Abstract
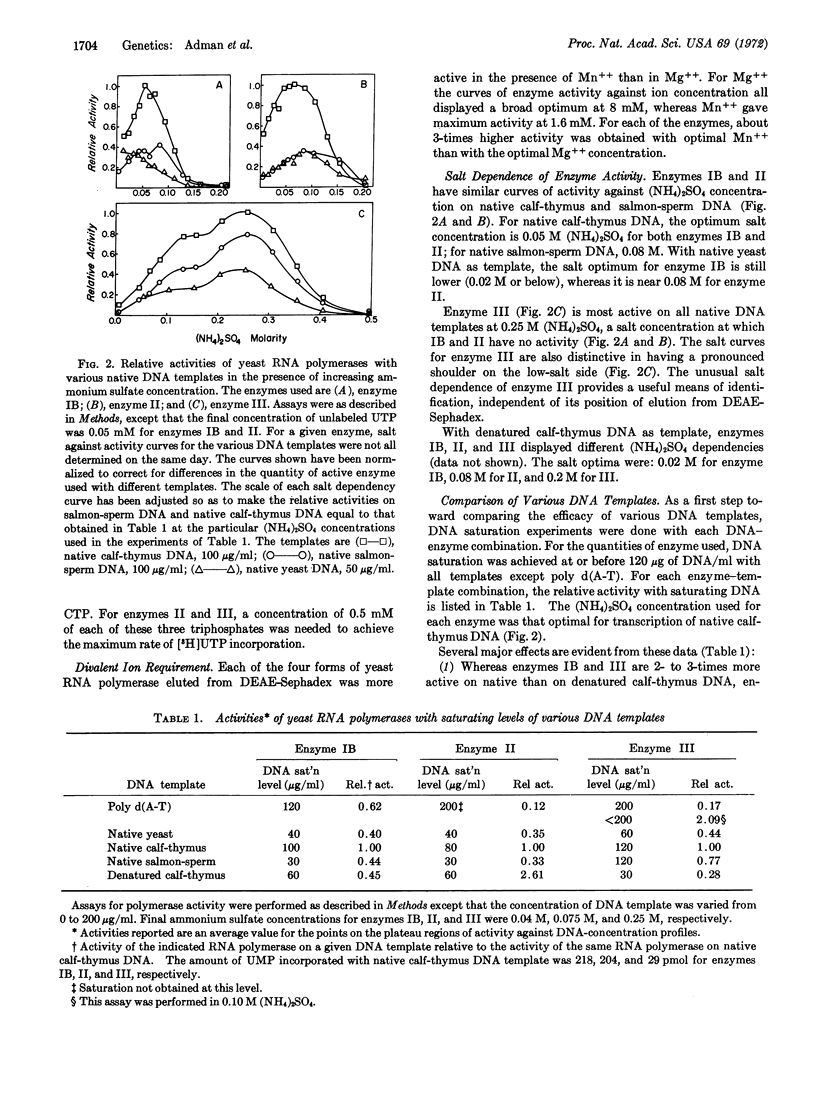
Four peaks of DNA-directed RNA polymerase activity are resolved by salt gradient elution of a sonicated yeast cell extract on DEAE-Sephadex. The enzymes, which are named IA, IB, II, and III in order of elution, all appear to come from cell nuclei. Only enzyme II is sensitive to α-amanitin. All enzymes are more active with Mn++ than with Mg++ as divalent ion. Enzymes IB and II have salt optima in the range 0.05-0.10 M (NH4)2SO4, whereas enzyme III is maximally active at 0.20-0.25 M (NH4)2SO4. With optimal salt concentration and saturating DNA, the template preference ratio, activity on native calfthymus DNA divided by activity on denatured calf-thymus DNA, is 2.2 for IB, 0.4 for II, and 3.5 for III. None of the yeast polymerases was inhibited by rifamycin SV. Rifamycin AF/013 effectively inhibited polymerases IB, II, and III.
Keywords: eukaryote, transcription, rifamycin AF, 013
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burgess R. R. Separation and characterization of the subunits of ribonucleic acid polymerase. J Biol Chem. 1969 Nov 25;244(22):6168–6176. [PubMed] [Google Scholar]
- Burgess R. R., Travers A. A., Dunn J. J., Bautz E. K. Factor stimulating transcription by RNA polymerase. Nature. 1969 Jan 4;221(5175):43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- Dezelee S., Sentenac A., Fromageot P. Role of DNA-RNA hybrids in eukaryots 1. Purification of yeast RNA polymerase B. FEBS Lett. 1972 Mar;21(1):1–6. doi: 10.1016/0014-5793(72)80148-0. [DOI] [PubMed] [Google Scholar]
- Esposito M. S., Esposito R. E. The genetic control of sporulation in Saccharomyces. I. The isolation of temperature-sensitive sporulation-deficient mutants. Genetics. 1969 Jan;61(1):79–89. doi: 10.1093/genetics/61.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawthorne D. C., Mortimer R. K. Genetic mapping of nonsense suppressors in yeast. Genetics. 1968 Dec;60(4):735–742. doi: 10.1093/genetics/60.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil A., Zillig W. Reconstitution of bacterial DNA-dependent RNA-polymerase from isolated subunits as a tool for the elucidation of the role of the subunits in transcription. FEBS Lett. 1970 Dec;11(3):165–168. doi: 10.1016/0014-5793(70)80519-1. [DOI] [PubMed] [Google Scholar]
- Horgen P. A., Griffin D. H. Specific inhibitors of the three RNA polymerases from the aquatic fungus Blastocladiella emersonii. Proc Natl Acad Sci U S A. 1971 Feb;68(2):338–341. doi: 10.1073/pnas.68.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman J., Cotman C., Mahler H. R., Sharp C. W. Biochemical correlates of respiratory deficiency. VII. Glucose repression. Arch Biochem Biophys. 1966 Sep 26;116(1):224–251. doi: 10.1016/0003-9861(66)90029-4. [DOI] [PubMed] [Google Scholar]
- Kedinger C., Nuret P., Chambon P. Structural evidence for two alpha-amanitin sensitive RNA polymerases in calf thymus. FEBS Lett. 1971 Jun 24;15(3):169–174. doi: 10.1016/0014-5793(71)80305-8. [DOI] [PubMed] [Google Scholar]
- Lindell T. J., Weinberg F., Morris P. W., Roeder R. G., Rutter W. J. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science. 1970 Oct 23;170(3956):447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- Losick R., Shorenstein R. G., Sonenshein A. L. Structural alteration of RNA polymerase during sporulation. Nature. 1970 Aug 29;227(5261):910–913. doi: 10.1038/227910a0. [DOI] [PubMed] [Google Scholar]
- Pulitzer J. F., Geiduschek E. P. Function of T4 gene 55. II. RNA synthesis by temperature-sensitive gene 55 mutants. J Mol Biol. 1970 Apr 28;49(2):489–507. doi: 10.1016/0022-2836(70)90259-7. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Termination factor for RNA synthesis. Nature. 1969 Dec 20;224(5225):1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- Roeder R. G., Rutter W. J. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969 Oct 18;224(5216):234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- Sonenshein A. L., Losick R. RNA polymerase mutants blocked in sporulation. Nature. 1970 Aug 29;227(5261):906–909. doi: 10.1038/227906a0. [DOI] [PubMed] [Google Scholar]
- Tsai M. J., Michaelis G., Criddle R. S. DNA-dependent RNA polymerase from yeast mitochondria. Proc Natl Acad Sci U S A. 1971 Feb;68(2):473–477. doi: 10.1073/pnas.68.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Blatti S. P., Rutter W. J. Molecular structures of DNA-dependent RNA polymerases (II) from calf thymus and rat liver. Proc Natl Acad Sci U S A. 1971 Dec;68(12):2994–2999. doi: 10.1073/pnas.68.12.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli W., Knüsel F., Schmid K., Staehelin M. Interaction of rifamycin with bacterial RNA polymerase. Proc Natl Acad Sci U S A. 1968 Oct;61(2):667–673. doi: 10.1073/pnas.61.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T., Igarashi K. RNA polymerase mutants of Escherichia coli. I. Mutants resistant to streptovaricin. Proc Natl Acad Sci U S A. 1968 Dec;61(4):1313–1319. doi: 10.1073/pnas.61.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]


