high-risk allele have about a doubling of diabetes risk, or to put it another way, about ~12% of them will have type 2 diabetes versus ~9% of heterozygotes and ~6% of noncarriers. The predisposing effect seems to be more pronounced among lean than obese individuals (Cauchi et al., 2008a; Watanabe et al., 2007).
TCF7L2, also known as TCF-4 or β-catenin interacting protein, is a high-mobility group box-containing transcription factor that is involved in the WNT signaling pathway, acting as a nuclear receptor for β-catenin (Yi et al., 2005; Prunier et al., 2004). Wnt signaling is critical for cell proliferation and involved in many aspects of embryogenesis, including adipogenesis (Prestwich and MacDougald, 2007), myogenesis (Cossu and Borello, 1999), and pancreatic islet development (McLin et al., 2007). TCF7L2 activation induces a variety of genes, including those for intestinal proglucagon and glucagon-like peptides-1 and -2 (Fehmann et al., 1995). The association with type 2 diabetes involves a common haplotype spanning part of intron 3, exon 4, and part of intron 4, but does not include any coding variants, suggesting an effect on gene expression as the most likely explanation for association with type 2 diabetes.
Clinically, carriers of the high-risk TCF7L2 genotype have reduced insulin secretion (Florez et al., 2006a), suggesting a possible role for TCF7L2 in the β cell dysfunction of type 2 diabetes. Overexpression studies in β cells, however, have given conflicting results, in one case showing a blunting of glucose-stimulated insulin secretion (Lyssenko et al., 2007) and in another a beneficial effect to protect islets from glucose- and cytokine-induced apoptosis and impaired function (Shu et al., 2008). Other studies suggest roles for TCF7L2 in T2D via control of the incretin axis, hepatic glucose production, and adipocyte function (Lyssenko et al., 2007; Cauchi et al., 2006). Actual expression data for TCF7L2 in humans are limited. One report indicates that islets from carriers of the risk genotypes have increased TCF7L2 mRNA compared to noncarriers (Lyssenko et al., 2007), but no significant difference in TCF7L2 expression was observed in islets or muscle in a study of unselected human T2D in the Diabetes Genome Anatomy Project (DGAP) (http://www.diabetesgenome.org) (Figure 2, bottom). TCF7L2 could exert its effect in a variety of ways, since TCF7L2 is expressed at high levels in a wide variety of tissues, including the hypothalamus (Figure 2, top).
Figure 2. Expression Profiles of Genes Placed at the Type 2 Diabetes Loci Identified to Date.
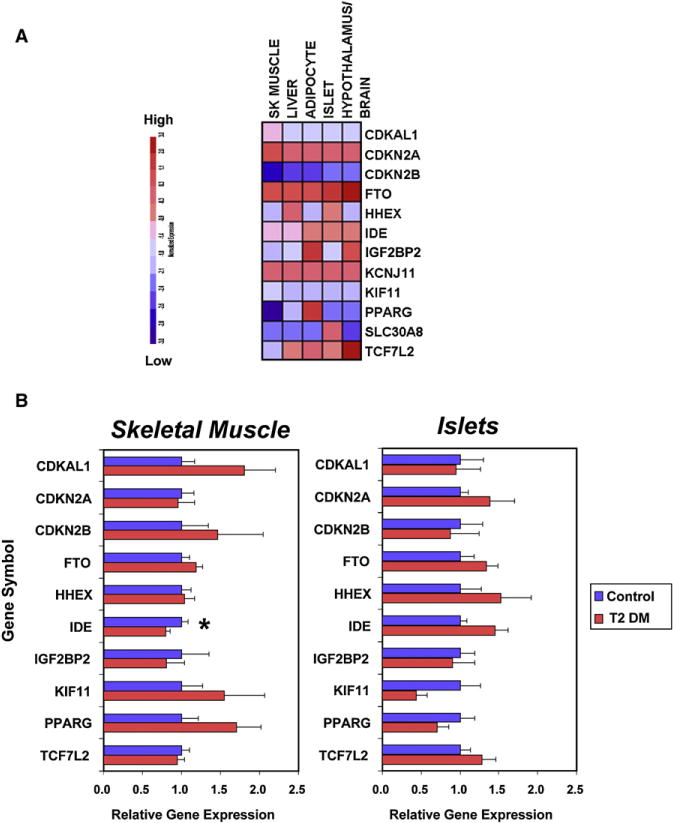
(A) Expression levels across different tissues.
(B) Relative expression levels in skeletal muscle and pancreatic islets from diabetic and nondiabetic subjects. Skeletal muscle expression data were derived from a cohort of metabolically characterized Mexican-American subjects with established DM2, treated with sulfonylureas or lifestyle only (n = 5), and compared with control individuals with normal glucose tolerance (n = 6) (Patti et al., 2003). The data on pancreatic islets were derived from isolated islets purified from five type 2 diabetic subjects and seven normoglycemic controls (Gunton et al., 2005). The mean duration of type 2 diabetes was 5.8 ± 2.1 years, and no subjects were insulin requiring. Mean HbA1c was 7.5 ± 0.5% in the diabetic subjects. RNA was extracted from at least 1000 islet equivalents per subject. RNA was prepared separately for each subject and hybridized to Affymetrix U133A and B microarrays. For each study, cRNA was prepared separately for each subject and hybridized to Affymetrix HuGene FL (muscle) and U133A and B (islet) microarrays. Complete microarray data sets are available on the Diabetes Genome Anatomy Project (DGAP) website (http://www.diabetesgenome.org).
SLC30A8 (8q24)
The next strongest SNP after those in TCF7L2 is in a 33kb LD block within the coding region of SLC30A8, a zinc membrane transporter (Zn-T8) that is highly expressed in pancreatic islets (Figure 3, top) (Chimienti et al., 2004). This gene first emerged as a type 2 diabetes locus in the McGill/Imperial study (Sladek et al., 2007). Although this polymorphism ranked only 34th in the screening GWA set with ORs of 1.18 and 1.53 for heterozygotes and homozygotes, respectively, this was one of only a handful of SNPs for which the association with type 2 diabetes was confirmed in the replication sets. Replication was also obtained in the Decode, WTCCC, Fusion, and DGI studies (Zeggini et al., 2007; Scott et al., 2007; Saxena et al., 2007; Steinthorsdottir et al., 2007).
Figure 3. Gene-Gene Interaction in the Etiology of Type 2 Diabetes as Exemplified by Animal Models.
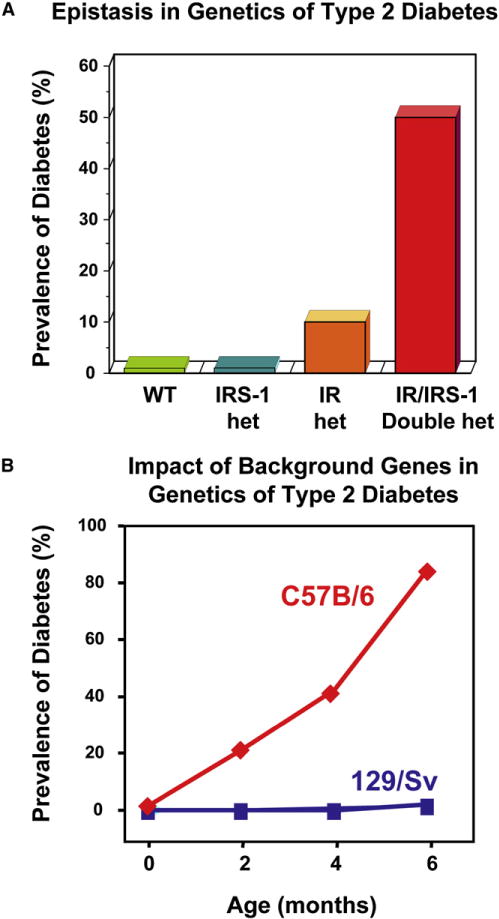
(A) Epistasis between insulin receptor (IR) and insulin receptor substrate 1 (IRS1) gene targeting in the development of type 2 diabetes.
