Abstract
The development of clustered, regularly interspaced, short palindromic repeats (CRISPR)/CRISPR-associated (Cas) technologies promises a quantum leap in genome engineering of model organisms. However, CRISPR-mediated gene targeting reports in Drosophila melanogaster are still restricted to a few genes, use variable experimental conditions, and vary in efficiency, questioning the universal applicability of the method. Here, we developed an efficient two-step strategy to flexibly engineer the fly genome by combining CRISPR with recombinase-mediated cassette exchange (RMCE). In the first step, two sgRNAs, whose activity had been tested in cell culture, were co-injected together with a donor plasmid into transgenic Act5C-Cas9, Ligase4 mutant embryos and the homologous integration events were identified by eye fluorescence. In the second step, the eye marker was replaced with DNA sequences of choice using RMCE enabling flexible gene modification. We applied this strategy to engineer four different locations in the genome, including a gene on the fourth chromosome, at comparably high efficiencies. Our data suggest that any fly laboratory can engineer their favorite gene for a broad range of applications within approximately 3 months.
Keywords: Drosophila, CRISPR/Cas9, homologous recombination, RMCE, muscle
Reverse genetics is currently booming with the establishment of TALEN- and CRISPR-mediated genome engineering (Hsu et al. 2014; Sander and Joung 2014; Joung and Sander 2013). In particular, the CRISPR/Cas9 technology appears to efficiently and specifically introduce double strand DNA breaks in the genome of the organism, which can then be utilized to either introduce point mutations by error-prone nonhomologous end-joining (NHEJ) or integrate heterologous DNA into the chromosome using the homology-directed repair (HDR) pathway (Sander and Joung 2014). In Drosophila, CRISPR-induced NHEJ has mainly been utilized to mutate genes that result in a visible, easily scored phenotype, such as white eyes or yellow body color, or to mutate GFP transgenes (Gratz et al. 2013a; Sebo et al. 2014; Ren et al. 2013; Bassett et al. 2013). Mutants in genes with no visible phenotype required PCR screening for their identification; therefore, high mutagenesis rates are important, which might be difficult to achieve at all positions in the fly genome (Kondo and Ueda 2013; Yu et al. 2013, 2014; Gokcezade et al. 2014; Port et al. 2014). Recently, this bottleneck was addressed by applying CRISPR-induced HDR to insert an attP-site together with a visible marker into the gene of interest (Baena-Lopez et al. 2013; Gratz et al. 2014; Xue et al. 2014). In some cases the visible marker was flanked by FRT or loxP sites allowing its excision to only leave one attP site (and one loxP or FRT site) within the gene. This attP site enables the introduction of any given DNA sequence into the gene of interest (Baena-Lopez et al. 2013; Gratz et al. 2014; Xue et al. 2014). However, the efficiency of reporter integration was rather low (Baena-Lopez et al. 2013) and only determined at a single genomic locus (Gratz et al. 2014; Xue et al. 2014), leaving the general applicability to the Drosophila genome an open question. Port et al. (2014) reported an alternative CRISPR-mediated HDR strategy by using a transgenic single guide RNA (sgRNA) source and crossing it to a transgenic Cas9 source. This method also enabled targeting of somatic cells in a tissue-specific manner, but it required the generation of new transgenic sgRNA lines for every locus (Port et al. 2014).
Here, we have developed a highly flexible two-step genome engineering platform that combines CRISPR-mediated HDR with ΦC31 recombinase-mediated cassette exchange (RMCE). In the first step, CRISPR is applied to integrate a splice acceptor and an SV40 terminator together with a 3xP3-dsRed eye reporter. This enables both the efficient identification of the targeted event and the creation of a strong loss of function allele. In the second step, two flanking attP sites are utilized to replace the inserted DNA by any DNA of choice using RMCE, an established standard technology in Drosophila (Venken et al. 2011). Together, this allows flexible cassette exchange to freely manipulate the gene of interest. We successfully applied this method to four different locations in the genome and efficiently generated several allele variants, including a conditional allele, from a single targeting event. Our streamlined CRISPR/Cas9-based and RMCE-based strategies make it practical to flexibly engineer any Drosophila gene of choice for a broad range of applications within approximately 3 months.
Materials and Methods
Fly strains and genetics
All fly work, unless otherwise stated, was performed at 25° under standard conditions. The Lig4[169] null allele (McVey et al. 2004) was obtained from the Bloomington Drosophila Stock Center, and y[1], M(Act5c-Cas9, [w+]) in M(3xP3-RFP.attP)ZH-2A, w[1118] was a gift from Fillip Port and Simon Bullock before publication (Port et al. 2014). Both markers (w+ and 3xP3-RFP) were removed by crossing to heat-shock-Cre. The y[1], M(Act5C-Cas9)ZH-2A, w[1118] flies were recombined with Lig4[169] to obtain y[1], M(Act5C-Cas9)ZH-2A, w[1118], Lig4[169].
Cell culture
Drosophila Schneider 2 (S2) cells stably expressing myc-Cas9 from a ubiquitin promoter were a gift from Klaus Förstemann before publication (Böttcher et al. 2014). S2 cells were cultured in Schneider’s Drosophila medium supplemented with 10% fetal calf serum (Life Technologies) and penicillin/streptomycin (GE Healthcare). sgRNA activities were tested by transfecting 1 µg sgRNA per 24 wells into the myc-Cas9 cells using Fugene HD (Promega), followed by DNA extraction and a T7-Endonuclease I assay (see supplied protocol for details).
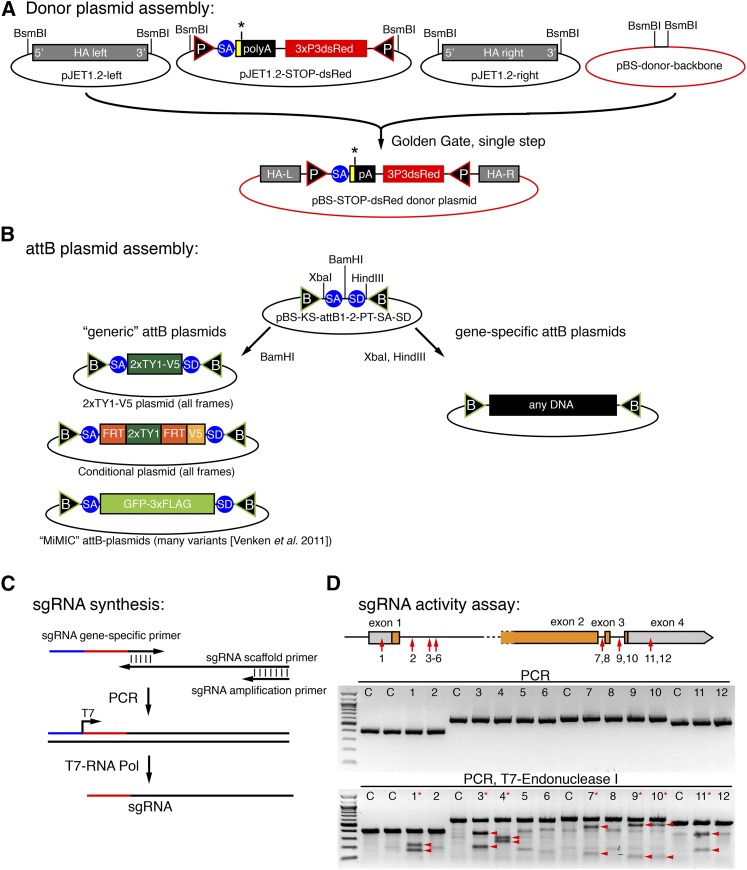
Plasmids
CC6-U6-gRNA_hsp70-Cas9 plasmid was a gift from Peter Duchek before publication (Gokcezade et al. 2014). pJET1.2-STOP-dsRed: attP1 and splicing acceptor (SA) were amplified with primers XZ82 and XZ83, SV40 terminator with XZ84 and XZ85, and attP2 with XZ88 and XZ89 from DNA extracted from a MiMIC fly line (Venken et al. 2011); 3xP3-dsRed was amplified from a fosmid fly line (Langer et al. 2010) with primers XZ86 and XZ87. These fragments were cloned into pLR-HD plasmid by Golden Gate cloning (Cermak et al. 2011). This assembled attP1-SA-STOP-SV40-3xP3-dsRed-attP2 cassette was amplified with primers XZ195 and XZ196 and blunt cloned into pJET1.2 to generate pJET1.2-STOP-dsRed. Because this STOP-dsRed cassette is flanked by two BsmBI sites, it can be easily assembled with both homology arms (Figure 3A): each homology arm of approximately 1 kb was amplified from genomic DNA of the target genotype with Phusion polymerase (NEB) and blunt-end cloned into pJET1.2 (CloneJET PCR Cloning Kit, Thermo Scientific). Primers used to amplify the homology arms have a 5′ BsmBI site enabling Golden Gate assembly with the STOP-dsRed cassette. All primers used are listed in Supporting Information, Table S1. pBS-donor-backbones pBS-GGAC-TTCT, pBS-GGAC-ATGC, and pBS-CGGA-GTGC were constructed by linearizing pBluescript with KpnI and SacII, followed by amplification with primer pairs XZ150 and XZ151, XZ156 and XZ151, and XZ161 and XZ162, respectively, and re-ligation. The generated pBS-donor-backbones harbor two BsmBI sites for donor plasmid assembly. pJET1.2-STOP-dsRed, pJET1.2-HA-left, pJET1.2-HA-right, and an appropriate pBS-backbone were assembled to the pBS-donor vector by Golden Gate cloning. attB plasmids FRT-2xTY1-FRT-V5 and 2xTY1-V5 fragments were synthesized as gBlocks (IDT) and cloned into the attB plasmid for all three reading frames. For construction of CC6-U6-gRNA_hsp70-Cas9-sgRNA1,3,4,7 and 9 the CC6-U6-gRNA_hsp70-Cas9 vector was cut with BbsI (NEB) and the annealed sgRNA targeting oligos were cloned into it. The vas-ΦC31(3xP3-EGFP.attB) plasmid was obtained from Johannes Bischof (Bischof et al. 2007). The attB site was removed by digestion with SpeI, followed by re-ligation.
Figure 3.
Cloning scheme and sgRNA activity tests. (A) Single-step Golden Gate assembly scheme of the STOP-dsRed donor vector cloned into a modified pBluescript backbone. (B) Scheme of the “generic” attB plasmids used in our study. A simple cloning step is sufficient to generate any gene-specific attB plasmid that can be used to replace an excised exon. (C) sgRNA synthesis scheme. (D) sgRNA activity assay of 12 different sgRNAs in S2 cells. PCR result with (bottom) or without (middle) T7-Endonuclease I treatment are shown. Digested products are marked by arrow heads and effective sgRNAs are marked by a red asterisk. C are controls without sgRNA.
All plasmids for embryo injections were purified with PureLink HiPure Plasmid Midiprep Kit (Life Technologies). Oligos are listed in Table S1.
sgRNA synthesis
The sgRNA dsDNA template was produced using overlap PCR with a small amount of a common sgRNA scaffold primer, a shorter sgRNA amplification primer, and a sgRNA gene-specific primer that includes the T7 promoter (Figure 3C) (Böttcher et al. 2014). All sgRNA primer sequences are listed in Table S1. The PCR product was cleaned by Qiagen MinElute kit (Qiagen). The sgRNAs were transcribed with T7-MEGAshortscript Kit (Life Technologies) and purified with MEGAclear Transcription Clean-Up Kit (Life Technologies).
Embryo injection
Preblastoderm embryos of the appropriate genotype were de-chorionated and injected with a FemtoJet apparatus (Eppendorf) using self-pulled glass needles (Harvard Apparatus) under standard conditions at room temperature. Injected embryos were kept for 2 d at 18° and the hatched larvae were collected and grown at 25°. For step 1 injections, pBS-donor plasmid, two sgRNAs, and (optionally) the CC6-U6-gRNA_hsp70-Cas9 plasmid were mixed and diluted in water. Lig4[169] embryos were injected with CC6-U6-gRNA_hsp70-Cas9 plasmid (100 ng/µl) and pBS-donor plasmid (500 ng/μl). y[1], M(Act5C-Cas9)ZH-2A, w[1118], Lig4[169] embryos were injected with both sgRNAs (60–70 ng/μl each) and pBS-donor plasmid (500 ng/μl). For step 2 injections, the attB plasmid (150 ng/μl) was mixed with vasa-ΦC31 plasmid (200 ng/μl).
Immunolabeling of IFMs:
Hemi-thoraces of adult Drosophila were prepared and stained as described (Weitkunat and Schnorrer 2014). Rabbit anti-Salm was used at 1:50 (Kühnlein et al. 1994), mouse anti-Flag (Sigma), mouse anti-V5 (Abcam), and rhodamine phalloidin (Invitrogen) were all used at 1:500. Nuclei were visualized by embedding in Vectashield plus DAPI (Vector Laboratories) and images were acquired on a Zeiss LSM780 confocal and processed with FIJI and Photoshop.
Detailed Drosophila genome engineering protocol by CRISPR-RMCE
-
CRISPR-sgRNA design and donor plasmid cloning ∼10 d
1.1 Verify sequence of the planned targeting regions for the sgRNAs in the fly strain used and in the S2 cells by sequencing to identify potential polymorphisms compared with the published sequence.
1.2 For designing the sgRNA targeting sites, choose one of the web tools (Beumer and Carroll 2014). We used an interface designed by the Zhang laboratory (http://crispr.mit.edu).
1.3 sgRNA production
1.3.1 Generate the dsDNA template for sgRNA in vitro transcription as described by Böttcher et al. (2014).
1.3.2 Transcribe sgRNA by T7-MEGAshortscript Kit (AM1354; Life Technologies). Use 150- to 250-ng template for a 20-μl reaction at 37° overnight.
1.3.3 Purify sgRNA by MEGAclear Transcription Clean-Up Kit (Life Technologies). Follow the manufacturer’s protocol and, in step 3, add an equal volume of 100% ethanol to the sample.
1.3.4 Check the sgRNA integrity on a gel and measure the concentration using a photometer (Nanodrop, Thermo Scientific). The expected yield is 50–100 µg, which is enough for the S2 cell assay and the fly injections.
1.4 sgRNA activity assay in S2 cells
1.4.1 Grow S2 cells in Schneider medium with 10% FCS (Life Technologies) to 5–10 × 106 cells/ml at 25°.
1.4.2 Dilute cells to 0.7 × 106/ml and plate 1 ml cells in S2 medium with 10% FCS per well in a 24-well plate for each transfection.
1.4.3 Prepare the transfection mix by diluting 1 μg sgRNA in 50 µl serum free medium and 4 µl Fugene HD mix plus 46 μl serum free medium, mix both, and incubate for 45 min at room temperature.
1.4.4 Add the Fugene/RNA mix to each well and mix gently by pipetting.
1.4.5 After 48–60 hr at 25°, harvest the cells and extract the genomic DNA by QIAamp DNA mini kit (Qiagen).
1.4.6 For the T7 Endonuclease I assay, amplify an approximately 500-bp fragment, which harbors the sgRNA targeting site with Phusion polymerase (NEB), and denature and anneal the PCR product as described by Zhang et al. (2014).
1.4.7 Mix on ice 10 µl annealed PCR product with 10 μl T7 Endonuclease I master mix [2 μl T7 endonuclease I buffer, 0.5 μl T7 endonuclease I (5 units, NEB) and 7.5 μl water].
1.4.8 Digest at 37° for 15–20 min using a PCR machine and load on 1.5% agarose gel immediately.
1.4.9 Estimate the efficiency of different sgRNAs by comparing the band intensities of the digested and nondigested bands.
1.5 Generation of donor plasmid (can be performed in parallel with steps 1.3 and 1.4 to save time)
1.5.1 Amplify left and right homology arms (approximately 1 kb, start as close to the sgRNA cutting site as possible) with Phusion polymerase (NEB) from the fly strain that is used for HDR and clone them into pJET-1.2 according to the CloneJET PCR Cloning Kit (Thermo Scientific).
1.5.2 Assemble the Golden Gate Cloning reaction [50 ng pBS-backbone, 80 ng pJET1.2-HA-left, 80 ng pJET1.2-HA-right, 80 ng pJET1.2-STOP-dsRed, 1.5 μl 10x T4 ligation buffer, 1 μl BsmBI (NEB, R0580), 1 μl T4 ligase (NEB, M0202) add water to 15 μl].
1.5.3 Ligate in PCR machine using the following cycles: 15 cycles of 37°, 5 min/16°, 10 min/37°, 15 min/50°, 5 min/80°, and 5 min/4°.
1.5.4 Assemble the Plasmid-safe nuclease reaction (15 μl ligation reaction 3 μl 10× Plasmid-safe buffer 1.2 μl 25 mM ATP 1 μl Plasmid-safe nuclease (Epicentre) 9.8 μl water).
1.5.5 Incubate at 37° for 60 min in PCR machine and transform 5–10 µl in bacteria. Most growing colonies will be correct.
-
Fly step 1 - CRISPR-mediated HDR ∼6 wk
2.1 Inject 600–800 Act5C-Cas9, lig4[169] embryos with pBS-donor (500 ng / μl) and two sgRNAs (each 60–70 ng/μl, targeting close to the chosen homology arms). Collect at least 50 fertile mosaic G0 flies.
2.2 Cross G0 flies individually (at least 50 vials) either to yw flies or appropriate balancer flies and screen all the F1 progeny for fluorescent red eyes using a fluorescent binocular (Leica MZ16-FA). Keep track of how many independent G0 founders lead to how many F1 carrier flies.
2.3 Generate stocks from an individual F1 carrier by crossing to balancer flies resulting in an isogenized stock for the engineered chromosome. Verify the targeting event by PCR and sequencing.
-
Fly step 2: ΦC31-mediated RMCE ∼6 wk
3.1 Inject a “generic” plasmid generated by Venken et al. (2011) or this study or your own custom-made gene-specific attB plasmid (150 ng/μl) mixed with vasa-ΦC31 plasmid (200 ng/μl) into approximately 200 embryos from an amplified stock generated at 2.3.
3.2 Cross G0 flies individually to an appropriate balancer and screen all F1 progeny for nonfluorescent eyes using a fluorescent binocular (Leica MZ16-FA).
3.3 Generate stocks from an individual F1 carrier by crossing to balancer flies, resulting in an isogenized stock for the engineered chromosome. Verify the correct orientation of the RMCE by PCR (will be ∼50%).
Results
Strategy overview
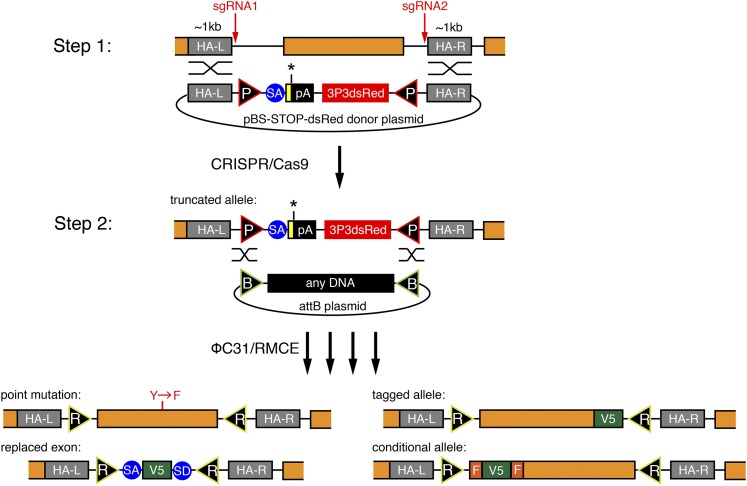
We aimed to develop a versatile and efficient strategy to modify the Drosophila genome that would allow various genome modifications such as the introduction of single point mutations, protein tags, exon deletions, or other desired changes in the gene of choice. Despite the suggested higher efficiency of CRISPR/Cas9-induced HDR as compared with Zn-finger–induced or TALEN-induced HDR, the identification of successfully targeted carrier flies is still a limiting step in the process. PCR-based screening or melting curve analysis methods require DNA extraction (Beumer et al. 2013a), which can be inconvenient for efficient stock generation. Therefore, we decided to develop a two-step strategy as illustrated in Figure 1, which enables efficient identification of the carrier flies and allows entirely flexible genome engineering. In the first step, we insert a 3xP3-dsRed marker enabling easy identification of the HDR event. A strong splice acceptor, followed by STOP codons and an SV40 polyA terminator, precedes the dsRed cassette. The inserted DNA is flanked by two attP sites in opposite orientations, a strategy that we adapted from the popular MiMIC system (Venken et al. 2011). If this cassette is inserted into an intron or replaces an endogenous exon, then it results in truncated mRNA of the targeted gene. Thus, step 1 can be used to create a loss-of-function allele (Figure 1).
Figure 1.
A two-step method to flexibly engineer the fly genome. Overview of the two-step procedure. In step 1, a donor vector consisting of two attP sites (P), a splice acceptor (SA), and STOP codons (yellow box, black asterisk) followed by an SV40 polyadenylation signal (pA) and a 3xP3dsRed marker are inserted on Cas9 cleavage with two sgRNAs. The orange coding exon is excised. In step 2, ΦC31-mediated RMCE inserts any DNA sequence between the two attB sites (B). Examples for various engineered exons are given, resulting in attR sites (R) in introns. F stands for FRT.
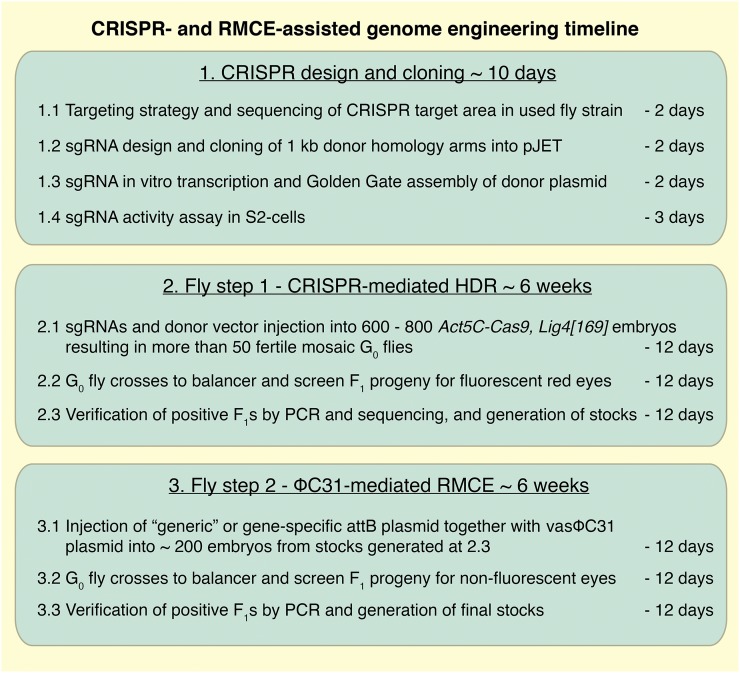
In the second step, RMCE is applied to replace the DNA between both attP sites by any DNA of choice, leaving a minimal scar of two attR sites, preferably in introns. RMCE has been used very efficiently in the MiMIC system, demonstrating that attR sites in introns generally do not interfere with gene function (Venken et al. 2011). Hence, our strategy enables the generation of various alleles, like a defined point mutation, a tagged allele, an exon replaced by a tag, or a conditional allele, from a single HDR carrier (Figure 1). This strategy should allow flexible editing of any Drosophila gene within approximately 3 months (Figure 2).
Figure 2.
Two-step genome engineering timeline. Schematic overview of the major steps of the genome engineering procedure. Details are provided in Materials and Methods.
CRISPR design and cloning
In step 1, we aimed to insert a STOP-3xP3-dsRed cassette flanked by two attP sites using a donor plasmid (Figure 1). Because the same strategy should be applicable to any gene, we established a single-step Golden Gate protocol to assemble the STOP-dsRed donor plasmid containing approximately 1-kb homology arms on each side, which has been shown to be of sufficient length for efficient HDR (Beumer et al. 2013b) and can be easily amplified by PCR. Cloning of the homology arms into the donor vector is thus very straightforward and takes only a few days for the gene of choice (Figure 3A, see Materials and Methods). This donor vector is the template for the HDR in step 1.
In step 2, RMCE exchanges the STOP-dsRed by any sequence located between two attB-sites in a provided donor plasmid (Figure 1). RMCE works very reliably and a large collection of plasmids to tag genes or insert reporters for various applications is available (Venken et al. 2011). These plasmids are fully compatible with our step 2 design. We have generated additional “generic” attB plasmids that can be used to tag any gene with a 2xTY1-V5 tag or to engineer a conditional allele using an FRT flanked 2xTY1 cassette followed by a V5 tag (Figure 3B). Flp-mediated deletion of the 2xTY1 cassette will lead to a frame shift and thus can be used to create loss-of-function clones at very high efficiency, as the flip-out will occur in cis (Hadjieconomou et al. 2011). The TY1 tag is a convenient affinity tag (Sarov et al. 2012). We have generated both constructs in all three reading frames.
CRISPR activity assay in cell culture
Many search algorithms exist to predict sgRNA target sequences for a given gene region (Beumer and Carroll 2014). However, to date there is no simple way of confirming if any of the predicted sgRNAs work efficiently. We developed such a selection assay to be able to only inject effective sgRNAs into fly embryos. We designed 12 different sgRNAs targeting different regions in the salm gene and synthesized the sgRNAs by a standard PCR and in vitro transcription reaction (Figure 3C). These sgRNAs were then individually transfected into Cas9 expressing S2 cells (Böttcher et al. 2014), and their cleavage efficiency was determined with a simple T7-Endonuclease I assay (see Materials and Methods) (Zhang et al. 2014). On average, approximately half of the tested sgRNAs work efficiently in this assay (Figure 3D), strongly suggesting that such a preselection test is useful to improve the in vivo success rates.
Step 1: HDR in Lig4 mutant embryos
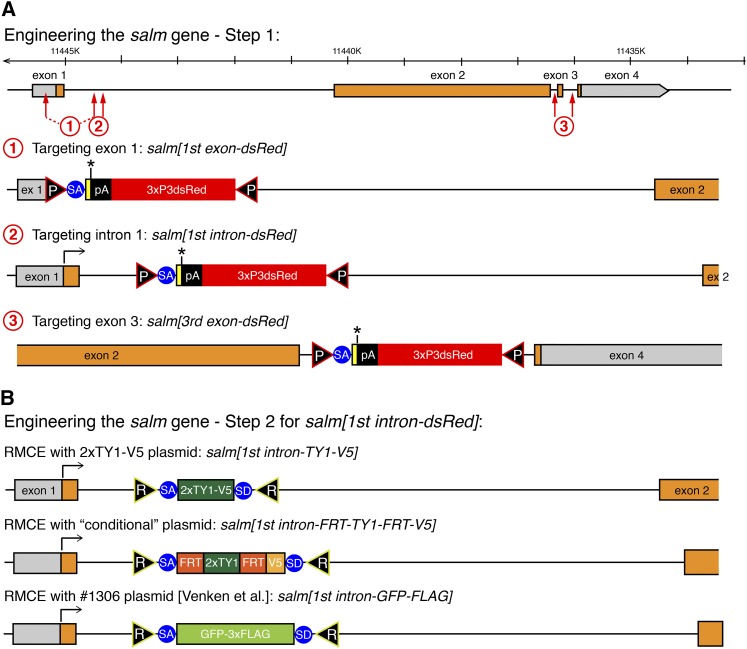
To test the efficiency of inserting our STOP-dsRed cassette, we designed three donor constructs targeting different regions in the salm gene (Figure 4A): the first, deleting parts of exon 1; the second, inserting the cassette into intron 1; and the third, deleting exon 3. For each construct approximately 1-kb homology arms were cloned in the STOP-dsRed donor vector. We injected the STOP-dsRed donor as circular plasmid together with two plasmids each containing a U6 promoter-driven sgRNA verified in S2 cells and a hsp70-Cas9 source (see Materials and Methods). We injected into Ligase4 mutant embryos, which were reported to exhibit a higher rate of HDR than wild-type embryos (Beumer et al. 2013a,b). We injected between 700 and 1500 embryos for each of the three constructs and were able to recover 11 red-eyed F1 carriers from two independent founders for the first intron construct and 72 red-eyed F1s from four independent founders for the third exon deletion construct (Table 1). This demonstrated that our strategy works in principle, but because we failed to recover the first exon deletion allele, we wanted to further improve the efficiency by using a different Cas9 source.
Figure 4.
Engineering of the salm gene. (A) Step 1 engineering of the salm gene. The genomic salm organization is depicted with coding exons in orange. The sgRNA targeting sites are indicated by red arrows and the resulting salm[1st exon-dsRed], [1st intron-dsRed] and [3rd exon-dsRed] alleles are shown. (B) Step 2 engineering of the salm gene. RMCE products of the salm[1st intron-dsRed] with three different exon cassettes are shown.
Table 1. Summary of the transformation efficiencies for the four different genomic locations modified in this study.
| Location | Injected Genotype | sgRNAs | Injected Embryos | Larvae | G0 Adults | Fertile G0 Adults | Independent G0 Founders | Total Red-Eyed F1 | Independent Founders Per Fertile G0s |
|---|---|---|---|---|---|---|---|---|---|
| salm[1st exon-dsRed] | Lig4−/− | sgRNA1 + sgRNA3 | 1500 | 396 | 166 | ND | 0 | 0 | ND |
| salm[1st intron-dsRed] | Lig4−/− | sgRNA3 | 700 | 171 | 149 | ND | 2 | 11 | ND |
| salm[3rd exon-dsRed] | Lig4−/− | sgRNA7 + sgRNA9 | 1500 | 312 | 150 | ND | 4 | 72 | ND |
| salm[1st exon-dsRed] | Act5C-Cas9, Lig4−/− | sgRNA1 + sgRNA3 | 700 | 124 | 48 | 30 | 1 | 13 | 3.3% |
| salm[1st intron-dsRed] | Act5C-Cas9, Lig4−/− | sgRNA3 + sgRNA4 | 700 | 200 | 122 | 64 | 9 | 54 | 14.1% |
| salm[3rd exon-dsRed] | Act5C-Cas9, Lig4−/− | sgRNA7 + sgRNA9 | 700 | 291 | 150 | 99 | 4 | 59 | 4.0% |
| bent | Act5C-Cas9, Lig4−/− | bent -sg1 + bent -sg3 | 700 | 204 | 118 | 56 | 1 | 5 | 1.8% |
Step 1: Transgenic Cas9 improves HDR efficiency
A number of transgenic Cas9 flies have been generated recently, and some of which have been used successfully (Ren et al. 2013; Xue et al. 2014; Gratz et al. 2014; Port et al. 2014). To test whether a transgenic Cas9 source is more efficient for HDR than a source from an injected plasmid, we targeted the same positions as above, but now using Act5C-Cas9, Lig4 flies. For this, we recombined an Act5C-Cas9 transgene expressing Cas9 ubiquitously, including maternally in the germline (Port et al. 2014), with the Lig4[169] null allele. Additionally, we removed the white and 3xP3dsRed markers from the Act5C-Cas9 transgene to obtain a Act5C-Cas9, Lig4[169] chromosome that is useful for the injection of our donor plasmids (see Materials and Methods). We injected 700 Act5C-Cas9, Lig4[169] embryos with two in vitro transcribed sgRNAs targeting either the first exon, the first intron, or the third exon of salm (using the same sgRNA target sequences as used above). We obtained one, nine, and four independent founders producing 13, 54, and 59 F1 carriers, respectively, demonstrating that all three locations were targeted successfully with frequencies between 3% and 14% per fertile G0 (Table 1). To verify that the targeted insertion occurred correctly, we tested a total of nine independent carriers from the three locations by PCR and sequencing. We were able to confirm that all of these targeted correctly by “ends-out” homologous recombination. We did not detect any “ends-in” insertions, which were reported to occur occasionally (Yu et al. 2014) (Figure S1 shows salm[1st intron-dsRed] as an example).
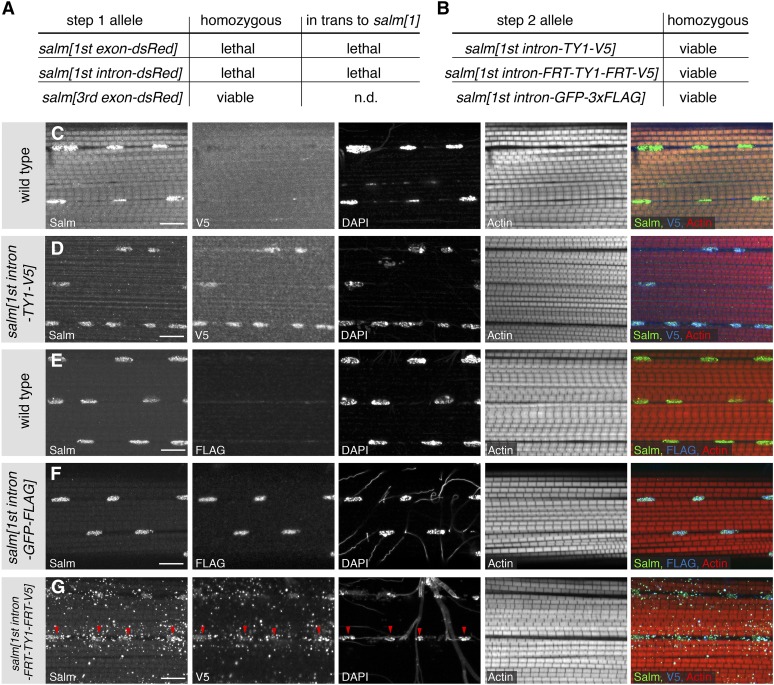
The first step of our gene-targeting strategy inserts a strong splice acceptor followed by a STOP cassette into the gene and thus should terminate transcription at this position. By design, the salm[1st exon-dsRed] allele additionally has a deleted ATG. As expected, the salm[1st exon-dsRed] allele is homozygous lethal, as well as lethal in trans to salm[1], demonstrating that we created a strong salm loss-of-function allele (Figure 5A). The salm[1st intron-dsRed] allele harbors an insertion in the first intron (Figure 4A). This allele is also homozygous lethal, and lethal in trans to salm[1], suggesting that the splice acceptor and STOP cassette are used efficiently to create a strong loss-of-function allele (Figure 5A). The salm[3rd exon-dsRed] allele only deletes the last 36 amino acids of the long SalmPA isoform, including 10 amino acids of the last zinc finger (Figure 4A). This allele is homozygous viable (Figure 5A). Taken together, these data suggest that our CRISPR-mediated step 1 strategy works efficiently to isolate targeted carrier flies at a practical frequency for routine use. Conveniently, these step 1 alleles are generally loss-of-function alleles if the insertion is located within the gene.
Figure 5.
Phenotypic analysis of the engineered salm alleles. (A) Lethality assay of the salm-dsRed alleles as homozygous or in trans to salm[1]. (B) All tagged salm[1st intron] alleles regain homozygous viability after step 2. (C–G) Localization of the tagged Salm proteins. Untagged Salm is located in the nucleus of wild-type IFMs (C, E), whereas V5 tagged Salm is only detected in the salm[1st intron-TY1-V5] and the salm[1st intron-FRT-TY1-FRT-V5] alleles (D, G). FLAG is found in the IFM nuclei of salm[1st intron-GFP-FLAG] adults (F). The Salm-FRT-TY1-FRT-V5 protein is found in the nuclei (red arrow heads) and also located in dots in the cytoplasm (G). Note the normal fibrillar morphology of the myofibrils in all the homozygous salm[1st intron] alleles (D, F, G). Actin was stained with phalloidin and the scale bars are 10 µm.
Step 2: Flexible gene editing by RMCE
A major benefit of our editing strategy is the flexible step 2 that enables the near-seamless insertion of any DNA sequence with only two remaining attR sites (Figure 1). To test the feasibility of step 2, we chose the salm[1st intron-dsRed] allele. We exchanged the STOP-dsRed cassette with a short 2xTY1-V5 exon, a FRT-2xTY1-FRT-V5 conditional exon, and a large GFP-3xFLAG exon from Venken et al. (2011) (Figure 4B). As expected, in all three cases the cassette exchange worked routinely and, typically, injection of approximately 200 embryos is sufficient to obtain two or more RMCE events in the correct orientation (see Materials and Methods). Importantly, the salm[1st intron-dsRed] lethality was reverted by RMCE in all three cases (Figure 5B). This demonstrates that our editing protocol generally does not result in any unwanted lethal mutations on the edited chromosome.
Salm protein is expressed in indirect flight muscles (IFMs) and is essential for fibrillar IFM fate specification (Schönbauer et al. 2011). Thus, we should detect the tagged Salm protein versions in the IFM nuclei of adult flies. Tagged protein from all three alleles, salm[1st intron-TY1-V5], salm[1st intron-FRT-TY1-FRT-V5], and salm[1st intron-GFP-FLAG] is expressed in IFMs. Salm-TY1-V5 and Salm-GFP-FLAG are readily detected in the IFM nuclei, and Salm-FRT-TY1-FRT-V5 shows an additional dotty pattern in the cytosol, which might be caused by the FRT sequence translated into protein (Figure 5C–G). The fibrillar IFM morphology is normal in all three homozygous salm alleles, showing that the tagged Salm proteins are indeed functional. Each IFM fiber contains several hundred nuclei. The conditional salm[1st intron-FRT-TY1-FRT-V5] should now enable a clonal loss-of-function analysis of salm in muscle only, as flip-out in cis is highly efficient (Hadjieconomou et al. 2011). Thus, this strategy should generally be versatile for the genetic analysis of muscle in the future.
Gene editing on the fourth chromosome
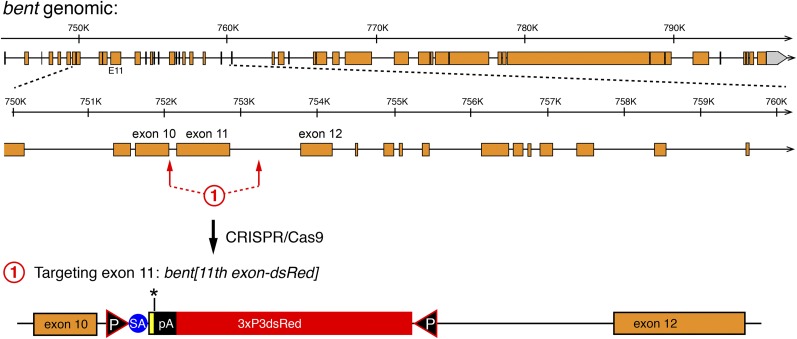
To demonstrate the general applicability of our gene editing strategy, we decided to apply it to an additional locus. We chose the bent gene, located on chromosome four, which is highly heterochromatic and thus difficult to manipulate by standard genetic tools. To our knowledge, there is only a single case reported in the literature that targeted a gene located on the fourth chromosome by classical ends-out mediated homologous recombination using long homology arms (Rodriguez-Jato et al. 2011). bent is a very large gene composed of at least 46 exons that are spread across more than 51 kb of genomic DNA (Figure 6). bent encodes for Projectin, a titin-like protein that is specifically expressed in muscles and essential for correct sarcomeric organization (Fyrberg et al. 1992; Ayme-Southgate et al. 1995; Schnorrer et al. 2010). It is supposedly silent in germ cells, in which the targeting event must happen. We chose to delete exon 11, an exon at the beginning of the PEVK domain of Projectin (Ayme-Southgate and Southgate 2006), using two sgRNAs flanking the exon. Both sgRNAs tested positively in the S2 cell assay (data not shown). We again used approximately 1 kb homology arms and injected the donor vector into 700 Act5C-Cas9, Lig4[169] embryos. We isolated five carriers from 1 founder out of a total of 56 fertile G0 flies, resulting in an HDR efficiency of 1.8%. We confirmed the bt[11th intron-dsRed] allele by sequencing of the locus. As expected, bt[11th intron-dsRed] is homozygous lethal and also lethal in trans to bt[I-b], a strong bent allele (Ayme-Southgate et al. 1995), again suggesting that the inserted splice acceptor is used effectively and transcription is prematurely terminated. Together, these results demonstrate that our CRISPR-mediated targeting strategy also works efficiently on the fourth chromosome, suggesting it can be generally applied to any locus of choice in the fly genome.
Figure 6.
Engineering of the bent gene. The entire genomic bent organization is shown at the top with a 10-kb zoom-in below. Coding exons are in orange. The sgRNA targeting sites flanking exon 11 are indicated by red arrows and the resulting bent[11th exon-dsRed] allele is shown at the bottom.
Discussion
CRISPR/Cas9 has been used successfully in many model organisms to generate mutants or to introduce targeted changes by HDR (Hsu et al. 2014). In Drosophila, there has been no general agreement regarding which strategy works most effectively to engineer the genome. To simply mutate a gene by CRISPR/Cas9-induced NHEJ, Cas9 was either injected as mRNA (Bassett et al. 2013; Yu et al. 2013), provided from an injected plasmid (Gratz et al. 2013a; Baena-Lopez et al. 2013), or provided from a transgenic source (Kondo and Ueda 2013; Ren et al. 2013; Sebo et al. 2014). Similarly, the sgRNA was either injected as in vitro transcribed sgRNA or provided by an injected plasmid or a transgenic source. A standard protocol has not yet emerged, although several genes have been mutated.
NHEJ can only induce small insertions or deletions. In contrast, HDR allows the defined engineering of a given gene and thus is suitable for a much wider range of applications. CRISPR/Cas9-mediated HDR has been used in Drosophila to insert short attP or tag sequences from single-strand oligonucleotides as donors (Gratz et al. 2013a) or larger cassettes including a dsRed marker cassette from a plasmid donor (Baena-Lopez et al. 2013; Yu et al. 2014; Gratz et al. 2014; Xue et al. 2014), again using various ways of injected or transgenic sources of sgRNAs or Cas9. The injected genotype was variable; sometimes Lig4 mutants were used (Baena-Lopez et al. 2013; Yu et al. 2014; Gratz et al. 2014; Xue et al. 2014), sometimes they were not used (Baena-Lopez et al. 2013; Yu et al. 2014; Gratz et al. 2014; Xue et al. 2014; Gokcezade et al. 2014; Port et al. 2014). Often the detection of the targeted event required laborious fly screening by PCR (Gratz et al. 2013b; Yu et al. 2014; Gokcezade et al. 2014).
Here we aimed to develop a universal and efficient CRISPR-based strategy that enables flexible genome engineering, including the insertion of large tags into the coding region of a gene or the generation of conditional alleles. This strategy should be generally applicable to most Drosophila genes. Our results confirmed that approximately 1-kb homology arms are of sufficient length to insert a large marker cassette, as has been suggested before for other loci (Gratz et al. 2014; Yu et al. 2014; Xue et al. 2014; Port et al. 2014). Thus, we could develop an efficient donor plasmid assembly protocol that facilitates cloning of the donor vector for any gene within a few days. Additionally, our data support the value of a quick pretesting strategy of predicted sgRNAs in S2 cells to eliminate inefficient sgRNAs, which would likely reduce targeting efficiency in vivo. However, we have not tested how well sgRNA efficiencies in S2 cells correlate with efficiencies in vivo. Conveniently, the same in vitro transcribed RNAs can be used for both S2 cell transfections and embryo injections. Our results suggest that a transgenic Cas9 source mediates HDR effectively in Ligase4 mutant germline cells. Although Act5C-Cas9 expression is not restricted to the germline, injections of the donor vector together with two verified sgRNAs led to a targeting efficiency of 2%–14% of fertile G0 flies for the incorporation of the large STOP-dsRed cassette, even for the bent locus on the heterochromatic fourth chromosome. This suggests that approximately 50–100 fertile G0 flies should be sufficient in most cases to identify positive carriers. We and others (Gokcezade et al. 2014) have observed relatively high developmental lethality of the injected G0 flies, which might be caused by somatic knock-out of the targeted gene. Thus, survival rate of the injected embryos and larvae might be increased when using a germline-restricted Cas9 source, such as nos-Cas9. However, nos-Cas9 was reported to be less efficient in germline transmission compared with Act5C-Cas9 (Port et al. 2014). We thus far have deleted up to approximately 1 kb of genomic sequence by HDR. Larger deletions would likely occur at reduced efficiencies; however, the dsRed marker should still make it practical to find them. The straightforward identification of carriers together with our simple cloning scheme should easily facilitate the insertion of the STOP-dsRed cassette into the gene of choice.
Recent reports using a transgenic sgRNA source (Port et al. 2014) or an injected sgRNA source (Ren et al. 2014) report very effective HDR rates with more than 50% of the fertile G0 flies being positive founders and, thus, screening by PCR-based methods were practical in these cases. However, both studies used only a single locus to insert the GFP tagging cassette; hence, a direct comparison with the efficiencies that we report here is difficult.
Our two-step strategy combines the advantages of both CRISPR and RMCE, thus allowing very flexible modifications of a particular gene region with minimal effort. Multiple fluorescent and affinity tags can be easily inserted or a deleted exon can effectively be replaced by various engineered exon versions. In principle, larger gene parts consisting of multiple exons can also be deleted and replaced by modified versions. This method is particularly valuable for genes that harbor complex transcriptional control and function in many tissues such as salm or for genes that are exceptionally large and exhibit complex alternative splicing patterns such as bent. The two-step strategy allows structure–function analysis at the endogenous locus without interfering with the regulatory regions included in introns, which cannot be achieved by simply inserting a cDNA at the transcriptional start site. The functionality of our method was verified by the reversion of the lethality for the step 2 alleles in the first intron of salm. This furthermore suggests that both steps do not generate additional unintended changes on the chromosome. Therefore, we hope that our strategy will promote the wide application of CRISPR-mediated HDR in Drosophila, making it a routine tool used in every fly laboratory like EMS mutagenesis or P-element–mediated transformation was in the past century.
Supplementary Material
Acknowledgments
We thank Klaus Förstemann for the Cas9 expressing S2 cells, Fillip Port and Simon Bullock for the Act5C-Cas9 flies, Peter Duchek for the CC6-U6-gRNA_hsp70-Cas9 plasmid, Johannes Bischof for the vasa-ΦC31 plasmid, and the Bloomington Drosophila Stock Center for fly strains. We are grateful to Reinhard Fässler for generous support and to Bettina Stender, Nicole Plewka, and Christiane Barz for excellent technical assistance. We thank Cornelia Schönbauer and Maria Spletter for critical comments regarding this manuscript. Our work was supported by the Max Planck Society, a Career Development Award from the Human Frontier Science Program (F.S.), the EMBO Young Investigator Program (F.S.), and the European Research Council under the European Union’s Seventh Framework Program (FP/2007-2013)/ERC Grant 310939.
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.013979/-/DC1
Communicating editor: B. J. Andrews
Literature Cited
- Ayme-Southgate A., Southgate R., 2006. Projectin, the elastic protein of the C-filaments, pp. 167–176 in Nature’s Versatile Engine: Insect Flight Muscle Inside and Out, edited by JVigoreaux . