Abstract
The TGFβ signaling pathway is essential to epithelial homeostasis and is often inhibited during progression of esophageal squamous cell carcinoma. Recently, an important role for TGFβ signaling has been described in the crosstalk between epithelial and stromal cells regulating squamous tumor cell invasion in mouse models of head-and-neck squamous cell carcinoma (HNSCC). Loss of TGFβ signaling, in either compartment, leads to HNSCC however, the mechanisms involved are not well understood. Using organotypic reconstruct cultures (OTC) to model the interaction between epithelial and stromal cells that occur in dysplastic lesions, we show that loss of TGFβ signaling promotes an invasive phenotype in both fibroblast and epithelial compartments. Employing immortalized esophageal keratinocytes established to reproduce common mutations of esophageal squamous cell carcinoma, we show that treatment of OTC with inhibitors of TGFβ signaling (A83–01 or SB431542) enhances invasion of epithelial cells into a fibroblast-embedded Matrigel/collagen I matrix. Invasion induced by A83–01 is independent of proliferation but relies on protease activity and expression of ADAMTS-1 and can be altered by matrix density. This invasion was associated with increased expression of pro-inflammatory cytokines, IL1 and EGFR ligands HB-EGF and TGFα. Altering EGF signaling prevented or induced epithelial cell invasion in this model. Loss of expression of the TGFβ target gene ROBO1 suggested that chemorepulsion may regulate keratinocyte invasion. Taken together, our data show increased invasion through inhibition of TGFβ signaling altered epithelial-fibroblasts interactions, repressing markers of activated fibroblasts, and altering integrin-fibronectin interactions. These results suggest that inhibition of TGFβ signaling modulates an array of pathways that combined promote multiple aspects of tumor invasion.
Keywords: Invasion, ADAMTS-1, MMP14, ESCC, cell matrix interaction
Introduction
Esophageal squamous cell carcinomas (ESCC) arise from a succession of alterations in the epithelium, leading to loss of tissue architecture and modification of the underlying stroma. In the progression from hyperplasia to dysplasia and carcinoma in situ, normal tissue homeostasis is lost and the basal cell layer, previously quiescent, becomes proliferative and expands in the upper layer of the epithelium while immune cells accumulate around the lesion [1,2]. In ESCC, invasion into the underlying tissue requires the initial growth of neoplastic tissue. Progression is characterized by poorly differentiated invading tumor cells as well as broad collective leading fronts retaining high differentiation [3].
As a potent regulator of tissue homeostasis and cell proliferation TGFβ1 induced signaling is often altered during early stages of cancer progression [4]. In later stages, TGFβ1 participates in the induction of myofibroblasts and fibrosis in the surrounding stroma and promotes tumor invasion in cells, in which its antiproliferative effect has been subverted [4,5].
TGFβ signaling is induced by the binding of TGFβ1 to TGFβRII followed by the heterodimerization of TGFβRI and TGFβRII and subsequent phosphorylation of the R-Smads Smad2/3, which are then translocated to the nucleus in association with Smad4. HNSCC often presents with TGFβ signaling alterations that affect receptors, co-activators, or repressors of the signaling pathway [6,7]. Inhibition of TGFβ in mouse models targeting epithelia of the head-and-neck region by knock-out of TGFβRII or Smad4 alone has little impact on tissue homeostasis. However, introducing a second hit through oncogene overexpression or inhibition of tumor suppressor genes leads to tumor formation [8,9]. Interestingly, knockout of TGFβ signaling members in fibroblasts is sufficient to induce tumor formation [10,11]. A common phenotype of these different models is the increase of local inflammation as a result of TGFβ signaling inhibition [9,10].
Based on these observations, it has been proposed that the activated stroma stimulates proliferation and epithelial cell invasion through increased secretion of chemotactic factors, notably TGFα, HGF, and SDF-1 [9–11]. However, the mechanisms underlying the crosstalk between epithelial and fibroblast compartments that induce invasion are still poorly understood. Using organotypic three-dimensional reconstruct cultures as a platform to study ESCC invasion, we have previously shown that expression of dominant-negative mutants of E-cadherin and TGFβRII in immortalized esophageal keratinocytes results in fibroblast-dependent epithelial cell invasion [12]. To elucidate the mechanisms underlying TGFβ-mediated control of tissue homeostasis and cell invasion, we used chemical TGFβ inhibitors in the present study. We show that inhibition of TGFβ signaling in fibroblasts and epithelial cells results in enhanced invasion in organotypic reconstruct cultures. Loss of TGFβ signaling elicits an inflammatory response in both compartments mediated by IL1, TGFα and HB-EGF and induces the expression of the matrix metalloproteases ADAMTS-1 and MMP14. Moreover, loss of TGFβ signaling inhibits ROBO1 expression in epithelial cells, promoting invasion in the underlying stroma. In conclusion these findings identify multiple detrimental downstream effectors of TGFβ inhibition.
Material and methods
Cell culture and reagents
Primary esophageal epithelial cells (keratinocytes) from normal human esophagus were established as described previously [13]. Fetal esophageal fibroblasts were grown in DMEM with 5% fetal bovine serum (FBS, Hyclone, Thermo Fisher Scientific, Waltham, MA), 100 units/mL penicillin, and 100 µg/mL streptomycin (Gibco, Invitrogen, Carlsbad, CA). A83-01 was used at a final concentration of 1µM (Tocris, Bristol U.K.), SB431542 at 10µM (Tocris), GM6001 at 1µM (Tocris), Erlotinib at 25nM (LC Laboratories, Woburn, MA), rEGF at 10ng/mL (Life technologies, Grand Island, NY). shRNA against ADAMTS-1 were purchased from Qiagen (Qiagen, Germantown, MD) and used according to manufacturer instructions.
OrganoTypic Culture
Organotypic culture (OTC) were grown as previously described [14] with the exception that for 2 more days before the cultures were harvested medium was changed to serum free Epi3 to allow for analysis of the conditioned media. To alter the matrix composition to higher collagen I, we used Collagen I (BD Biosciences, San Jose, CA) at the final concentration of 4.6 mg/mL either mixed with Matrigel or applied as collagen I layer on top of the matrix. Conditioned media were snap frozen and conserved at −80C. Each cultures was used for protein analysis by formalin fixation and paraffin embedding, as well as protein extraction by mechanically peeling the epithelium. The protein were extracted with lysis buffer containing NP40 and Tween20 as previously described [12]. Similarly, total RNA from the epithelium and stroma was isolated using miRNeasy kit (Qiagen). Quantification of invasion was done by measuring the area of epithelial cells invading in the underlying matrix per 1µm length of tissue and statistically analyzed using unpaired Student’s t-test.
Histology
Five micron sections were applied to Probe-on Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA). Slides were stained with hematoxylin and eosin, and images were captured on a Zeiss microscope with a Zeiss Hrc5 camera (Carl Zeiss Microscopy, Thornwood, NY).
Immunohistochemistry was performed with the Vecta Elite kit (Vector Laboratories, Burlingame, CA) following the manufacturer’s protocol using their reagents. Antigen retrieval was performed by heating paraffin sections in a pressure cooker for 12 minutes followed by a one-hour incubation in the pressure cooker. Primary antibody was incubated overnight at 4°C and secondary antibody for 30 minutes at 37°C. Then, the signal was subsequently developed using athe DAB substrate kit for peroxidase. Quantification of proliferation was done by calculating the ratio of Ki67-positive cells to the number of cells in the basal layer.
Immunofluorescence
Formalin-fixed paraffin embedded organotypic culture tissues were sectioned at 5µm, deparaffinized and heated in 1xTE buffer in a pressure cooker for 12 min for antigen retrieval. Samples were blocked in 1xPBS containing 5% Bovine Serum Albumin (BSA; Sigma-Aldrich, Saint- Louis, MO) for 1 hour before incubation with primaries antibody in 1xPBS-BSA overnight at 4°C. Tissues were then rinsed in 1xPBS and incubated with secondary antibodies in 1xPBS-BSA for 1 hour at room temperature. Finally, the sections were mounted with Vectashield mounting medium containing Dapi (Vector Laboratories, Burlingame, CA). Pictures were taken on a Zeiss microscope, using Axiocam and Axiovision software (Carl Zeiss Microscopy, Thornwood, NY).
Proliferation assay
Cells were plated at 1000 cells per well in a 96-well plate, WST-1 reagent was added to each well for an hour at 37°C to estimate cell proliferation (Roche, Nutley, NJ). Absorbance measurements were taken in 24-hour increments at 450nm using a BioTek Synergy 4 plate reader (BioTek, Winooski, VT).
Western Blot
Western blots were performed as previously described. All results are representative of at least 3 experiments [12].
Cytokine array
Experiments were done according to the manufacturer’s protocol, using the RayBio® Human Cytokine Antibody Array 8 (RayBiotech, Norcross, GA). One mL of conditioned medium from the organotypic culture was used per membrane. The signal intensity was measured in Image J (National Institutes of Health) [15] and results were normalized to the provided positive controls.
Real time PCR
RNA extracted from organotypic culture cells was reversed transcribed using SuperScript III kit according to manufacturer’s instructions (LifeTechnologies, Grand Island, NY). For quantitative real-time PCR, QPCR, TaqMan® Array Human Extracellular Matrix & Adhesion Molecules plates were used with TaqMan Fast Universal Mastermix (LifeTechnologies) according to the manufacturer’s instructions, with 10ng of cDNA. Results were normalized to GAPDH expression. The primers used were ROBO1 forward 5’-GAC AAA ACC CTT CGG ATG TCA-3’, ROBO1 Reverse 5’-CCA GTG GAG AGC CAT CTT TCT-3’, SLIT2 forward 5’-AGC TTA GAC GAA TTG ACC TGA GC-3’, SLIT2 reverse 5’-CCG AAG GCA GTT TAT CTT GTT GG-3’, IL1α Forward, 5’-TGG TAG TAG CAA CCA ACG GGA-3’, IL1α reverse 5’-ACT TTG ATT GAG GGC GTC ATT C-3’, IL1β forward 5’-ATG ATG GCT TAT TAC AGT GGC AA-3’, IL1β reverse 5’-GTC GGA GAT TCG TAG CTG GA-3’, SDF-1 forward 5’-TCT GAG AGC TCG CTT GAG TG-3’, SDF-1 reverse 5’-GTG GAT CGC ATC TAT GCA TG-3’, HB-EGF forward 5’- GGG CAT GAC TAA TTC CCA CTG A-3’, HB-EGF reverse 5’-GCC CAA TCC TAG ACG GCA AC3’, EGF forward 5’-CAA CCA GTG GCT GGT GAG GA-3’, EGF reverse 5’-GAG CCC TTA CTG GAT ACT GGA A-3’, AREG forward 5’-GTG GTG CTG TCG CTC TTG ATA CTC-3’, AREG reverse 5’-TCA AAT CCA TCA GCA CTG TGG TC-3’, TGFα forward 5’-AGA TAG ACA GCA GCC AAC CCT GA-3’, TGFα reverse 5’-CTA GGG CCA TTC TGC CCA TC-3’, HGF forward 5’-CAA TAG CAT GTC AAG TGG AG-3’, HGF reverse 5’-CTG TGT TCG TGT GGT ATC AT-3’, ADAMTS-1 forward 5’-ACT GGA AGC ATA AGA AAG AAG CG-3’, ADAMTS-1 reverse 5’-AAT TCT GCC ATC GAC TGG TCT-3’, MMP2 forward 5’-TAC AGG ATC ATT GGC TA CACA CC-3’, MMP2 reverse 5’-GGT CAC ATC GCT CCA GAC T-3’, MMP9 forward 5’-TGT ACC GCT ATG GTT ACA CTC G-3’, MMP9 reverse 5’-GGC AGG GAC AGT TGC TTC T-3’, MMP14 forward 5’-GGC TAC AGC AAT ATG GCT ACC-3’, MMP14 reverse 5’-GAT GGC CGC TGA GAG TGA C-3’. Relative fold expression was calculated after normalization to GAPDH. Histograms represent fold expression ±standard error calculated from standard deviation of mean ΔCt. Statistical analysis was performed between ΔCt mean using unpaired Student’s t-test. Commercial QPCR arrays for Human Extracellular Matrix Adhesion Molecules were purchased from Applied Biosystems (Foster City, CA) and used according to the manufacturer’s instructions.
Antibodies
Antibodies against Vimentin, and αSMA were purchased from Sigma-Aldrich. N-cadherin, p44/42 MAPK, Phospho-p44/42 MAPK, EGFR antibodies were from Cell Signaling Technology (Danvers, MA); α-tubulin and ROBO1 from Abcam (Cambridge, MA). Fibronectin, Integrin αV, Integrin α2, Integrin α5, Integrin β1, Integrin β4 (BD Biosciences); MMP14 (Epitomics, Burlingam, CA); Ki67 Vector Laboratories (Burlingame, CA).
Zymography
Zymography was performed as previously described [16]. Briefly, conditioned medium was separated by SDS PAGE containing gelatin at 4°C washed and incubated overnight at 37°C in a development buffer (0.05M Tris-HCl pH 8.8, 5mM CaCl2, 0.02% NaN3). The gels were then stained with 0.1% Coomassie Blue R-250 for an hour and destained in methanol/acetic/acid/water solution (10%, 20%, 70% v:v:v). Images were taken on Gel Doc XR system (Bio-Rad, Hercules, CA).
Affymetrix data analysis
Esophageal organotypic cultures were laser dissected and purified total RNA was Biotin-labelled and hybridized onto Affymetrix GeneChip U133 Plus 2.0 oligonucleotide array, Accession No. GSE19472. Transfac enrichment analysis was done using Enrichr software http://amp.pharm.mssm.edu/Enrichr/ (Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool) with the position weight matrices (PWMs) from TRANSFAC [17] and JASPAR [18]. Gene ontologies analysis was conducted on WebGestalt http://bioinfo.vanderbilt.edu/webgestalt/ [19].
Biostatistical analysis
Biostatistical analysis was performed using Prism version 6.00 for Mac (GraphPad Software, La Jolla, CA). In vitro and in vivo experiments were analyzed using Student’s t-tests or one-way ANOVAs. Statistical significance was set at p<0.05. All experiments were done in triplicates with at least 3 biological replicates.
Results
Esophageal keratinocytes expressing dominant-negative forms of E-cadherin and TGFβRII show an inflammatory signature in OTC
We have previously shown that immortalized esophageal epithelial cells expressing dominant-negative E-cadherin and dominant-negative TGFβRII (ECdnT) were more invasive than esophageal keratinocytes expressing wild-type or mutant E-cadherin alone when grown in a model of organotypic culture (OTC) [12]. The observed invasion was shown to be fibroblast-dependent but could be induced with fibroblast-conditioned media suggesting a role for secreted cytokines and chemotactic factors. To identify a cytokine-induced gene signature, messenger RNA from epithelial cells in OTC was extracted by laser dissection and an expression profile was established using a gene expression array [20]. Comparison of gene expression in ECdnT cells with control E-cadherin-overexpressing cells (E) using enrichment analysis of potential transcription factors showed an enrichment of genes upregulated by NFκB (NFKB1 p-value: 0.00001246, z-Score: 1.65, combined score 9.79); notably we found upregulation of S100A7, S100A7A, IL8 and CD14 (Table 1). Similarly, gene ontology analysis, using WebGestalt [19], indicated enrichment in inflammatory and defense response pathways (p=0.0006, p=8.78e-05 respectively).
Table 1.
Affymetrix array analysis based on laser dissected epithelial cells from OTC
| List of upregulated genes ECdnT versus E (change fold >1.5 ,p<0.05) | ||||||
| S100A7 | S100A7A | SRGN | ASS1 | IL8 | ECM2 | NPL |
| PDZK1IP1 | SAA2 | GPNMB | ASNS | ALDH1L2 | DAPL1 | CD14 |
| CXCL1 | RSAD2 | KYNU | IFIT2 | CXCL10 | TRPM6 | LHX2 |
| TUBE1 | SYTL5 | GPNMB | CFB | SERPINA3 | NELL2 | GCH1 |
| CHI3L2 | SGPP2 | PCK2 | RASGRP1 | PSAT1 | ZBTB24 | RSAD2 |
| List of downregulated genes ECdnT versus E (change fold >1.5, p<0.05) | ||||||
| IL36RN | IFNK | AKT3 | KRT23 | IGFBP3 | SERPINE1 | NUAK1 |
| ABAT | PTHLH | GALNT5 | S1PR3 | EGLN3 | SPOCK1 | PMEPA1 |
| C11orf96 | LOXL2 | FST | GPR155 | CYP26B1 | SLITRK6 | COCH |
| TNS1 | MFAP5 | SLC2A14 | DIXDC1 | ENO2 | MFAP5 | CA9 |
| IGFBP3 | KRT24 | PLD5 | KANK4 | |||
To detect secreted proteins from both compartments, epithelium and fibroblasts, we analyzed conditioned medium (CM) using a cytokine array and identified a 1.5-fold increase of Angiogenin (ANG), BMP4, IL1α and IL1RN and several other inflammatory cytokines in CM from invasive ECdnT OTCs compared to non-invasive control cultures overexpressing E-cadherin (Table 2). To determine the origin of the increased chemokine expression, we analyzed mRNA expression in both, epithelial and fibroblast cells extracted from invasive ECdnT and non-invasive E OTC. Amongst the highest upregulated chemotactic factors we detected SDF-1 with a 4–fold increase in fibroblasts (Figure 1 A, stroma) and IL1α and TGFα with a 2-fold increase. HGF was increased by 2.5-fold in the epithelial compartment of ECdnT OTC (Figure 1A). These results highlight that invasion of ECdnT cells in OTC is associated with an inflammatory gene expression Signature.
Table 2.
Cytokines highly expressed in ECdnT OTC conditioned medium (in bold fold change>1.5)
| ANG | BDNF | CKβ 8–1 | BMP4 | CNTF | Fractalkine | GCP-2 |
| GNDF | i-309 | IL1RN | IL1α | IL1β | IL3 | LIGHT |
| MDC | MIG | MIP-3α | TNF-β | SCF |
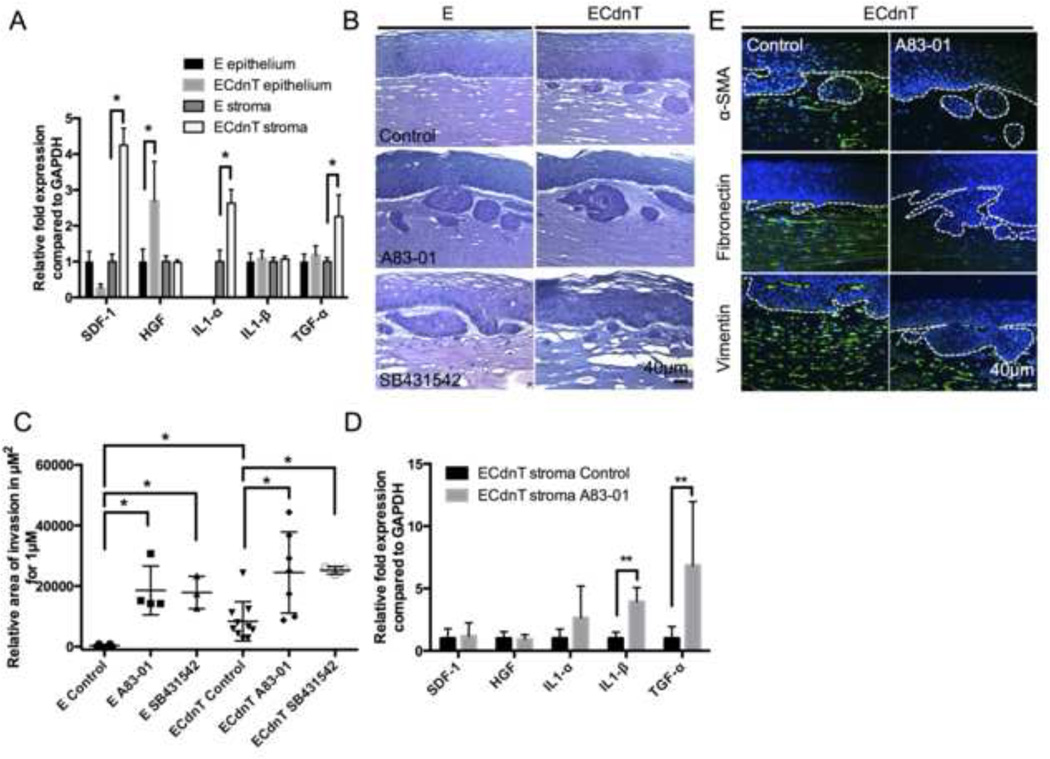
Figure 1. Loss of TGFβ promotes pro-inflammatory cytokines gene expression and collective invasion.
A: The expression of major chemotactic factors was measured in both epithelial cells and fibroblasts (stroma) extracted from organotypic culture (OTC) of control cells expressing wild-type E-cadherin (E) or cells expressing dominant-negative mutants for E-cadherin and TGFβRII (ECdnT). Values are obtained from three technical replicates. The relative expression was normalized to control E cells set at 1 and values were corrected using GAPDH expression. B: Representative pictures of OTC for E and ECdnT cells with no treatment (control), or with treatment of TGFβ signaling inhibitors A83–01 (1µM) or SB431542 (10µM) showing a pro-invasive effect of the inhibitors. C: Graphic representation of quantified invasion areas from OTC. Measurements were taken along 1mm length of OTC sections from the described conditions. Each mark is representative of a biological replicate. D: Expression of chemotactic factors expressed by fibroblasts (stroma) cultured in OTC with ECdnT cells from 4 independent experiments. Untreated ECdnT stroma (control) conditions are normalized to 1 and values are corrected using GAPDH expression. E: Inhibition of TGFβ signaling prevented stromal activation in ECdnT OTC as shown by representative immunofluorescence stainings for markers of stromal activation, α-SMA, fibronectin and vimentin (* p<0.05 as determined by a Student’s t test, data are represented by mean ± SD).
Chemical inhibition of TGFβ signaling advances invasion of esophageal keratinocytes
As we observed that the disruption of TGFβ signaling using dominant-negative mutant of TGFβRII together with functional loss of E-cadherin promotes cell invasion and the secretion of pro-inflammatory cytokines in esophageal keratinocytes, we set out to further explore the contributions by TGFβ.
TGFβ1 is a known regulator of epithelial proliferation and a modulator of the inflammatory response in tumor tissues. To better understand the effect of the crosstalk between epithelial cells and fibroblasts on epithelial cell invasion, we inhibited TGFβ signaling in OTC using A83–01 and SB431542 inhibitors of ALK5, 4 and 7 (TGFβRI, ACVR1B, ACVR1C). As the dominant-negative mutant TGFβRII only partially inhibits TGFβ signaling, we added these compounds to completely abolish signaling in ECdnT cells.
Inhibition of TGFβ1 signaling, using either compound not only enhanced collective cell invasion in ECdnT cells, but also induced cell invasion in control E cells (Figure 1B, C). The observed invasion was reminiscent of ESCC carcinoma in situ with highly differentiated keratin pearls. Analysis of cytokines expressed in the fibroblasts upon treatment with A83–01 in the ECdnT OTC showed an upregulation of IL1β and TGFα similarly to the differences observed between ECdnT and E cells (Figure 1D), suggesting an association with the invasive phenotype.
As we previously linked invasion to the activation of the stroma [13], we stained OTC sections for αSMA, a marker of myofibroblasts and carcinoma associated fibroblasts. αSMA was downregulated following A83–01 treatment. Similarly, the deposition of fibronectin into the matrix was reduced in the presence of A83–01, while the expression of vimentin in the fibroblasts was comparable between the different growth conditions (Figure 1E).
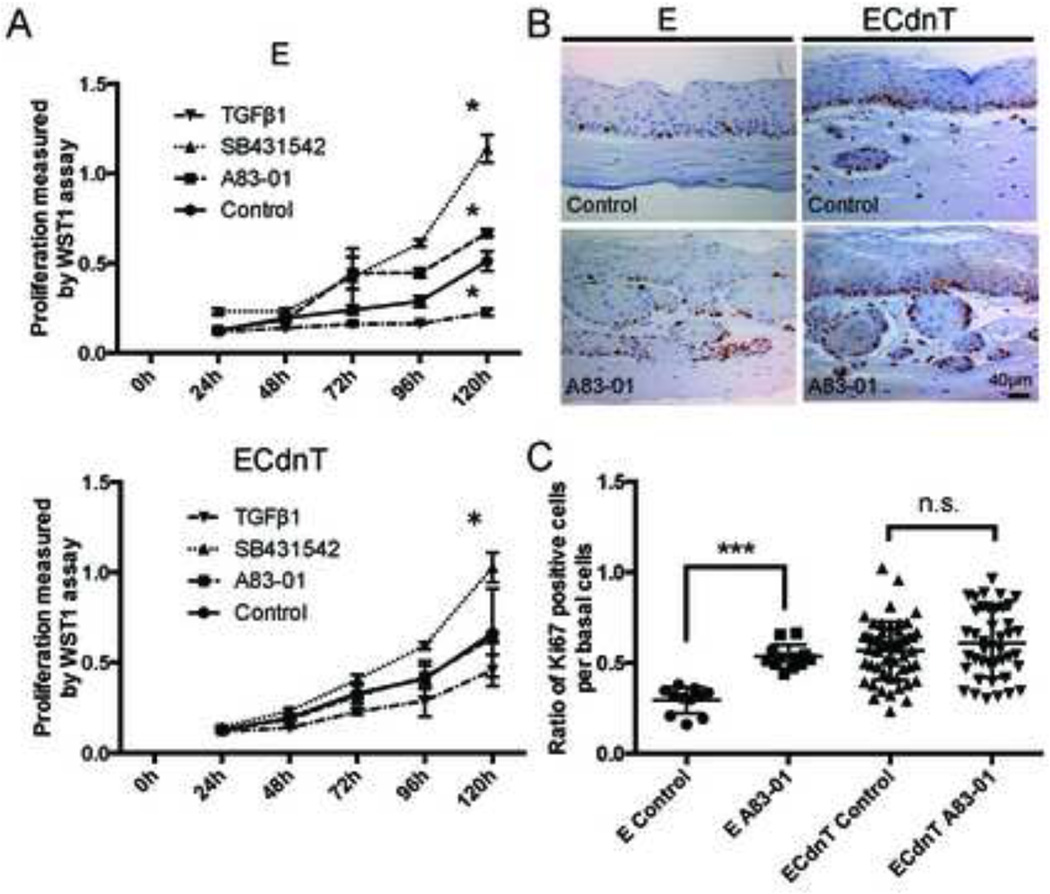
As TGFβ1 is known to have anti-proliferative function in normal epithelial cells, we wanted to determine the contributions of TGFβ1 to the regulation of proliferation in our model. We show that ECdnT cells grown as a monolayer on plastic were resistant to the growth inhibitory effect of TGFβ1 (Figure 2A). The addition of A83–01 had no effect on the proliferation of ECdnT. Similarly SB431542 induced only a slight increase of proliferation possibly indicating that inhibition of TGFβ signaling was not complete after expression of the dominant-negative TGFβRII in ECdnT cells. When grown in OTC, we confirmed that the inhibition of TGFβ1 signaling did not lead to a significant increase in proliferation in ECdnT cells, but induced higher proliferation of E cells (Figure 2B and C). Similarly, we determined the effects of A83–01 on apoptosis without any discernable differences (data not shown). We conclude that the increased epithelial cell invasion of ECdnT upon treatment with A83–01 is independent of their proliferative capacity.
Figure 2. Inhibition of TGFβ signaling promotes invasion in OTC independently of proliferation in ECdnT.
A: ECdnT cells are protected from the growth inhibitory effect of TGFβ1. Proliferation index was measured by WST1 assay using E and ECdnT cells grown on plastic without treatment (control), treated with TGFβ1 (5ng/mL) or TGFβ signaling inhibitors A83–01 (1µM) and SB431542 (10µM). Values are representative of 3 independent experiments. (*p<0.05, as determined by a Student’s t test, data are represented by mean ± SD). B: Ki67 staining of E and ECdnT cells in OTC comparing control condition and treatment with A83–01 (1µM) to determine proliferation. C: Quantification of Ki67-positive cells per basal cells visible in 100x field shows that A83–01 did not affect proliferation in ECdnT OTC. Values were obtained from three independent experiments (*** p<0.001 as determined by a Student’s t test, data are represented by mean ± SD).
ECdnT collective cell invasion is protease dependent
Tumor cell invasion is essentially driven by the ability of the epithelial cells to secrete proteases that allow degradation and invasion into the underlying matrix. It is also dependent on the capacity of the cells to establish interactions with matrix components and the generation of migratory forces.
Inhibition of matrix metalloproteases using the chemical compound GM6001 led to a complete suppression of invasion for cells untreated or treated with A83–01 (Figure 3 A and B). To determine if the pro-invasive effect of the TGFβ1 inhibitor A83–01 could be resulting from changes in the extracellular matrix, we modified the density of the OTC matrix by increasing the collagen concentration to 4.6 mg/mL and therefore consequently increasing the dependency on protease secretion to facilitate invasion. Using a higher concentrated collagen matrix suppressed the invasive potential of ECdnT cultures without treatment, however, addition of A83–01 could still induce epithelial cell invasion into the high collagen matrix (Figure 3C and D). Analyzing the proliferation index by counting Ki67-positive cells showed that the higher collagen concentration in the matrix had no effect on proliferation (Figure 3C). Altering the concentration of the collagen throughout the matrix affects not only epithelial cell invasion, but the stromal compartment as well. To analyze effects of the matrix on the epithelium and epithelial-fibroblast interaction without altering the matrix around the fibroblasts, we added a collagen I layer onto the matrix to separate the epithelial cells from the fibroblast-embedded matrix. The collagen I layer prevented invasion of ECdnT, but similarly the higher overall concentration did not inhibit invasion of culture treated with A83–01 suggesting a direct effect of epithelial-matrix and/or epithelial-stromal interaction on invasion. These data suggest a necessary role for fibroblast-secreted factors as well as protease activity in ECdnT (Figure 3C).
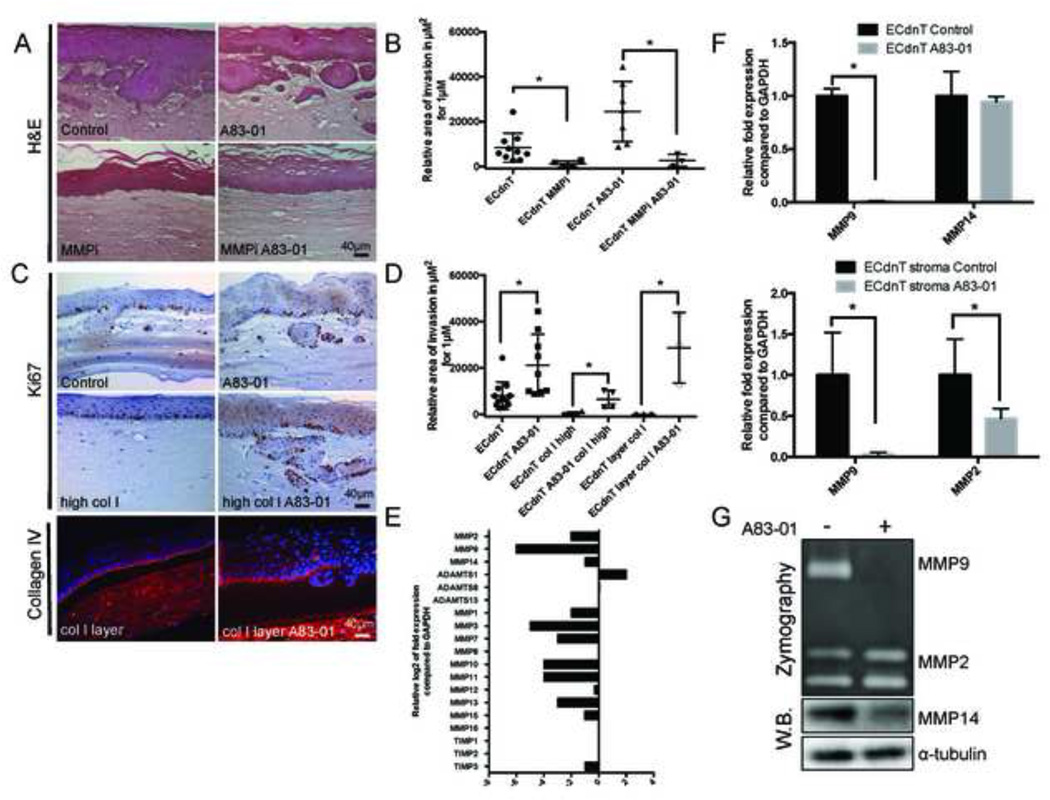
Figure 3. Collective invasion of ECdnT cells induced by A83–01 is dependent on MMP activity.
A: Treatment with the MMP inhibitor GM6001 illustrates the protease activity requirement for ECdnT cells invasion in OTC. Representative sections of OTC in control condition, after treatment with TGFβ signaling inhibitor A83–01 (1µM), pan-proteases inhibitor MMPi (1µM GM6001) or combination of both. B: Quantification and graphical representation of the invasion area under the described conditions along 1µm length of three OTC sections. C: Increased collagen concentrations (high col I) do not inhibit invasion of A83–01 treated ECdnT in OTC. Representative images of Ki67 immunohistochemistry staining of ECdnT OTCs comparing control and high collagen I concentration (high col I). Immunofluorescence staining for the basement membrane marker collagen IV in OTC with overlay of collagen I (col I layer) on top of the fibroblast matrix was performed. D: Quantification of the invasion area as described for (C) along 1mm length of OTC sections. E: Inhibition of TGFβ signaling leads to downregulation of proteases expression. Expression of proteases was measured by Human Extracellular Matrix Adhesion Molecules QPCR array in ECdnT cells extracted from OTC shown as the log2 ratio between A83–01 treated and control cells. F: Treatment with A83–01 induces inhibition of MMP9 expression. Relative expression of MMP9 and MMP14 in epithelial cells, and MMP9 and MMP2 from the respective fibroblasts isolated from ECdnT OTCs as measured by QPCR. Expression is normalized to untreated ECdnT OTC (control) and corrected using GAPDH expression G. Quantification of MMP9 and MMP2 secretion in OTC using conditioned medium as measured by zymography. MMP14 expression was analyzed by Western Blot of protein lysates from ECdnT cells extracted from OTC. (* p<0.05 as determined by a Student’s t test, data are represented by mean ± SD).
To determine if matrix metalloproteases were differentially expressed in the presence of TGFβ signaling inhibition, we measured the gene expression of matrix proteins and proteases associated with invasion in the epithelial cells by qPCR using an array for human extracellular matrix associated protein (Figure 3E).
Inhibition of TGFβ1 signaling was associated with a reduction in expression of many proteases; most associated with Smad regulation, with the notable exception of ADAMTS-1 (Figure 3E). We confirmed loss of MMP9 expression, a TGFβ target gene, in ECdnT cells and show that MMP14 expression, a membrane-bound MMP involved in matrix degradation by epithelial cells, was unaffected. Similarly, in fibroblasts, expression of MMP9 was downregulated whereas expression of MMP2 (Figure 3F), which can participate in the activation of MMP9 with MMP14, was unaffected. To analyze the secretion of matrix metalloproteases, we used OTC conditioned medium and show that MMP9 secretion is inhibited upon treatment with A83–01, whereas secretion of MMP14 in epithelial cells was only slightly affected (Figure 3G). Although the inhibition of invasion by GM6001 indicates the dependency of proteases activity for ECdnT invasion, we show that following treatment with A83–01 most metalloproteases examined were inhibited and therefore not likely to function in cell invasion downstream of TGFβ inhibition.
Analysis of protease expression showed an upregulation of ADAMTS-1 in cells treated with A83–01 (Figure 3E). We confirmed this result by qPCR (Figure 4A) and studied the effect of ADAMTS-1 expression in ECdnT cells to determine its involvement in promoting invasion of ECdnT cells through shRNA-mediated inhibition (Figure 4B, C). We generated ECdnT cell lines with suppression of ADAMTS-1 using 4 different shRNA clones targeting ADAMTS-1 and a non-silencing shRNA control. ECdnT expressing ADAMTS-1 shRNA were less invasive (Figure 4B, C).
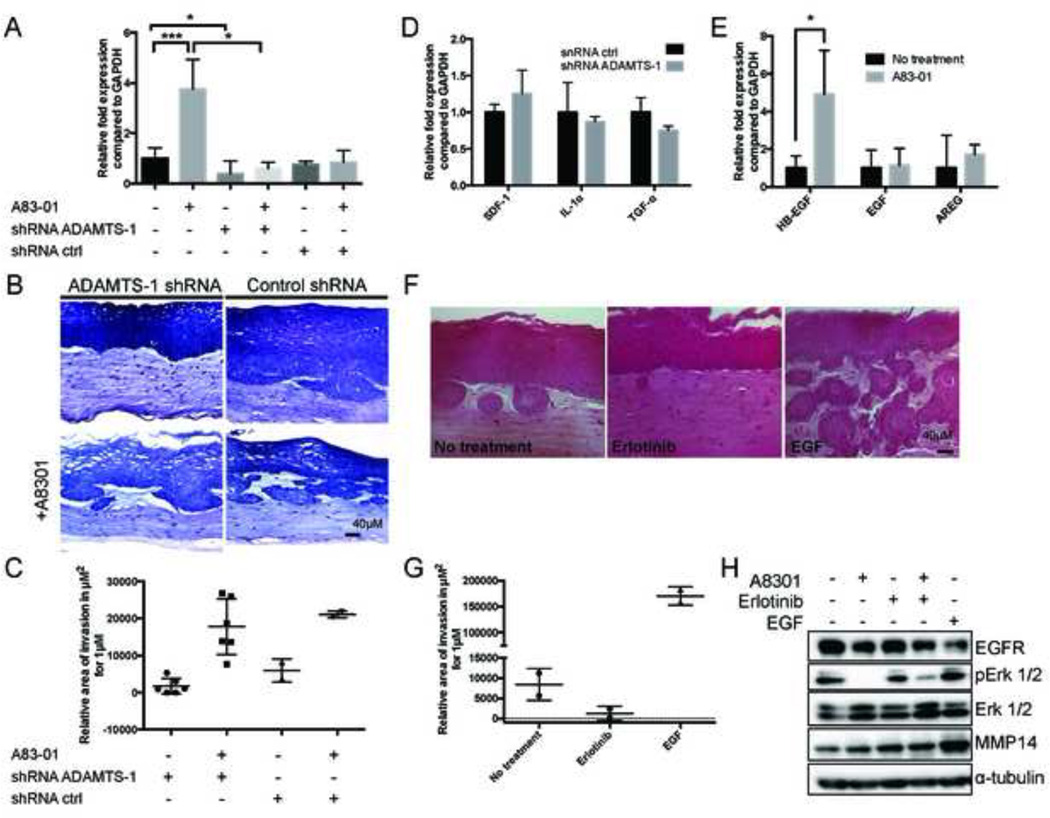
Figure 4. Inhibition of TGFβ signaling promotes EGFR ligand expression and activation in OTC.
A: ADAMTS-1 expression is induced after A83–01 treatment. Expression of ADAMTS-1 in ECdnT cells and cells with stable knockout of ADAMTS-1 (shRNA ADAMTS1) was measured by QPCR in samples from 3 independent experiments. Values are normalized to ECdnT control and corrected using GAPDH expression. B: Inhibition of ADAMTS-1 in ECdnT leads to reduce invasion. Representative pictures of ECdnT ADAMTS-1 shRNA in control conditions and after treatment with A83–01. C: Quantification of the relative invasion in ECdnT with shRNA ADAMTS-1 and control. Each mark represents an independent experiment. D: In the absence of ADAMTS-1 TGF-α expression is not increased in the fibroblasts. Relative expression of SDF-1, IL1α and TGF-α in fibroblasts extracted from non-invasive OTC after shRNA-mediated ADAMTS-1 knock-down was measured by QPCR. E: Inhibition of TGFβ signaling induces expression of the EGFR ligand HB-EGF in fibroblasts. Relative expression of the EGFR ligands HB-EGF, EGF and AREG in fibroblasts extracted from OTC was measured by QPCR. Values were normalized to no treatment condition and representative of 4 independent experiments F: EGF signaling induces invasion of ECdnT in OTC. Representative pictures of OTC after treatment with the EGF inhibitor Erlotinib (25nM) or EGF at (10ng/mL). G: Quantification of invasion after treatment with EGFR inhibitor or EGF. Invasive areas of OTC for EGF and Erlotinib were measured using four sections from two independent experiments. H: EGF signaling induces MMP14 expression. Representative Western Blot of pERK 1/2 and MMP14 expression after treatment with A83–01, Erlotinib or EGF. (* p<0.05 as determined by a Student’s t test, data are represented by mean ± SD).
Loss of TGFβ signaling promotes EGF-mediated invasion of ECdnT cells
ADAMTS-1 has been shown to activate HB-EGF and TGFα in cooperation with MMP1 [21]. Incidentally, as amplification of EGFR is a frequent occurrence in ESCC, we aimed to identify the role of EGF family members in epithelial cell invasion in this model. The unique opportunity provided by OTCs is to analyze the epithelial and stromal compartments. We already established that fibroblasts expressed more TGFα upon treatment with A83–01 (Figure 1D). Analyzing the expression of SDF-1, IL1α and TGF-α in the fibroblasts of non-invasive organotypic cultures following shRNA-mediated suppression of ADAMTS-1 revealed no significant induction of TGFα (Figure 4D). As EGF signaling is frequently upregulated in ESCC, we studied expression of others EGFR ligands in the fibroblasts. Analysis by qPCR showed an increase of HB-EGF (p<0.05), but not EGF or AREG (Figure 4E).
To elucidate the role of EGFR signaling in this context, we treated OTCs with the EGFR inhibitor, Erlotinib, and compared its effect with untreated ECdnT or EGF-stimulated OTCs as a control for EGFR-induced signaling. ECdnT OTCs grown in the presence of Erlotinib presented with suppressed invasion compared to untreated cells (Figure 4F, G). EGF was used a positive control and induced a strong increase of invasion (Figure 4F, G). Western Blot analysis of the epithelial cells isolated from OTC showed a reduction in EGFR signaling following A8301 treatment, as measured by phosphorylation of ERK1/2 and by reduced level of EGFR protein, which is expected after internalization and degradation following stimulation of the receptor. We also noted a strong increase in MMP14 expression after EGF treatment (Figure 4H).
Inhibition of TGFβ alters the integrin expression profile
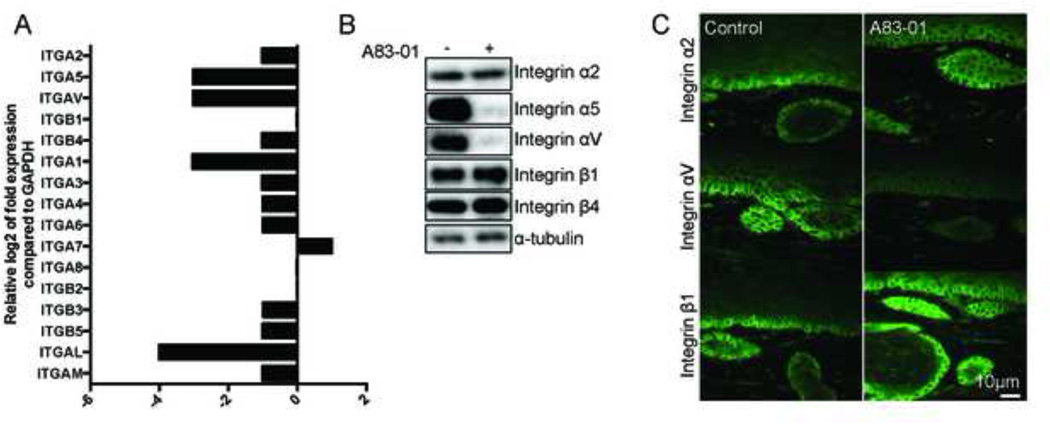
Tumor invasion is the result of matrix degradation and remodeling as migration occurs following cellular attachment to the matrix. Matrix attachment is mediated by expression of integrins that direct cellular cytoskeleton contraction and directional migration. Integrins have been shown to interact with and promote EGFR signaling [22,23]. The integrin subunit β1 is a central node in the regulation of collagen binding and an essential mediator of invasion in OTC [24], while integrin αV, and to a lesser extend α2, mediates binding to RGD peptides and fibronectin. They also have been shown to be upregulated in later stages of tumor progression [25]. Analysis of integrin expression by qPCR showed an overall downregulation of expression in particular of integrin αV, α5 and α1 following A83–01 treatment (Figure 5A). We confirmed protein expression of the integrins β1, αV, α5, and α2 on protein level by immunofluorescence and Western Blot (Figure 5B and C). While αV and α5 were downregulated, the integrin subunit β1 was unaffected by treatment with A83–01. Like Integrin β1, the β4 subunit is involved in laminin binding, while the α2 subunit also participates in collagen binding. The inhibition of integrins that are known targets of TGFβ signaling suggests a direct transcriptional repression effect of the inhibitor and indicates that these integrins are dispensable for epithelial cell invasion.
Figure 5. Inhibition of TGFβ signaling represses expression of integrins involved in fibronectin binding.
A: Inhibition of TGFβ signaling leads to the downregulation of integrin subunits involved in fibronectin binding. Expression of integrin subunits in ECdnT cells extracted from OTC measured by Human Extracellular Matrix Adhesion Molecules QPCR array. B: Inhibition of TGFβ signaling induces loss of integrin subunits α5 and αV that are involved in fibronectin binding. Representative Western blot of ECdnT cells extracted from OTC with or without A83–01 treatment. C: Representative pictures of immunofluorescence stainings with antibodies against integrin α2, αV and β1 of untreated (control) OTC or treated with A83–01 show loss of integrin αV.
The TGFβ signaling target ROBO1 is downregulated in the invasive epithelial compartment
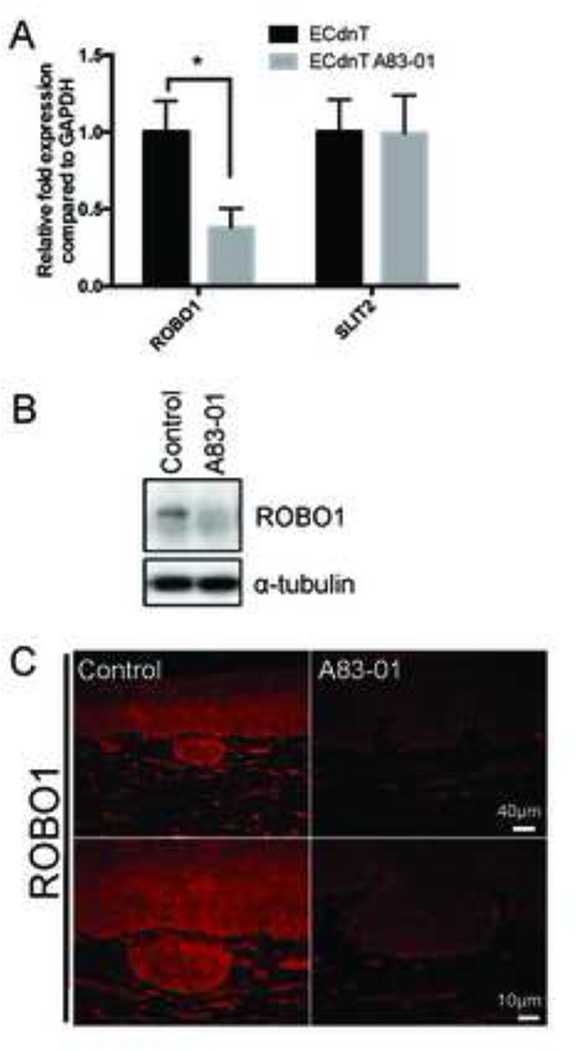
ROBO1 is a target of TGFβ signaling in breast epithelial cells and its expression is controlled by myofibroblasts during morphogenesis [26]. Interestingly, loss of chromosome 3p, which contains ROBO1 and TGFBR2 genes, is frequently observed in HNSCC [27]. Moreover, ROBO1, in concert with integrins, has been shown to be responsible for cardioblast polarization [28] and regulation of responsiveness to Slit- repellent signals [29]. We therefore aimed to determine the expression of ROBO1 in invasive ECdnT cultures after TGFβ inhibition. We show by qPCR that expression of ROBO1 is inhibited upon treatment with A83–01 (Figure 6A). We further confirmed loss of protein expression for ROBO1 after TGFβ inhibition by Western Blot and immunofluorescence (Figure 6B, C) suggesting a link to loss of chemorepulsion between epithelial cells and fibroblasts following loss of TGFβ signaling.
Figure 6. Loss of TGFβ signaling leads to ROBO1 downregulation in ECdnT.
A: ROBO1 expression is dependent on TGFβ signaling. Relative expression of ROBO1 and SLIT2 in ECdnT cells extracted from OTC was measured by QPCR. Values were normalized to ECdnT control and corrected with GAPDH expression. Values were obtained from 3 independent experiments B: Representative Western Blot for ROBO1 protein expression in ECdnT cells extracted from OTC. C: Representative immunofluorescence staining of ROBO1 in ECdnT OTC without treatment (control) or A83–01. (* p<0.05 as determined by a Student’s t test, data are represented by mean ± SD).
Discussion
In the present study we show that loss of TGFβ signaling leads to collective invasion of epithelial cells through a complex mechanism requiring EGFR signaling, changes in integrin expression and loss of ROBO1 with an associated change in inflammatory gene expression. The alteration of multiple regulatory pathways involved in cell invasion suggests that TGFβ signaling controls a complex array of genes involved in multiple aspects of invasion. Much of this complex regulation is mediated by epithelial-stromal crosstalk, suggesting that loss of the TGFβ signaling pathway in tumors has both cell autonomous and non-autonomous roles.
Role of TGFβ signaling in inflammation
TGFβ and NFkB signaling are both frequently altered in tumorigenesis. TGFβ signaling inhibition induces NFkB signaling in the gut and protects the colon in early stage colitis [30,31]. In HNSCC cell lines, reduced levels of TGFβ signaling are associated with activation of nuclear NFkB [32], while on the other hand, TGFβ1 can induce nuclear NFkB in later stages of HNSCC progression through the upregulation of TGFβ-activated kinase 1 (TAK1) [33]. We show that immortalized esophageal cell lines expressing dominant-negative forms of TGFβRII and E-cadherin display upregulation of inflammatory associated genes, notably overexpression of the pro-inflammatory cytokine IL8 [34–36] and S100A7/Psoriasin, both markers which are associated with ESCC and HNSCC progression [37–39]. Other pro-inflammatory cytokines enriched were IL1α and IL1β. IL1β inhibits TGFβ signaling pathway through upregulation of Smad7 [40,41], inhibition of Smad3 phosphorylation through IL1β binding to TAK1 [42], or in chondrocytes, induction of TGFβRII degradation [43]. In agreement with these results and observations made in mouse models, we show that fibroblasts grown with ECdnT display an increased expression of IL1α, SDF-1 and TGFα. Moreover, we show that inhibition of TGFβ signaling induces expression of IL1β in OTC fibroblasts, which is consistent with the observation that fibroblasts expressing a dominant-negative mutant of TGFβRII also produce more IL1β and stimulate invasion [44]. The role of TGFβ in promoting secretion of chemotactic factors is controversial as it has been shown to either induce expression of HGF [5], another potent inducer of squamous cell carcinoma invasion in OTC [45], or inhibit expression of HGF and SDF-1 [46,47]. We did not observe increased SDF-1 or HGF upon loss of TGFβ signaling in our experimental data. Interestingly, loss of TGFβ signaling through targeted deletion of SMAD4 in T cells resulted in the onset of head-and-neck SCC supporting that the imbalance of TGFβ signaling between epithelial cells and the stroma could be a key factor in the initiation of cancer [48].
Role of TGFβ signaling in matrix remodeling
Tumor cells are dependent on the expression of proteases to degrade the basement membrane allowing migration of epithelial cells through underlying stroma. Similarly, invasion in OTC is dependent upon three parameters: matrix stiffness, attachment to the matrix mainly through integrin β1 and expression of proteases [24]. Treatment with a pan-inhibitor of matrix metalloproteases, GM6001, confirms the protease-dependency of ECdnT cells in our system to invade into the underlying stroma.
A stiffer matrix is associated with tumor progression in breast cancer and promotes integrin binding and cell invasion [49,50]. Lysyl oxidase, which mediates collagen crosslinking, is also increased in the breast stroma of a PyMT-driven knock-out of TGFβRII [51]. Moreover the stiffer matrix is also susceptible to favor fibroblasts activation through Yap1 activation [52]. Increasing matrix stiffness in OTC shifts cell migration towards proteolytic invasion [53]. In our model, increasing matrix stiffness or laying a denser collagen I layer inhibited invasion of ECdnT, but did not prevent invasion induced by TGFβ inhibition, which was associated with a degradation of the de novo synthesized basement membrane.
Intriguingly, many MMPs known to digest the extracellular matrix to facilitate invasion were suppressed by TGFβ inhibition. Notably, MMP9, a known target of TGFβ signaling [54] and a protease we had previously identified as being potentially important in mediating ECdnT invasion through its interaction with CD44 and cathepsin B [16], was strongly inhibited in epithelial and fibroblasts cells. Although MMP9 has been implicated in in oral dysplasia and HNSCC in vivo, [55–58], our data suggest that other proteases are also critical regulators of invasion. MMP14 expression, which has previously been shown to be involved in invasion in organotypic culture [59], appeared slightly downregulated, although its protein level was unaffected after treatment with A83–01 suggesting that ECdnT maintained enough matrix degrading capabilities for them to invade. However, MMP function is not restricted to matrix degradation, but can lead to the release and activation of secreted factors involved in motility and invasion.
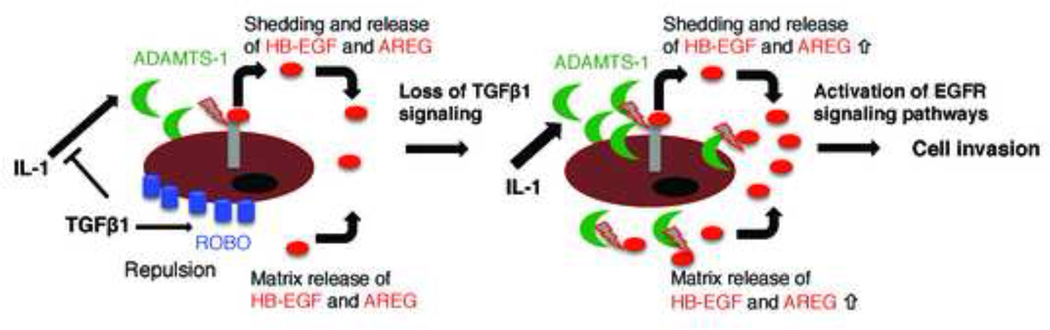
Our work has implicated an additionally protease, ADAMTS-1, involved in esophageal cell invasion. ADAMTS-1 cleaves the proteoglycans aggrecan and versican and has been shown to have a potential collagenase activity [60,61]. ADAMTS-1 also participates in stromal activation and has been shown to be upregulated by IL1β and downregulated by TGFβ in decidual stromal cells [62,63]. Notably, ADAMTS-1 has been shown to participate in the cleavage of HB-EGF and TGFα with MMP1 and to induce EGF signaling [21]. We found that inhibition of TGFβ signaling increased TGFα, HB-EGF and IL1β expression, suggesting that TGFβ signaling may regulate activation of EGFR signaling via ADAMTS-1 expression (Figure 7).
Figure 7. Proposed working model.
In the presence of TGFβ, cytokines such as IL1 that have been shown to upregulate ADAMTS-1 are inhibited and therefore little ADAMTS-1-dependent shedding and release of HB-EGF and AREG from the cell or matrix occurs. ROBO1 as a TGFβ target is expressed highly and functions in chemorepulsion suppressing cell invasion. Consequently, in the absence of TGFβ, ROBO1 expression is lost. At the same time, ADAMTS-1 expression is increased and the higher levels of EGFR ligands result in increased cell invasion.
Role of TGFβ signaling in EGFR regulation
Previous studies have illustrated a role of either TGFα [64] or EGF [65] in promoting invasion of epithelial cells in organotypic culture. Fibroblasts present in the underlying stroma are essential to promote invasion of epithelial cells in OTC models [66]. Increased expression of several EGF family ligands in the fibroblasts following treatment with A83–01 prompted us to determine the effect of EGF signaling on ECdnT invasion. Moreover, we had shown in a previous study that ECdnT cells exposed to conditioned medium obtained from organotypic culture demonstrated increased phosphorylation of ERK 1/2 signaling [12]. We show that after treatment of ECdnT with an inhibitor of EGFR, Erlotinib, invasion was inhibited. As a control, we treated the culture with EGF and observed a strong increase of invasion that was associated with higher expression of MMP14 and MMP2 secretion.
The role of TGFβ signaling in matrix attachment
TGFβ signaling has been described to regulate the expression of many integrins [67]. In turn, integrins themselves can regulate the activation of TGFβ1 ligands, promote expression of MMP9 [25,67], control matrix stiffness [68] and regulate EGFR signaling [22,23]. The expression of integrins can be under the regulation of fibroblasts [69]. Moreover, migration of epithelial cells is either inhibited or promoted depending on the nature of integrins expressed [70,71]. We show that inhibition of TGFβ signaling is associated with downregulation of integrins α5 and αv, both participating in fibronectin binding, which can promote migration in three-dimensional matrices [24].
While inhibition of TGFβ signaling can result in the upregulation of pro-invasive factors, it can also induce loss of chemorepellents. One such chemorepellent pathway is elicited by ROBO1, which has been shown to participate with SLIT2 in axon guidance and to control breast morphogenesis and tumor invasion [26,72]. ROBO1, a target of TGFβ signaling [26,73], is a receptor that together with integrins can regulate cardioblast polarization [28] and the responsiveness to SLIT repellent signals [29]. Moreover ROBO1 prevents the chemotactic effect of HGF and SDF-1 in epithelia cells [74,75]. We show that loss of TGFβ signaling inhibits the expression of ROBO1 and membrane localization, suggesting that the pro-invasive effect of loss of TGFβ signaling could be regulated by the loss of chemorepulsion in the absence of increased secretion of chemotactic factors (Figure 7).
In conclusion, we show that disruption of TGFβ signaling alters multiple pathways that all impinge upon cellular invasion. These pathways regulate multiple aspects of cell-matrix and cell-cell interactions including matrix and integrin rearrangements, possibly downstream of inflammatory cues, culminating in the secretion of EGFR ligands by stromal cells. These interactions promote both the cell autonomous and non-autonomous changes needed for invasion of epithelial cells into underlying tissues.
Highlights.
Chemical inhibition of TGFβ signaling advances collective invasion of esophageal keratinocytes
Collective cell invasion is protease dependent and ADAMTS-1 is upregulated
Inhibition of TGFβ signaling promotes EGF-mediated invasion
Loss of TGFβ signaling promotes collagen binding and induces alterations in integrin expression
The TGFβ target ROBO1 is downregulated in cell invasion implying mechanisms of chemorepulsion
Acknowledgements
CDA is supported by the National Institute of Health (DK094900 and DK091491). HAL is the recipient of NIH T32-CA009593 (Microenvironmental Influences in Cancer). We wish to thank the Vanderbilt Core facilities supported through the Digestive Disease Research Center (P30-DK058404) and the Vanderbilt Ingram Cancer Center Support grant (P30-CA068485. Further support for this study was provided through CTSA award No. UL1TR000445 from the National Center for Advancing Translational Sciences. We would also like to thank Dr. Anna Means for helpful discussions and editing. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- Ecad
E-cadherin
- EC
dominant negative E-cadherin
- MMP
matrix metalloprotease
- Ctrl
control
- TGFβRII
TGF beta-receptor II
- SCC
squamous cell carcinoma
- ESCC
esophageal squamous cell carcinoma
- HNSCC
head and neck squamous cell carcinoma
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest Statement:
The authors have no conflict of interest to declare.
References
- 1.Speight PM. Update on Oral Epithelial Dysplasia and Progression to Cancer. Head Neck Pathol. 2007;1:61–66. doi: 10.1007/s12105-007-0014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gannot G, Gannot I, Vered H, Buchner A, Keisari Y. Increase in immune cell infiltration with progression of oral epithelium from hyperkeratosis to dysplasia and carcinoma. Br. J. Cancer. 2002;86:1444–1448. doi: 10.1038/sj.bjc.6600282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabbert HE, Shimoda T, Hainaut P, Nakamura Y, Field JK, Inoue H. Pathol. Genet. Tumours Dig. Syst., rld Health Organization Classification of Tumours. Lyon: IARC Press; 2000. Squamous cell carcinoma of the oesophagus; pp. 31–36. [Google Scholar]
- 4.Massagué J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis MP, Lygoe KA, Nystrom ML, Anderson WP, Speight PM, Marshall JF, et al. Tumour-derived TGF-β1 modulates myofibroblast differentiation and promotes HGF/SF-dependent invasion of squamous carcinoma cells. Br. J. Cancer. 2004;90:822–832. doi: 10.1038/sj.bjc.6601611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy L, Hill CS. Alterations in components of the TGF-p superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006;17:41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Prime SS, Davies M, Pring M, Paterson IC. The Role of TGF-β in Epithelial Malignancy and its Relevance to the Pathogenesis of Oral Cancer (Part II) Crit. Rev. Oral Biol. Med. 2004;15:337–347. doi: 10.1177/154411130401500603. [DOI] [PubMed] [Google Scholar]
- 8.Bian Y, Hall B, Sun Z-J, Molinolo A, Chen W, Gutkind JS, et al. Loss of TGF-β signaling and PTEN promotes head and neck squamous cell carcinoma through cellular senescence evasion and cancer-related inflammation. Oncogene. 2012;31:3322–3332. doi: 10.1038/onc.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E. Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell. 2007;12:313–327. doi: 10.1016/j.ccr.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhowmick NA, Chytil A, Plieth D, Gorska AE, Dumont N, Shappell S, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 11.Cheng N, Bhowmick NA, Chytil A, Gorksa AE, Brown KA, Muraoka R, et al. Loss of TGF-beta type II receptor in fibroblasts promotes mammary carcinoma growth and invasion through upregulation of TGF-alpha-, MSP- and HGF-mediated signaling networks. Oncogene. 2005;24:5053–5068. doi: 10.1038/sj.onc.1208685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andl CD, McCowan KM, Allison GL, Rustgi AK. Cathepsin B is the driving force of esophageal cell invasion in a fibroblast-dependent manner. Neoplasia N. Y. N. 2010;12:485–498. doi: 10.1593/neo.10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andl CD, Fargnoli BB, Okawa T, Bowser M, Takaoka M, Nakagawa H, et al. Coordinated functions of E-cadherin and transforming growth factor beta receptor II in vitro and in vivo. Cancer Res. 2006;66:9878–9885. doi: 10.1158/0008-5472.CAN-05-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andl CD, Mizushima T, Nakagawa H, Oyama K, Harada H, Chruma K, et al. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J. Biol. Chem. 2003;278:1824–1830. doi: 10.1074/jbc.M209148200. [DOI] [PubMed] [Google Scholar]
- 15.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Bras GF, Allison GL, Richards NF, Ansari SS, Washington MK, Andl CD. CD44 upregulation in E-cadherin-negative esophageal cancers results in cell invasion. PloS One. 2011;6:e27063. doi: 10.1371/journal.pone.0027063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadaba R, Birke H, Wang J, Hooper S, Andl CD, Di Maggio F, et al. Imbalance of desmoplastic stromal cell numbers drives aggressive cancer processes. J. Pathol. 2013;230:107–117. doi: 10.1002/path.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu X, Wang Q, Hu G, Van Poznak C, Fleisher M, Reiss M, et al. ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev. 2009;23:1882–1894. doi: 10.1101/gad.1824809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivaska J, Heino J. Cooperation Between Integrins and Growth Factor Receptors in Signaling and Endocytosis. Annu. Rev. Cell Dev. Biol. 2011;27:291–320. doi: 10.1146/annurev-cellbio-092910-154017. [DOI] [PubMed] [Google Scholar]
- 23.Yu X, Miyamoto S, Mekada E. Integrin alpha 2 beta 1-dependent EGF receptor activation at cell-cell contact sites. J. Cell Sci. 2000;113:2139–2147. doi: 10.1242/jcs.113.12.2139. [DOI] [PubMed] [Google Scholar]
- 24.Zaman MH, Trapani LM, Sieminski AL, MacKellar D, Gong H, Kamm RD, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas GJ, Lewis MP, Hart IR, Marshall JF, Speight PM. αvβ6integrin promotes invasion of squamous carcinoma cells through up-regulation of matrix metalloproteinase-9. Int. J. Cancer. 2001;92:641–650. doi: 10.1002/1097-0215(20010601)92:5<641::aid-ijc1243>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 26.Macias H, Moran A, Samara Y, Moreno M, Compton JE, Harburg G, et al. SLIT/ROBO1 Signaling Suppresses Mammary Branching Morphogenesis by Limiting Basal Cell Number. Dev. Cell. 2011;20:827–840. doi: 10.1016/j.devcel.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh S, Ghosh A, Maiti GP, Alam N, Roy A, Roychoudhury S, et al. Alterations of ROBO1/DUTT1 and ROBO2 loci in early dysplastic lesions of head and neck: clinical and prognostic implications. Hum. Genet. 2009;125:189–198. doi: 10.1007/s00439-008-0610-9. [DOI] [PubMed] [Google Scholar]
- 28.Vanderploeg J, Vazquez Paz LL, MacMullin A, Jacobs JR. Integrins are required for cardioblast polarisation in Drosophila. BMC Dev. Biol. 2012;12:8. doi: 10.1186/1471-213X-12-8. doi:10.1186/1471-213X-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens A, Jacobs JR. Integrins regulate responsiveness to slit repellent signals. J. Neurosci. Off. J. Soc. Neurosci. 2002;22:4448–4455. doi: 10.1523/JNEUROSCI.22-11-04448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engle SJ, Ormsby I, Pawlowski S, Boivin GP, Croft J, Balish E, et al. Elimination of colon cancer in germ-free transforming growth factor beta 1-deficient mice. Cancer Res. 2002;62:6362–6366. [PubMed] [Google Scholar]
- 31.Monteleone G, Mann J, Monteleone I, Vavassori P, Bremner R, Fantini M, et al. A Failure of Transforming Growth Factor-β1 Negative Regulation Maintains Sustained NF-κB Activation in Gut Inflammation. J. Biol. Chem. 2004;279:3925–3932. doi: 10.1074/jbc.M303654200. [DOI] [PubMed] [Google Scholar]
- 32.Cohen J, Chen Z, Lu S-L, Yang XP, Arun P, Ehsanian R, et al. Attenuated Transforming Growth Factor β Signaling Promotes Nuclear Factor-κB Activation in Head and Neck Cancer. Cancer Res. 2009;69:3415–3424. doi: 10.1158/0008-5472.CAN-08-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freudlsperger C, Bian Y, Contag Wise S, Burnett J, Coupar J, Yang X, et al. TGF-β and NF-κB signal pathway cross-talk is mediated through TAK1 and SMAD7 in a subset of head and neck cancers. Oncogene. 2013;32:1549–1559. doi: 10.1038/onc.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Arbieva ZH, Guo S, Marucha PT, Mustoe TA, DiPietro LA. Positional differences in the wound transcriptome of skin and oral mucosa. BMC Genomics. 2010;11:471. doi: 10.1186/1471-2164-11-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knowles LM, Stabile LP, Egloff AM, Rothstein ME, Thomas SM, Gubish CT, et al. HGF and c-Met Participate in Paracrine Tumorigenic Pathways in Head and Neck Squamous Cell Cancer. Clin. Cancer Res. 2009;15:3740–3750. doi: 10.1158/1078-0432.CCR-08-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren Y, Cao B, Law S, Xie Y, Lee PY, Cheung L, et al. Hepatocyte Growth Factor Promotes Cancer Cell Migration and Angiogenic Factors Expression: A Prognostic Marker of Human Esophageal Squamous Cell Carcinomas. Clin. Cancer Res. 2005;11:6190–6197. doi: 10.1158/1078-0432.CCR-04-2553. [DOI] [PubMed] [Google Scholar]
- 37.Ji J, Zhao L, Wang X, Zhou C, Ding F, Su L, et al. Differential expression of S100 gene family in human esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2004;130:480–486. doi: 10.1007/s00432-004-0555-x. [DOI] [PubMed] [Google Scholar]
- 38.Kaur J, Matta A, Kak I, Srivastava G, Assi J, Leong I, et al. S100A7 overexpression is a predictive marker for high risk of malignant transformation in oral dysplasia. Int. J. Cancer J. Int. Cancer. 2014;134:1379–1388. doi: 10.1002/ijc.28473. [DOI] [PubMed] [Google Scholar]
- 39.Tripathi SC, Matta A, Kaur J, Grigull J, Chauhan SS, Thakar A, et al. Nuclear S100A7 Is Associated with Poor Prognosis in Head and Neck Cancer. PLoS ONE. 2010;5:e11939. doi: 10.1371/journal.pone.0011939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bitzer M, von Gersdorff G, Liang D, Dominguez-Rosales A, Beg AA, Rojkind M, et al. A mechanism of suppression of TGF-β/SMAD signaling by NF-κB/RelA. Genes Dev. 2000;14:187–197. [PMC free article] [PubMed] [Google Scholar]
- 41.Yan X, Liu Z, Chen Y. Regulation of TGF-β signaling by Smad7. Acta Biochim. Biophys. Sin. 2009;41:263–272. doi: 10.1093/abbs/gmp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benus GFJD, Wierenga ATJ, de Gorter DJJ, Schuringa JJ, van Bennekum AM, Drenth-Diephuis L, et al. Inhibition of the Transforming Growth Factor β (TGFβ) Pathway by Interleukin-1β Is Mediated through TGFβ-activated Kinase 1 Phosphorylation of SMAD3. Mol. Biol. Cell. 2005;16:3501–3510. doi: 10.1091/mbc.E04-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baugé C, Legendre F, Leclercq S, Elissalde JM, Pujol JP, Galéra P, et al. Interleukin-1beta impairment of transforming growth factor beta1 signaling by down-regulation of transforming growth factor beta receptor type II and up-regulation of Smad7 in human articular chondrocytes. Arthritis Rheum. 2007;56:3020–3032. doi: 10.1002/art.22840. [DOI] [PubMed] [Google Scholar]
- 44.Kiskowski MA, Jackson 2nd RS, Banerjee J, Li X, Kang M, Iturregui JM, et al. Role for stromal heterogeneity in prostate tumorigenesis. Cancer Res. 2011;71:3459–3470. doi: 10.1158/0008-5472.CAN-10-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grugan KD, Miller CG, Yao Y, Michaylira CZ, Ohashi S, Klein-Szanto AJ, et al. Fibroblast-secreted hepatocyte growth factor plays a functional role in esophageal squamous cell carcinoma invasion. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11026–11031. doi: 10.1073/pnas.0914295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daly AJ, McIlreavey L, Irwin CR. Regulation of HGF and SDF-1 expression by oral fibroblasts - Implications for invasion of oral cancer. Oral Oncol. 2008;44:646–651. doi: 10.1016/j.oraloncology.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 47.McKeown STW, Hyland PL, Locke M, Mackenzie IC, Irwin CR. Keratinocyte growth factor and scatter factor expression by regionally defined oral fibroblasts. Eur. J. Oral Sci. 2003;111:42–50. doi: 10.1034/j.1600-0722.2003.00002.x. [DOI] [PubMed] [Google Scholar]
- 48.Kim B-G, Li C, Qiao W, Mamura M, Kasprzak B, Kasperczak B, et al. Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature. 2006;441:1015–1019. doi: 10.1038/nature04846. [DOI] [PubMed] [Google Scholar]
- 49.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Provenzano PP, Inman DR, Eliceiri KW, Knittel JG, Yan L, Rueden CT, et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6:11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pickup MW, Laklai H, Acerbi I, Owens P, Gorska AE, Chytil A, et al. Stromally Derived Lysyl Oxidase Promotes Metastasis of Transforming Growth Factor-β-Deficient Mouse Mammary Carcinomas. Cancer Res. 2013;73:5336–5346. doi: 10.1158/0008-5472.CAN-13-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, et al. Mechano-transduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer associated fibroblasts. Nat. Cell Biol. 2013;15 doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ehrbar M, Sala A, Lienemann P, Ranga A, Mosiewicz K, Bittermann A, et al. Elucidating the role of matrix stiffness in 3D cell migration and remodeling. Biophys. J. 2011;100:284–293. doi: 10.1016/j.bpj.2010.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krstic J, Santibanez JF. Transforming Growth Factor-Beta and Matrix Metalloproteinases: Functional Interactions in Tumor Stroma-Infiltrating Myeloid Cells. Sci. World J. 2014;2014 doi: 10.1155/2014/521754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurahara S, Shinohara M, Ikebe T, Nakamura S, Beppu M, Hiraki A, et al. Expression of MMPS, MT-MMP and TIMPs in squamous cell carcinoma of the oral cavity: correlations with tumor invasion and metastasis. Head Neck. 1999;21:627–638. doi: 10.1002/(sici)1097-0347(199910)21:7<627::aid-hed7>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 56.Patel BP, Shah PM, Rawal UM, Desai AA, Shah SV, Rawal RM, et al. Activation of MMP-2 and MMP-9 in patients with oral squamous cell carcinoma. J. Surg. Oncol. 2005;90:81–88. doi: 10.1002/jso.20240. [DOI] [PubMed] [Google Scholar]
- 57.Rosenthal EL, Matrisian LM. Matrix metalloproteases in head and neck cancer. Head Neck. 2006;28:639–648. doi: 10.1002/hed.20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith J, Rattay T, McConkey C, Helliwell T, Mehanna H. Biomarkers in dysplasia of the oral cavity: a systematic review. Oral Oncol. 2009;45:647–653. doi: 10.1016/j.oraloncology.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 59.Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J. Cell Biol. 2009;185:11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Arao Tan I, Ricciardelli C, Russell DL. The metalloproteinase ADAMTS1: A comprehensive review of its role in tumorigenic and metastatic pathways. Int. J. Cancer. 2013;133:2263–2276. doi: 10.1002/ijc.28127. [DOI] [PubMed] [Google Scholar]
- 61.Lind T, Birch MA, McKie N. Purification of an insect derived recombinant human ADAMTS-1 reveals novel gelatin (type I collagen) degrading activities. Mol. Cell. Biochem. 2006;281:95–102. doi: 10.1007/s11010-006-0637-y. [DOI] [PubMed] [Google Scholar]
- 62.Ng YH, Zhu H, Pallen CJ, Leung PCK, MacCalman CD. Differential effects of interleukin-1beta transforming growth factor-beta1 on the expression of the inflammation-associated protein, ADAMTS-1, in human decidual stromal cells in vitro. Hum. Reprod. Oxf. Engl. 2006;21:1990–1999. doi: 10.1093/humrep/del108. [DOI] [PubMed] [Google Scholar]
- 63.Rocks N, Paulissen G, Quesada-Calvo F, Munaut C, Gonzalez M-LA, Gueders M, et al. ADAMTS-1 metalloproteinase promotes tumor development through the induction of a stromal reaction in vivo. Cancer Res. 2008;68:9541–9550. doi: 10.1158/0008-5472.CAN-08-0548. [DOI] [PubMed] [Google Scholar]
- 64.Turksen K, Choi Y, Fuchs E. Transforming growth factor alpha induces collagen degradation and cell migration in differentiating human epidermal raft cultures. Cell Regul. 1991;2:613–625. doi: 10.1091/mbc.2.8.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shiozaki H, Kadowaki T, Doki Y, Inoue M, Tamura S, Oka H, et al. Effect of epidermal growth factor on cadherin-mediated adhesion in a human oesophageal cancer cell line. Br. J. Cancer. 1995;71:250–258. doi: 10.1038/bjc.1995.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007;9:1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 67.Margadant C, Sonnenberg A. Integrin-TGF-? crosstalk in fibrosis cancer and wound healing. EMBO Rep. 2010;11:97–105. doi: 10.1038/embor.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mierke CT. The integrin alphav beta3 increases cellular stiffness and cytoskeletal remodeling dynamics to facilitate cancer cell invasion. New J. Phys. 2013;15:015003. [Google Scholar]
- 69.Kaur P, Carter WG. Integrin expression and differentiation in transformed human epidermal cells is regulated by fibroblasts. J. Cell Sci. 1992;103:755–763. doi: 10.1242/jcs.103.3.755. [DOI] [PubMed] [Google Scholar]
- 70.Jacquemet G, Green DM, Bridgewater RE, von Kriegsheim A, Humphries MJ, Norman JC, et al. RCP-driven a5p1 recycling suppresses Rac and promotes RhoA activity via the RacGAP1-IQGAP1 complex. J. Cell Biol. 2013;202:917–935. doi: 10.1083/jcb.201302041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim JP, Zhang K, Kramer RH, Schall TJ, Woodley DT. Integrin receptors and RGD sequences in human keratinocyte migration: unique anti-migratory function of alpha 3 beta 1 epiligrin receptor. J. Invest. Dermatol. 1992;98:764–770. doi: 10.1111/1523-1747.ep12499947. [DOI] [PubMed] [Google Scholar]
- 72.Chang P-H, Hwang-Verslues WW, Chang Y-C, Chen C-C, Hsiao M, Jeng Y-M, et al. Activation of Robo1 signaling of breast cancer cells by Slit2 from stromal fibroblast restrains tumorigenesis via blocking PI3K/Akt/?-catenin pathway. Cancer Res. 2012;72:4652–4661. doi: 10.1158/0008-5472.CAN-12-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Labbé E, Lock L, Letamendia A, Gorska AE, Gryfe R, Gallinger S, et al. Transcriptional Cooperation between the Transforming Growth Factor-β and Wnt Pathways in Mammary and Intestinal Tumorigenesis. Cancer Res. 2007;67:75–84. doi: 10.1158/0008-5472.CAN-06-2559. [DOI] [PubMed] [Google Scholar]
- 74.Marlow R, Strickland P, Lee JS, Wu X, PeBenito M, Binnewies M, et al. SLITs Suppress Tumor Growth In vivo by Silencing Sdf1/Cxcr4 within Breast Epithelium. Cancer Res. 2008;68:7819–7827. doi: 10.1158/0008-5472.CAN-08-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stella MC, Trusolino L, Comoglio PM. The Slit/Robo System Suppresses Hepatocyte Growth Factor-dependent Invasion and Morphogenesis. Mol. Biol. Cell. 2009;20:642–657. doi: 10.1091/mbc.E08-03-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]