(B) Impact of genetic background on the development of type 2 diabetes in mice with genetic insulin resistance.
Of all of the new potential type 2 diabetes genes, SLC30A8 is one of the few involving a nonsynonymous polymorphism—an arginine to tryptophan substitution at amino acid 325 (Sladek et al., 2007). SLC30A8-overexpressing cells display enhanced glucose-stimulated insulin secretion (Chimienti et al., 2006), suggesting that the risk allele might act by impairing transporter function, thereby decreasing the amount of zinc available for cocrystallization with insulin in the secretory vesicles of β cells. Reduced pancreatic and islet zinc levels have been observed in some animal models of type 2 diabetes (Taylor, 2005), and zinc supplementation has been shown to improve glucose tolerance in db/db mice (Simon and Taylor, 2001). On the other hand, studies have not identified any deficiency of pancreatic zinc content in patients with type 2 diabetes (Taylor, 2005), and at least one study suggests that dietary zinc supplementation in humans with diabetes further decreased glucose tolerance (Raz et al., 1989). Since zinc has many roles in the cell in addition to its role in insulin crystallization, and since Zn-T8 is only one member of a large family of zinc transporters, more mechanistic studies are clearly warranted. SLC30A8 has also recently been identified as an autoantigen in human type 1 diabetes (Wenzlau et al., 2007).
HHEX/IDE/KIF11 (10q23)
A locus on 10q23 ranked third for association with T2D (after TCF7L2 and SLC30A8) in the McGill scan, and was independently replicated in the WTCCC, Fusion, and DGI studies giving a small p value (p = 5.7 × 10−10), but a combined OR of only 1.13. SNPs at this locus have been also found to be associated with measures of insulin secretion in a large multicentric study from Europe and in the DPP study (Pascoe et al., 2007; Moore et al., 2008). The association signal lies in a 295 Kb block of LD that includes at least three potential type 2 diabetes genes: HHEX (a homeobox transcription factor), KIF11 (a kinesin interacting factor), and IDE (insulin degrading enzyme) genes (Table 3). Which, if any, of these genes actually contribute to type 2 diabetes risk is not known. Each of these genes shows rather broad tissue expression (Figure 2, top). Based on its provocative name and data suggesting roles in both insulin signaling and islet function (Farris et al., 2003), IDE would probably be viewed as the strongest biological candidate gene at this locus. However, a previous large study of IDE as a candidate for type 2 diabetes was negative (Florez et al., 2006b). Reanalysis of the DGAP data reveals no difference in expression of IDE in islets between the individuals with diabetes and healthy controls (Figure 2, bottom), while in muscle, expression of IDE is modestly reduced in T2 DM (21% decrease, p = 0.05). Despite its name, IDE is not an insulin-specific protease and has been shown to play roles in degradation of glucagon, brain amyloid proteins, and viral glycoproteins (Qiu et al., 1998). HHEX, on the other hand, is a transcriptional repressor, active in cardiac and pancreas development (Tanaka et al., 1999; Foley and Mercola, 2005), as well as WNT signaling (McLin et al., 2007), while KIF11 is involved in centrosome migration and mitosis (Kapitein et al., 2005).
CDKAL1 (6p22)
An association signal at 6p22 maps to a 15 kb linkage disequilibrium block in intron 5 of the gene for CDKAL1 (CDK5 regulatory subunit-associated protein 1-like 1). The strongest association of 6p22 with T2D was observed in the Decode study (OR = 1.20) (Steinthorsdottir et al., 2007), with weaker association in the WTCCC, Fusion, and DGI study (combined OR = 1.12), and no association in the McGill/Imperial study (Sladek et al., 2007; Zeggini et al., 2007; Scott et al., 2007; Saxena et al., 2007).
The CDKAL1 gene encodes a 65 kD protein that is expressed in a broad range of tissues (Figure 2, top) and is believed to be an inhibitor of CDK5 (cyclin-dependent kinase 5). CDK5 itself has been shown to blunt insulin secretion in response to glucose and to play a permissive role in the decrease of insulin gene expression that results from glucotoxicity (Ubeda et al., 2006). Thus, one can speculate that reduced expression of CDKAL1 would result in enhanced activity of CDK5 in β cells, and this would lead to decreased insulin secretion; in agreement, this locus was significantly associated with small decreases in insulin response to a glucose load (Steinthorsdottir et al., 2007; Saxena et al., 2007; Pascoe et al., 2007; Palmer et al., 2008, Stancáková et al., 2008). However, in the analysis of human islet and muscle expression data from DGAP, there does not appear to be a significant difference in the level of expression of CDKAL1 between subjects with diabetes and controls (Figure 2, bottom).
CDKN2A/2B (9p21)
Two signals of association with T2D have been localized to chromosome 9p21. The first one—represented by SNP rs10811661—is supported by the DGI, WTCCC, and Fusion scans with a combined OR of 1.20 and a p = 7.8 × 10−15 (Zeggini et al., 2007; Scott et al., 2007; Saxena et al., 2007). A second weaker signal in this region was detected by the WTCCC and Fusion studies about 100 kb in a telomeric direction (SNP rs564398, p = 1.3 × 10−6 and p = 0.039, respectively). The two association signals, involving noncoding polymorphisms, are separated by a recombination hotspot and thus could be independent. The centromeric signal (SNP rs10811661) is close to the CDKN2A and CDKN2B genes (cyclin-dependent kinase inhibitor 2a and 2b) and the telomeric signal includes both of these genes. Both genes code for inhibitors of CDK4 (cyclin-dependent kinase 4). CDKN2A encodes two molecules, p16INK4a and p14 (ARF), while CDKN2B encodes p15INK4b (Kamb et al., 1994; Pomerantz et al., 1998).
CDK4 is involved in cell-cycle regulation in a wide variety of cells. Interestingly, mice with targeted disruption of this gene have small islets and develop insulin-deficient diabetes, while mice expressing a CDK4 form insensitive to physiological inhibitors exhibit β cell hyperplasia (Rane et al., 1999). However, a study of 1276 healthy individuals failed to observe any association between CDKN2A or CDKN2B polymorphisms and insulin secretion (Pascoe et al., 2007). These two genes, however, are widely expressed and could have effects in many other tissues (Figure 2, top). Furthermore, this region includes other genes, including a mitochondrial RNA processing endoribonuclease and the gene for methylthioadenosine phosphorylase (Nobori et al., 1996), as well as several ESTs. Interestingly, a “major” locus for coronary artery disease, abdominal aneurysms, and peripheral vascular disease has also been identified in this region (Helgadottir et al., 2007; McPherson et al., 2007, Helgadottir et al., 2008), suggesting a common genetic link between diabetes and vascular disorders (Stern, 1995).
IGF2BP2 (3q27)
Association signals at 3q27 were observed in the WTCCC, Fusion, and DGI scans, although the combined OR is only 1.14 (p = 8.9 × 10−16) (Zeggini et al., 2007; Scott et al., 2007; Saxena et al., 2007). This is consistent with previous studies showing linkage at this location with quantitative metabolic traits and type 2 diabetes (Kissebah et al., 2000; Vionnet et al., 2000). The SNPs displaying the strongest association with type 2 diabetes lie in a 50 kb region within intron 2 in the gene coding for IGF-2BP2. IGF-2BP2 is not an IGF binding protein, but a protein that binds to the 5′UTR of the insulin-like growth factor 2 (IGF-2) mRNA, thereby regulating its translation (Nielsen et al., 1999). Several other genes with important metabolic functions are also within the radius of a potentially regulatory effect by the risk variants, including PPP1R2 (protein phosphatase 1, regulatory subunit 2), MAP3K13 (mitogen-activated protein kinase kinase kinase), LIPH (lipase H), DGKG (diacylglycerol kinase gamma 1), AHSG (alpha-2-HS-glycoprotein, a putative inhibitor of insulin receptor signaling), and ADIPOQ, the insulin-sensitizing adipokine adiponectin. Reanalysis of the DGAP human islet and muscle data shows no difference in expression of IGF2BP2 in diabetes (Figure 2A), so whether the association with type 2 diabetes is mediated by an effect on the expression of IGF2BP2 or these other genes will have to be addressed by further studies.
FTO (16q12)
In the WTCCC scan, the association signal at this locus (OR = 1.27, p = 7.3 × 10−14) was second in magnitude only to that of TCF7L2 (Zeggini et al., 2007). The diabetes-associated alleles at this location, however, were also strongly associated with increased BMI, and the association with type 2 diabetes was lost when the analysis was adjusted for body weight, suggesting that the effect on T2D risk is mediated by an effect on adiposity. Indeed, this locus has been associated with obesity, with ORs ranging from 1.3 to 1.9 (Frayling et al., 2007; Dina et al., 2007). Adults homozygous for the FTO risk allele weigh about 3 kg more than individuals not carrying this allele. About 16% of white Europeans are homozygous for the high-risk A allele, and these individuals are 1.7 times more likely to be obese than those homozygous for the low-risk T allele. The SNPs showing the strongest association lie in a 47 kb linkage disequilibrium block encompassing parts of the first two introns and exon 2 of the FTO gene.
The function of the FTO (fat and obesity associated) gene is still unclear. FTO shares sequence motifs with Fe(II)- and 2-oxo-glutarate-dependent oxygenases and is localized in the nucleus, where it catalyzes the demethylation of 3-methylthymine in single-stranded DNA (Gerken et al., 2007). In mice FTO mRNA is most abundant in the brain, particularly in hypothalamic nuclei governing energy balance (Figure 2, top). FTO levels in the arcuate nucleus are regulated by feeding and fasting (Gerken et al., 2007). Other genes in close proximity to the FTO polymorphism include an Akt interacting protein (AKTIP), two members of the ATP binding cassette subfamily C (ABCC), and KIAA1005/RPGRIP1L (a gene of unknown function). Functional studies based on knockout and overexpression models will be needed to understand the pathways through which variants at this locus control body weight and glucose homeostasis.
KCNJ11 and PPARγ
Most of the genes previously identified through the candidate gene approach did not rank high for association with T2D in the GWA studies. KCNJ11 and PPARG are two exceptions. Both were found to be associated with type 2 diabetes in the WTCCC, Fusion, and DGI studies with a combined OR of 1.14 (p = 6.7 × 10−11 and p = 1.7 × 10−6, respectively) (Zeggini et al., 2007; Scott et al., 2007; Saxena et al., 2007), and both involve nonsynonymous polymorphisms. The association with the KCNJ11 gene concerns a common glutamate to lysine substitution at position 23 (E23K). The lysine allele has been shown to reduce the sensitivity of β cell ATP-sensitive K+ channels toward inhibitory ATP4−, thereby increasing the threshold for insulin release (Schwanstecher et al., 2002). Consistent with this, this polymorphism has been associated with an insulin secretion defect in multiple studies (Nielsen et al., 2003, Florez et al., 2004b). However, the β cell may not be the only important target organ for KCNJ11. A transgenic mouse expressing a mutant Kir6.2 subunit (encoded by the KCNJ11 gene) in the hypothalamus driven by the POMC promoter exhibits impaired whole-body glucose disposal and a defect in glucose sensing in POMC neurons when placed on a high-fat diet (Parton et al., 2007). Thus, the KCNJ11polymorphisms could contribute to loss of glucose sensing in both the β cell and POMC neurons, leading to impairment in insulin action, as well as insulin secretion.
The identified polymorphism in PPARγ, which appears to modify susceptibility to type 2 diabetes mellitus and obesity, was identified a decade ago and produces a Pro12-to-Ala (P12A) change in the PPARγ2 gene (Deeb et al., 1998; Beamer et al., 1998). This was initially confirmed in a meta-analysis (Altshuler et al., 2000) and reconfirmed in the GWA studies. Resistance to diabetes is associated with the minor (Ala12) allele and susceptibility with the major allele (Pro12), which has a prevalence of about 85% among nondiabetic individuals and 88% among diabetic subjects. Exactly how this change in amino acid produces this effect still remains unclear. However, this change occurs specifically in the PPARγ2 isoform of the gene, which is the form specifically expressed in adipose tissue and which is the target of the insulin sensitizing thiazolidinediones (TZDs). Mutations in other functional regions of this gene have also been found in individuals with severe insulin resistance due to familial partial lipodystrophy type 3 (Barroso et al., 1999).
JAZF1, CDC123-CAMK1D, TSPAN8-LGR5, THADA, ADAMTS9, and NOTCH2
Genomic loci near these six genes did not reach significance in the original GWA screens, but their association with type 2 diabetes was confirmed in a large replication study including more than 50,000 individuals, with OR ranging from 1.09 to 1.15 (Zeggini et al., 2008). JAZF1, CDC123/CAMK1D, and TSPAN8-LGR5 are associated with small alterations in insulin secretion, whereas the mechanisms linking the other three loci to type 2 diabetes remain uncertain (Grarup et al., 2008).
Additional Loci Associated with Quantitative Metabolic Traits
Mutations in the melanocortin-4 receptor (MC4R) have previously been shown to account for up to 5% of childhood-onset severe obesity (O’Rahilly, 2007), and analysis of the GWAS data has shown a modest, but consistent, association of fat mass and waist circumference with a locus immediately downstream of this gene (Loos et al., 2008; Chambers et al., 2008). Analysis of quantitative traits among the nondiabetic controls of type 2 diabetes GWA studies has also identified variants in the region of the glucose-6-phosphatase catalytic subunit 2 (G6PC2) that function as modulators of fasting glucose, although this effect does not appear to translate into an increased risk of type 2 diabetes (Chen et al., 2008). No consistent associations have been thus far reported with measures of insulin secretion and insulin sensitivity, but further insights are expected from the meta-analyses of the available GWA data.
The Emerging Genetic Architecture of Type 2 Diabetes
Taken together, what do these GWA association studies tell us about the genetics of type 2 diabetes? First, assuming our basic model is correct, these studies suggest that type 2 diabetes may be more heterogeneous and more polygenic than previously believed. While previous reviews had suggested there may be two or three major genetic forms of type 2 diabetes along with some minor forms, the GWA studies would suggest that from a genetic perspective type 2 diabetes represents a very large number of different disorders. This seems a bit at odds with the common clinical presentation of the disease and raises a question as to how these different genetic forms of type 2 diabetes might differ at a clinical level.
The second finding of these studies is the relatively high frequency of each of the risk variants in the population, ranging from 0.26 for TCF7L2 to 0.85 for PPARγ (Table 3). The high frequency of the risk variants together with the small risk of type 2 diabetes conferred by each allele translate into a very large number of individuals who carry several of the disease variants but do not develop type 2 diabetes. Indeed, one can estimate from the individual allele frequencies that 99% of the population carries nine or more risk alleles, whereas only 8%–10% of the population develops type 2 diabetes. Such low penetrance is consistent with the mild effect of the risk variants, requiring the presence of other factors, presumably environmental, for the development of hyperglycemia. The mild effect may also be explained by the fact that most of these variants are in noncoding regions of the gene, where they may produce only subtle differences in regulation of expression. This scenario contrasts sharply with that of Mendelian forms of diabetes, such as MODY, which are caused by rare mutations in the coding sequence resulting in significant amino acid substitutions or truncated proteins, leading to hyperglycemia even in the absence of other diabetogenic exposures.
The third finding is that the loci identified to date appear to explain only a small proportion of the familial clustering of type 2 diabetes. Indeed, in the WTCCC study, the nine strongest loci together account for only 7% of the 30%–60% increase in the risk of type 2 diabetes typically observed in siblings of type 2 diabetes probands as compared to the general population (Zeggini et al., 2007). If our current genetic models are correct (and this remains an important question), this would suggest that the loci identified to date are just the tip of the iceberg. In each GWA scan, other loci showed significant associations with type 2 diabetes but were not pursued because they were not replicated across multiple studies. While some of these are certainly false positives, some of these are likely genuine effects that need to be explored further.
At least four other hypotheses also need to be considered in interpreting these studies. First, it is possible that our previous estimates of the contribution of genetics to development of type 2 diabetes may be exaggerated by our inability to assess other factors in family and population studies, such as shared environment. Indeed, there are environmental risk factors that may not have been previously appreciated, and compared to our robust approaches to genetic analysis, our techniques for identifying environmental risk factors are still very primitive. Furthermore, critical environmental exposures (e.g., maternal obesity, intrauterine environment, nutritional composition, or differences in intestinal flora) may be subtle and begin as early as intrauterine life, thus altering patterns of development and programming metabolic responsiveness to environmental stimuli during later life. Such concepts are supported by studies of monozygotic twins, in which primary DNA sequence is identical, yet diabetes risk is discordant and linked to birth weight (Poulsen et al., 1999) and by the findings that low birth weight confers increased risk for diabetes equal in magnitude to the effect of the TCF7L2 polymorphism (Cauchi et al., 2006).
Second, our concepts about how genes interact with each other and with the environment may be too simplistic. For example, in a polygenic rodent model of type 2 diabetes created by heterozygous inactivation of both the insulin receptor and IRS-1, there is evidence of a very marked epistasis, such that neither gene defect alone can produce diabetes in more than 10% of mice, but when they occur together, more than 50% of animals develop diabetes at young age (Figure 3A) (Bruning et al., 1997). Furthermore, appearance of “clinical” diabetes in this and other rodent models can be markedly different depending on the genetic background of the mouse, demonstrating other important gene-gene interactions (Figure 3B) (Almind et al., 2003).
Third, it is possible that the current models of genetics are clouded by our underestimate of the role of epigenetics in disease. In rodent models that mimic low birth weight in humans by food restriction or restriction of placental flow of pregnant dams, it has now been clearly demonstrated that not only do the low birth weight offspring have an increased risk of diabetes, but this risk is passed on to the second generation, probably through effects on intrauterine and early postnatal nutritional effects on genetic imprinting and/or epigenetic regulation of gene expression or development (Sharif et al., 2007; Ozanne and Constancia, 2007; Jimenez-Chillaron et al., 2006). Moreover, these epigenetic marks, while resulting from early life exposures, can actually progress during postnatal life and thus contribute to age-dependent transcriptional repression of key metabolic genes, as has recently been demonstrated for the β cell transcription factor PDX1 (Park et al., 2008) and the insulin-sensitive glucose transporter GLUT4 (Raychaudhuri et al., 2008, Woo and Patti, 2008).
Finally, we need to consider the possibility that our model of the genetics of type 2 diabetes with a moderate number of relatively common polymorphisms causing disease, is wrong. If the genetic component of type 2 diabetes is represented by uncommon (<5% of the population), but highly penetrant, disease alleles, the GWA studies conducted to date would have been limited in their ability to find the disease genes since the current SNP arrays capture only a small fraction of all the rare variants that are estimated to exist in the human genome.
Insights into Pathogenesis from GWA Studies
The GWA association studies continue to open our eyes to the broad nature of molecules that might contribute to the pathogenesis of type 2 diabetes. Clearly many of the genes identified in both GWA and positional cloning studies would not be considered typical “candidate” genes. However, we do need to be cautious about interpreting what these studies tell us about the genes and tissues involved in the pathogenesis of type 2 diabetes. It is especially important to keep in mind that most of the observed associations are in large domains of strong linkage disequilibrium in noncoding regions of the genome. These association signals are usually named as if there are defects in the closest specific gene, but many of these domains contain multiple genes, and thus the “designated” type 2 diabetes gene may or may not be the gene whose expression or function is altered by the polymorphism. Also, in silico analyses predict that up to 50% of conserved cis-acting elements in the human genome are up to 1 Mb from target genes, sometimes in the introns of neighboring genes (Vavouri et al., 2006). These predictions have been confirmed by chIP-on-chIP experiments (Carroll et al., 2005) and RACE data from the Encode project (Birney et al., 2007). Thus, the true type 2 diabetes genes may be placed at some distance from the association signals.
With these caveats in mind, it is interesting to note that several of the genes placed in the proximity of GWA signals are expressed in β cells. This, together with the findings of association with glucose-stimulated insulin secretion (Steinthorsdottir et al., 2007; Florez et al., 2006a; Sladek et al., 2007), has led some to conclude that β cell defects have a more primal role in the etiology of type 2 diabetes than insulin resistance or that changes in muscle oxidative metabolic phenotypes are not genetic in origin. However, this may not be necessarily the case. The focus of the GWA studies on overt type 2 diabetes favored finding genes marking a limitation of β cell function, since ultimately all forms of diabetes could be viewed as having relative insulin deficiency. The identification of variants predisposing to diabetes through effects on insulin sensitivity will require other study designs, such as ones based on association between insulin sensitivity and genotype in the prediabetic state, i.e., before the onset of overt hyperglycemia. Another factor clouding the discovery of potential insulin resistance genes in GWA studies may be the very strong environment-gene interactions for insulin resistance with body weight, physical activity, and other factors, as compared to the potentially purer manifestation of genetic control on insulin secretion.
Implications of GWA Results for the Development of New Diagnostics and Therapeutics for Type 2 Diabetes
One of the major promises of genetic research has been the hope that this will lead us to new diagnostics and therapeutics for disease. The new GWA findings could represent a significant step in this direction, but it is still early to know. With regard to prediction of disease, most of the single loci identified by GWA mark only a 10%–20% increase in risk of disease, and even the strongest single association, TCF7L2, represents only about a doubling of disease risk in its homozygous state. Higher relative risks can be generated by combining multiple markers together. Indeed, about 97% of the population carry between 9 and 20 risk alleles at the 15 type 2 diabetes loci identified thus far (Figure 4A). If we assume that these genetic effects are additive, the ~1.5% of the population who have ≥20 risk alleles have a 6-fold increase in the odds of developing type 2 diabetes as compared to the 1.5% who carry ≤9 risk alleles (Figure 4B). In terms of predictive value, this translates into an increment of about 10 percentage points over the probability of type 2 diabetes before the genetic test (from 7%—the prevalence of type 2 diabetes in the population—to 17%, Figure 4C). However, for more common risk categories such as those defined by carrying 17 or 18 risk alleles, the increment over the prior probability is in the order of only 3–5 percentage points (Figure 4C). The predictive value of multiple positive markers could be higher if some of the 15 type 2 diabetes genes interact with each other as one recent report indicates (Cauchi et al., 2008b). However, other studies have not shown such synergistic effects. Thus, at present, even a panel of multiple genes has uncertain usefulness in genetic prediction, and it remains to be seen how this would compare to traditional clinical markers of prediction, such as presence of obesity, family history, ethnic background, history of low birth weight or gestational diabetes, etc.
Figure 4. Predictive Value of Multiple Genetic Markers for Type 2 Diabetes.
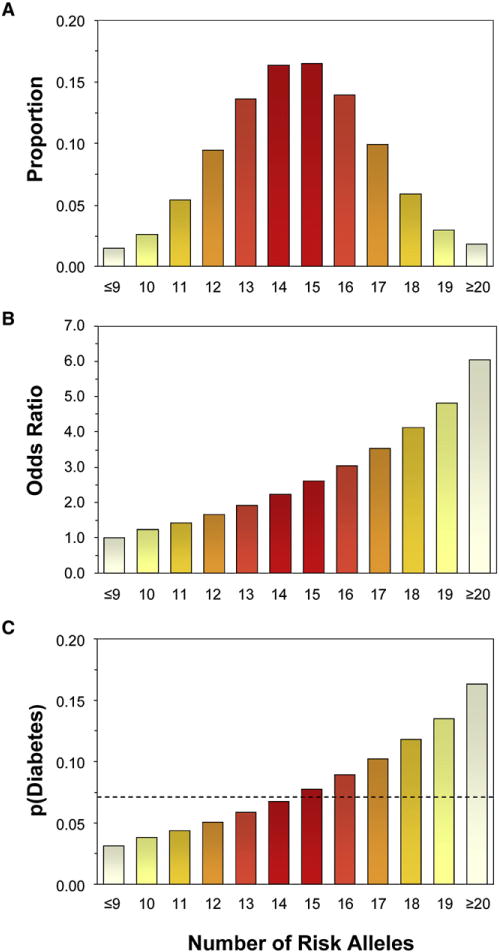
(A) Number of risk alleles carried by individuals from the general population at the 15 type 2 diabetes loci identified to date. Estimates are based on the risk allele frequencies reported in Table 3 and assume independent segregation of the 15 loci.
(B) Odds ratios of type 2 diabetes as a function of the number of risk alleles. The average OR associated with each number of allele was estimated under the assumption of a multiplicative (log-additive) model on the basis of the individual ORs listed in Table 3 and the relative frequencies of the risk alleles in each class.
(C) Probability of type 2 diabetes as a function of the number of risk alleles. Estimates assume an average (pretest) probability of type 2 diabetes of 0.07 (indicated by the dashed line).
With regard to therapeutic implications, these risk alleles do point to a number of previously uninvestigated pathways that might alter β cell function, insulin action, or metabolism. Even for variants that have a small effect on type 2 diabetes risk, it is conceivable that the activity of the cellular pathways in which these are placed can be modulated by drugs to a much larger extent than what is observed as the consequence of natural variation. On the other hand, some of these pathways, such as the Wnt and cell-cycle pathways, are so central to normal cellular growth and function that finding a selective drug target may be difficult.
Looking Forward
Our knowledge of the genetics of type 2 diabetes has come a long way since the days of a few candidate genes studied by means of one or two haphazardly chosen restriction fragment length polymorphisms. Yet, these new findings should be considered as a starting point rather than the arrival. There is little doubt that additional type 2 diabetes loci and genes will be identified through follow-up of “hot spots” in the existing scans (Zeggini et al., 2008) and GWA scans focused on certain ethnic groups and specific clinical or pathophysiological phenotypes of type 2 diabetes.
Also, the current analyses have only considered the effects of individual SNPs in isolation from the effect of other genes or environmental factors. As the effect of these variants becomes increasingly established, attention needs to be focused on genegene and gene-environment interactions. Prospective cohorts for which data are available on dietary exposures, physical activity, and other potential environmental modifiers will be needed to accomplish this task.
Finally, it should be emphasized again that the current GWA studies are based on the “common-disease/common-variant hypothesis” holding that genetic predisposition to common disorders is determined by common genetic variants with small or modest effects (Reich and Lander, 2001). As discussed above, the available genotyping arrays are not designed to investigate the alternative “heterogeneity hypothesis” maintaining that susceptibility to common disorders results from a large number of rare variants, each having a relatively large effect (Pritchard and Cox, 2002). Identification of such rare variants, if they exist, will require resequencing of the entire genome of type 2 diabetes cases and controls. Fortunately, this previously insurmountable task is becoming increasingly realistic with the ongoing development of new, powerful sequencing methods. Likewise, the contribution of structural variants, such as copy-number variants, insertions, deletions, and duplications, could also contribute to genetic susceptibility not explained by single nucleotide substitutions (Feuk et al., 2006). With these additional pieces in hand, we would have a complete view of the genetic architecture of type 2 diabetes. We do not know how the final picture will look, but these initial findings suggest that it will be different from anything we imagined.
Supplementary Material
Acknowledgments
Part of the research discussed in this paper was supported by NIH grants DK60837 (Diabetes Genome Anatomy Project), DK55523, and DK62948, the Graetz Foundation and a Research Grant from the American Diabetes Association (to M.E.P.).
Footnotes
SUPPLEMENTAL DATA
Supplemental Data include one table and can be found online at http://www.cellmetabolism.org/cgi/content/full/8/3/186/DC1/.
References
- Almind K, Inoue G, Pedersen O, Kahn CR. A common amino acid polymorphism in insulin receptor substrate-1 causes impaired insulin signaling. Evidence from transfection studies. J Clin Invest. 1996;97:2569–2575. doi: 10.1172/JCI118705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almind K, Delahaye L, Hansen T, Van Obberghen E, Pedersen O, Kahn CR. Characterization of the Met326Ile variant of phosphatidylinositol 3-kinase p85alpha. Proc Natl Acad Sci USA. 2002;99:2124–2128. doi: 10.1073/pnas.042688799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almind K, Kulkarni RN, Lannon SM, Kahn CR. Identification of Interactive Loci Linked to Insulin and Leptin in Mice With Genetic Insulin Resistance. Diabetes. 2003;52:1535–1543. doi: 10.2337/diabetes.52.6.1535. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, et al. The common PPARgamma Pro12Ala is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, et al. Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature. 1999;402:880–883. doi: 10.1038/47254. [DOI] [PubMed] [Google Scholar]
- Barroso I, Luan J, Middelberg RP, Harding AH, Franks PW, Jakes RW, Clayton D, Schafer AJ, O’Rahilly S, Wareham NJ. Candidate Gene Association Study in Type 2 Diabetes Indicates a Role for Genes Involved in beta-Cell Function as Well as Insulin Action. PLoS Biol. 2003;1:E20. doi: 10.1371/journal.pbio.0000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamer BA, Yen CJ, Andersen RE, Muller D, Elahi D, Cheskin LJ, Andres R, Roth J, Shuldiner AR. Association of the Pro12Ala variant in the peroxisome proliferator-activated receptor γ2 gene with obesity in two Caucasian populations. Diabetes. 1998;47:1806–1808. doi: 10.2337/diabetes.47.11.1806. [DOI] [PubMed] [Google Scholar]
- Bergman RN. Orchestration of glucose homeostasis: from a small acorn to the California oak. Diabetes. 2007;56:1489–1501. doi: 10.2337/db07-9903. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunham LR, Singaraja RR, Hayden MR. Variations on a gene:rare and common variants in ABCA1 and their impact on HDL cholesterol levels and atherosclerosis. Annu Rev Nutr. 2006;26:105–129. doi: 10.1146/annurev.nutr.26.061505.111214. [DOI] [PubMed] [Google Scholar]
- Bruning JC, Winnay J, Bonner-Weir S, Taylor SI, Accili D, Kahn CR. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell. 1997;88:561–572. doi: 10.1016/s0092-8674(00)81896-6. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Cauchi S, Meyre D, Dina C, Choquet H, Samson C, Gallina S, Balkau B, Charpentier G, Pattou F, Stetsyuk V, et al. Transcription factor TCF7L2 genetic study in the French population: expression in human beta-cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55:2903–2908. doi: 10.2337/db06-0474. [DOI] [PubMed] [Google Scholar]
- Cauchi S, Choquet H, Gutierrez-Aguilar R, Capel F, Grau K, Proenca C, Dina C, Duval A, Balkau B, Marre M, et al. Effects of TCF7L2 polymorphisms on obesity in European populations. Obesity (Silver Spring) 2008a;16:476–482. doi: 10.1038/oby.2007.77. [DOI] [PubMed] [Google Scholar]
- Cauchi S, Meyre D, Durand E, Proenca C, Marre M, Hadjadj S, Choquet H, De Graeve F, Gaget S, Allegaert F, et al. Post genome-wide association studies of novel genes associated with type 2 diabetes show gene-gene interaction and high predictive value. PLoS ONE. 2008b;3:e2031. doi: 10.1371/journal.pone.0002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, Froguel P, Balding D, Scott J, Kooner JS. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- Chen WM, Erdos MR, Jackson AU, Saxena R, Sanna S, Silver KD, Timpson NJ, Hansen T, Orru M, Grazia PM, et al. Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J Clin Invest. 2008;118:2620–2628. doi: 10.1172/JCI34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimienti F, Devergnas S, Favier A, Seve M. Identification and cloning of a beta-cell-specific zinc transporter, ZnT-8, localized into insulin secretory granules. Diabetes. 2004;53:2330–2337. doi: 10.2337/diabetes.53.9.2330. [DOI] [PubMed] [Google Scholar]
- Chimienti F, Devergnas S, Pattou F, Schuit F, Garcia-Cuenca R, Vandewalle B, Kerr-Conte J, Van Lommel L, Grunwald D, Favier A, Seve M. In vivo expression and functional characterization of the zinc transporter ZnT8 in glucose-induced insulin secretion. J Cell Sci. 2006;119:4199–4206. doi: 10.1242/jcs.03164. [DOI] [PubMed] [Google Scholar]
- Cossu G, Borello U. Wnt signaling and the activation of myogenesis in mammals. EMBO J. 1999;18:6867–6872. doi: 10.1093/emboj/18.24.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowie CC, Rust KF, Byrd-Holt DD, Eberhardt MS, Flegal KM, Engelgau MM, Saydah SH, Williams DE, Geiss LS, Gregg EW. Prevalence of diabetes and impaired fasting glucose in adults in the U.S. population: National Health And Nutrition Examination Survey 1999–2002. Diabetes Care. 2006;29:1263–1268. doi: 10.2337/dc06-0062. [DOI] [PubMed] [Google Scholar]
- Crunkhorn S, Dearie F, Mantzoros C, Gami H, da Silva WS, Espinoza D, Faucette R, Barry K, Bianco AC, Patti ME. Peroxisome proliferator activator receptor gamma coactivator-1 expression is reduced in obesity: potential pathogenic role of saturated fatty acids and p38 mitogen-activated protein kinase activation. J Biol Chem. 2007;282:15439–15450. doi: 10.1074/jbc.M611214200. [DOI] [PubMed] [Google Scholar]
- Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkänen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, Carlsson LM, Kiess W, Vatin V, Lecoeur C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Navas MA, Dufort D, Rossant J, Stoffel M. Regulation of a transcription factor network required for differentiation and metabolism. Science. 1998;281:692–695. doi: 10.1126/science.281.5377.692. [DOI] [PubMed] [Google Scholar]
- Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe DJ, Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta -protein, and the beta -amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci USA. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehmann HC, Goke R, Goke B. Cell and molecular biology of the incretin hormones glucagon-like peptide I and glucose-dependent insulin releasing polypeptide. Endocr Rev. 1995;16:390–410. doi: 10.1210/edrv-16-3-390. [DOI] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Flegal KM, Ezzati TM, Harris MI, Haynes SG, Juarez RZ, Knowler WC, Perez-Stable EJ, Stern MP. Prevalence of diabetes in Mexican-Americans, Cubans, and Puerto Ricans from the Hispanic Health and Nutrition Examination Survey 1982–1984. Diabetes Care. 1991;14:628–638. doi: 10.2337/diacare.14.7.628. [DOI] [PubMed] [Google Scholar]
- Florez JC, Sjogren M, Burtt N, Orho-Melander M, Schayer S, Sun M, Almgren P, Lindblad U, Tuomi T, Gaudet D, et al. Association testing in 9,000 people fails to confirm the association of the insulin receptor substrate-1 G972R polymorphism with type 2 diabetes. Diabetes. 2004a;53:3313–3318. doi: 10.2337/diabetes.53.12.3313. [DOI] [PubMed] [Google Scholar]
- Florez JC, Burtt N, de Bakker PI, Almgren P, Tuomi T, Holmkvist J, Gaudet D, Hudson TJ, Schaffner SF, Daly MJ, et al. Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes. 2004b;53:1360–1368. doi: 10.2337/diabetes.53.5.1360. [DOI] [PubMed] [Google Scholar]
- Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med. 2006a;355:241–250. doi: 10.1056/NEJMoa062418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florez JC, Wiltshire S, Agapakis CM, Burtt NP, de Bakker PI, Almgren P, Bengtsson BK, Tuomi T, Gaudet D, Daly MJ, et al. High-density haplotype structure and association testing of the insulin-degrading enzyme (IDE) gene with type 2 diabetes in 4,206 people. Diabetes. 2006b;55:128–135. [PubMed] [Google Scholar]
- Florez JC, Manning AK, Dupuis J, McAteer J, Irenze K, Gianniny L, Mirel DB, Fox CS, Cupples LA, Meigs JB. A 100K genome-wide association scan for diabetes and related traits in the Framingham Heart Study: replication and integration with other genome-wide datasets. Diabetes. 2007;56:3063–3074. doi: 10.2337/db07-0451. [DOI] [PubMed] [Google Scholar]
- Foley AC, Mercola M. Heart induction by Wnt antagonists dependsonthe homeodomain transcription factor Hex. Genes Dev. 2005;19:387–396. doi: 10.1101/gad.1279405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloyn AL, Pearson ER, Antcliff JF, Proks P, Bruining GJ, Slingerland AS, Howard N, Srinivasan S, Silva JM, Molnes J, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350:1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- Grarup N, Andersen G, Krarup NT, Albrechtsen A, Schmitz O, Jorgensen T, Borch-Johnsen K, Hansen T, Pedersen O. Association testing of novel type 2 diabetes risk-alleles in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, ADAMTS9, and NOTCH2 loci with insulin release, insulin sensitivity and obesity in a population-based sample of 4,516 glucose-tolerant middle-aged Danes. Diabetes. 2008 doi: 10.2337/db08-0436. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunton JE, Kulkarni RN, Yim S, Okada T, Hawthorne WJ, Tseng YH, Roberson RS, Ricordi C, O’Connell PJ, Gonzalez FJ, Kahn CR. Loss of ARNT/HIF1beta mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell. 2005;122:337–349. doi: 10.1016/j.cell.2005.05.027. [DOI] [PubMed] [Google Scholar]
- Hansen L, Pedersen O. Genetics of type 2 diabetes mellitus: status and perspectives. Diabetes Obes Metab. 2005;7:122–135. doi: 10.1111/j.1463-1326.2004.00396.x. [DOI] [PubMed] [Google Scholar]
- Hanson RL, Bogardus C, Duggan D, Kobes S, Knowlton M, Infante AM, Marovich L, Benitez D, Baier LJ, Knowler WC. A search for variants associated with young-onset type 2 diabetes in American Indians in a 100K genotyping array. Diabetes. 2007;56:3045–3052. doi: 10.2337/db07-0462. [DOI] [PubMed] [Google Scholar]
- Hayes MG, Pluzhnikov A, Miyake K, Sun Y, Ng MC, Roe CA, Below JE, Nicolae RI, Konkashbaev A, Bell GI, et al. Identification of type 2 diabetes genes in Mexican Americans through genome-wide association studies. Diabetes. 2007;56:3033–3044. doi: 10.2337/db07-0482. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- Hogan P, Dall T, Nikolov P. Economic costs of diabetes in the US in 2002. Diabetes Care. 2003;26:917–932. doi: 10.2337/diacare.26.3.917. [DOI] [PubMed] [Google Scholar]
- Jimenez-Chillaron JC, Hernandez-Valencia M, Lightner A, Faucette RR, Reamer C, Przybyla R, Ruest S, Barry K, Otis JP, Patti ME. Reductions in caloric intake and early postnatal growth prevent glucose intolerance and obesity associated with low birthweight. Diabetologia. 2006;49:1974–1984. doi: 10.1007/s00125-006-0311-7. [DOI] [PubMed] [Google Scholar]
- Kahn CR. Insulin action, diabetogenes, and the cause of type II diabetes (Banting Lecture) Diabetes. 1994;43:1066–1084. doi: 10.2337/diab.43.8.1066. [DOI] [PubMed] [Google Scholar]
- Kahn CR, Vicent D, Doria A. Genetics of non-insulin-dependent (type-II) diabetes mellitus. Annu Rev Med. 1996;47:509–531. doi: 10.1146/annurev.med.47.1.509. [DOI] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, III, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kissebah AH, Sonnenberg GE, Myklebust J, Goldstein M, Broman K, James RG, Marks JA, Krakower GR, Jacob HJ, Weber J, et al. Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci USA. 2000;97:14478–14483. doi: 10.1073/pnas.97.26.14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolewski AS, Warram JH. Epidemiology of late complications of diabetes: Abasis for the development and evaluation of preventive program. In: Kahn CR, Weir GC, King GL, Moses AC, Smith RJ, Jacobson AM, editors. Joslin’s Diabetes Mellitus. New York: Lippincott, Williams & Wilkins; 2005. [Google Scholar]
- Ling C, Poulsen P, Carlsson E, Ridderstrale M, Almgren P, Wojtaszewski J, Beck-Nielsen H, Groop L, Vaag A. Multiple environmental and genetic factors influence skeletal muscle PGC-1alpha and PGC-1beta gene expression in twins. J Clin Invest. 2004;114:1518–1526. doi: 10.1172/JCI21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Poulsen P, Simonsson S, Ronn T, Holmkvist J, Almgren P, Hagert P, Nilsson E, Mabey AG, Nilsson P, et al. Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J Clin Invest. 2007;117:3427–3435. doi: 10.1172/JCI30938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, Inouye M, Freathy RM, Attwood AP, Beckmann JS, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssenko V, Almgren P, Anevski D, Perfekt R, Lahti K, Nissen M, Isomaa B, Forsen B, Homstrom N, Saloranta C, et al. Predictors of and longitudinal changes in insulin sensitivity and secretion preceding onset of type 2 diabetes. Diabetes. 2005;54:166–174. doi: 10.2337/diabetes.54.1.166. [DOI] [PubMed] [Google Scholar]
- Lyssenko V, Lupi R, Marchetti P, Del Guerra S, Ortho-Melander M, Almgren P, Sjogren M, Ling C, Eriksson KF, Lethagen AL, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magre J, Delepine M, Khallouf E, Gedde-Dahl T, Jr, Van Maldergem L, Sobel E, Papp J, Meier M, Megarbane A, Bachy A, et al. Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat Genet. 2001;28:365–370. doi: 10.1038/ng585. [DOI] [PubMed] [Google Scholar]
- Martin BC, Warram JH, Krolewski AS, Bergman RN, Soeldner JS, Kahn CR. Role of glucose and insulin resistance in development of type 2 diabetes mellitus: results of a 25-year follow-up study. Lancet. 1992;340:925–929. doi: 10.1016/0140-6736(92)92814-v. [DOI] [PubMed] [Google Scholar]
- McAteer JB, Prudente S, Bacci S, Lyon HN, Hirschhorn JN, Trischitta V, Florez JC. The ENPP1 K121Q polymorphism is associated with type 2 diabetes in European populations: evidence from an updated meta-analysis in 42,042 subjects. Diabetes. 2008;57:1125–1130. doi: 10.2337/db07-1336. [DOI] [PubMed] [Google Scholar]
- McLin VA, Rankin SA, Zorn AM. Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development. 2007;134:2207–2217. doi: 10.1242/dev.001230. [DOI] [PubMed] [Google Scholar]
- McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AF, Jablonski KA, McAteer JB, Saxena R, Pollin TI, Franks PW, Hanson RL, Shuldiner AR, Knowler WC, Altshuler D, Florez JC. Extension of type 2 diabetes genome-wide association scan results in the diabetes prevention program Diabetes, in press. 2008 doi: 10.2337/db08-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen EM, Hansen L, Carstensen B, Echwald SM, Drivsholm T, Glumer C, Thorsteinsson B, Borch-Johnsen K, Hansen T, Pedersen O. The E23K variant of Kir6.2 associates with impaired post-OGTT serum insulin response and increased risk of type 2 diabetes. Diabetes. 2003;52:573–577. doi: 10.2337/diabetes.52.2.573. [DOI] [PubMed] [Google Scholar]
- Nobori T, Takabayashi K, Tran P, Orvis L, Batova A, Yu AL, Carson DA. Genomic cloning of methylthioadenosine phosphorylase: a purine metabolic enzyme deficient in multiple different cancers. Proc Natl Acad Sci USA. 1996;93:6203–6208. doi: 10.1073/pnas.93.12.6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rahilly S. Human obesity and insulin resistance: lessons from experiments of nature. Biochem Soc Trans. 2007;35:33–36. doi: 10.1042/BST0350033. [DOI] [PubMed] [Google Scholar]
- Ozanne SE, Constancia M. Mechanisms of disease: the developmental origins of disease and the role of the epigenotype. Nat Clin Pract Endocrinol Metab. 2007;3:539–546. doi: 10.1038/ncpendmet0531. [DOI] [PubMed] [Google Scholar]
- Palmer ND, Goodarzi MO, Langefeld CD, Ziegler J, Norris JM, Haffner SM, Bryer-Ash M, Bergman RN, Wagenknecht LE, Taylor KD, et al. Quantitative trait analysis of type 2 diabetes susceptibility loci identified from whole genome association studies in the Insulin Resistance Atherosclerosis Family Study. Diabetes. 2008;57:1093–1100. doi: 10.2337/db07-1169. [DOI] [PubMed] [Google Scholar]
- Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton LE, Ye CP, Coppari R, Enriori PJ, Choi B, Zhang CY, Xu C, Vianna CR, Balthasar N, Lee CE, et al. Glucose sensing by POMC neurons regulates glucose homeostasis and is impaired in obesity. Nature. 2007;449:228–232. doi: 10.1038/nature06098. [DOI] [PubMed] [Google Scholar]
- Pascoe L, Tura A, Patel SK, Ibrahim IM, Ferrannini E, Zeggini E, Wee-don MN, Mari A, Hattersley AT, McCarthy MI, et al. Common variants of the novel type 2 diabetes genes CDKAL1 and HHEX/IDE are associated with decreased pancreatic beta-cell function. Diabetes. 2007;56:3101–3104. doi: 10.2337/db07-0634. [DOI] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci USA. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson ER, Flechtner I, Njolstad PR, Malecki MT, Flanagan SE, Larkin B, Ashcroft FM, Klimes I, Codner E, Iotova V, et al. Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N Engl J Med. 2006;355:467–477. doi: 10.1056/NEJMoa061759. [DOI] [PubMed] [Google Scholar]
- Pizzuti A, Frittitta L, Argiolas A, Baratta R, Goldfine ID, Bozzali M, Ercolino T, Scarlato G, Iacoviello L, Vigneri R, et al. A polymorphism (K121Q) of the human glycoprotein PC-1 gene coding region is strongly associated with insulin resistance. Diabetes. 1999;48:1881–1884. doi: 10.2337/diabetes.48.9.1881. [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Pospisilik JA, Knauf C, Joza N, Benit P, Orthofer M, Cani PD, Ebersberger I, Nakashima T, Sarao R, Neely G, et al. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131:476–491. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance–a population-based twin study. Diabetologia. 1999;42:139–145. doi: 10.1007/s001250051131. [DOI] [PubMed] [Google Scholar]
- Prestwich TC, MacDougald OA. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol. 2007;19:612–617. doi: 10.1016/j.ceb.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant…or not? Hum Mol Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- Prunier C, Hocevar BA, Howe PH. Wnt signaling: physiology and pathology. Growth Factors. 2004;22:141–150. doi: 10.1080/08977190410001720860. [DOI] [PubMed] [Google Scholar]
- Qiu WQ, Walsh DM, Ye Z, Vekrellis K, Zhang J, Podlisny MB, Rosner MR, Safavi A, Hersh LB, Selkoe DJ. Insulin-degrading enzyme regulates extracellular levels of amyloid beta-protein by degradation. J Biol Chem. 1998;273:32730–32738. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- Rampersaud E, Damcott CM, Fu M, Shen H, McArdle P, Shi X, Shelton J, Yin J, Chang YP, Ott SH, et al. Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the Old Order Amish: evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes. 2007;56:3053–3062. doi: 10.2337/db07-0457. [DOI] [PubMed] [Google Scholar]
- Rane SG, Dubus P, Mettus RV, Galbreath EJ, Boden G, Reddy EP, Barbacid M. Loss of cyclin-dependent kinase (Cdk4) expression causes insulin-deficient diabetes and cdk4 activation results in β-islet cell hyperplasia. Nat Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri N, Raychaudhuri S, Thamotharan M, Devaskar SU. Histone code modifications repress glucose transporter 4 expression in the intrauterine growth-restricted offspring. J Biol Chem. 2008;283:13611–13626. doi: 10.1074/jbc.M800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz I, Karsai D, Katz M. The influence of zinc supplementation on glucose homeostasis in NIDDM. Diabetes Res. 1989;11:73–79. [PubMed] [Google Scholar]
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- Rich SS. Mapping genes in diabetes: Genetic epidemiological perspective. Diabetes. 1990;39:1315–1319. doi: 10.2337/diab.39.11.1315. [DOI] [PubMed] [Google Scholar]
- Ronn T, Poulsen P, Hansson O, Holmkvist J, Almgren P, Nilsson P, Tuomi T, Isomaa B, Groop L, Vaag A, Ling C. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia. 2008;51:1159–1168. doi: 10.1007/s00125-008-1018-8. [DOI] [PubMed] [Google Scholar]
- Salonen JT, Uimari P, Aalto JM, Pirskanen M, Kaikkonen J, Todorova B, Hypponen J, Korhonen VP, Asikainen J, Devine C, et al. Type 2 diabetes whole-genome association study in four populations: the DiaGen consortium. Am J Hum Genet. 2007;81:338–345. doi: 10.1086/520599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Schwanstecher C, Meyer U, Schwanstecher M. K(IR)6.2 polymorphism predisposes to type 2 diabetes by inducing overactivity of pancreatic beta-cell ATP-sensitive K(+) channels. Diabetes. 2002;51:875–879. doi: 10.2337/diabetes.51.3.875. [DOI] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton S, Lloyd DJ, Jackson SN, Evans R, Niermeijer MF, Singh BM, Schmidt H, Brabant G, Kumar S, Durrington PN, et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- Sharif J, Nakamura M, Ito T, Kimura Y, Nagamune T, Mitsuya K, Okamura K. Food restriction in pregnant mice can induce changes in histone modifications and suppress gene expression in fetus. Nucleic Acids Symp Ser (Oxf) 2007;51:125–126. doi: 10.1093/nass/nrm063. [DOI] [PubMed] [Google Scholar]
- Shu L, Sauter NS, Schulthess FT, Matveyenko AV, Oberholzer J, Maedler K. TCF7L2 regulates -cell survival and function in human pancreatic islets. Diabetes. 2008;57:645–653. doi: 10.2337/db07-0847. [DOI] [PubMed] [Google Scholar]
- Simon SF, Taylor CG. Dietary zinc supplementation attenuates hyperglycemia in db/db mice. Exp Biol Med (Maywood) 2001;226:43–51. doi: 10.1177/153537020122600107. [DOI] [PubMed] [Google Scholar]
- Skyler JS, Oddo C. Diabetes trends in the USA. Diabetes Metab Res Rev. 2002;18(Suppl 3):S21–S26. doi: 10.1002/dmrr.289. [DOI] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Stancáková A, Pihlajamaki J, Kuusisto J, Stefan N, Fritsche A, Haring H, Andreozzi F, Succurro E, Sesti G, Boesgaard TW, et al. Single-nucleotide polymorphism rs7754840 of CDKAL1 is associated with impaired insulin secretion in nondiabetic offspring of type 2 diabetic subjects and in a large sample of men with normal glucose tolerance. J Clin Endocrinol Metab. 2008;93:1924–1930. doi: 10.1210/jc.2007-2218. [DOI] [PubMed] [Google Scholar]
- Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- Stern MP. Diabetes and cardiovascular disease. The “common soil” hypothesis. Diabetes. 1995;44:369–374. doi: 10.2337/diab.44.4.369. [DOI] [PubMed] [Google Scholar]
- Stern MP. The search for type 2 diabetes susceptibility genes using whole-genome scans: an epidemiologist’s perspective. Diabetes Metab Res Rev. 2002;18:106–113. doi: 10.1002/dmrr.268. [DOI] [PubMed] [Google Scholar]
- Strachan T, Read A. Human Molecular Genetics. Oxford: Garland Science; 2003. [Google Scholar]
- Tanaka T, Inazu T, Yamada K, Myint Z, Keng VW, Inoue Y, Taniguchi N, Noguchi T. cDNA cloning and expression of rat homeobox gene, Hex, and functional characterization of the protein. Biochem J. 1999;339:111–117. [PMC free article] [PubMed] [Google Scholar]
- Taylor CG. Zinc, the pancreas, and diabetes: insights from rodent studies and future directions. Biometals. 2005;18:305–312. doi: 10.1007/s10534-005-3686-x. [DOI] [PubMed] [Google Scholar]
- Ubeda M, Rukstalis JM, Habener JF. Inhibition of cyclin-dependent kinase 5 activity protects pancreatic beta cells from glucotoxicity. J Biol Chem. 2006;281:28858–28864. doi: 10.1074/jbc.M604690200. [DOI] [PubMed] [Google Scholar]
- Vavouri T, McEwen GK, Woolfe A, Gilks WR, Elgar G. Defining a genomic radius for long-range enhancer action: duplicated conserved non-coding elements hold the key. Trends Genet. 2006;22:5–10. doi: 10.1016/j.tig.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Vionnet N, El Habib H, Dupont S, Gallina S, Francke S, Dotte S, De Matos F, Durand E, Lepretre F, Lecoeur C, et al. Genomewide search for type 2 diabetes-susceptibility genes in french whites: evidence for a novel susceptibility locus for early-onset diabetes on chrososome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21–q24. Am J Hum Genet. 2000;67:1470–1480. doi: 10.1086/316887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe RM, Allayee H, Xiang AH, Trigo E, Hartiala J, Lawrence JM, Buchanan TA. Transcription factor 7-like 2 (TCF7L2) is associated with gestational diabetes mellitus and interacts with adiposity to alter insulin secretion in Mexican Americans. Diabetes. 2007;56:1481–1485. doi: 10.2337/db06-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijnen CF, Rich SS, Meigs JB, Krolewski AS, Warram JH. Risk of diabetes in siblings of index cases with Type 2 diabetes: implications for genetic studies. Diabet Med. 2002;19:41–50. doi: 10.1046/j.1464-5491.2002.00624.x. [DOI] [PubMed] [Google Scholar]
- Wenzlau JM, Juhl K, Yu L, Moua O, Sarkar SA, Gottlieb P, Rewers M, Eisenbarth GS, Jensen J, Davidson HW, Hutton JC. The cation efflux transporter ZnT8 (Slc30A8) is a major autoantigen in human type 1 diabetes. Proc Natl Acad Sci USA. 2007;104:17040–17045. doi: 10.1073/pnas.0705894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo M, Patti ME. Diabetes risk begins in utero. Cell Metab. 2008;8:5–7. doi: 10.1016/j.cmet.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Yi F, Brubaker PL, Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by beta-catenin and glycogen synthase kinase-3beta. J Biol Chem. 2005;280:1457–1464. doi: 10.1074/jbc.M411487200. [DOI] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


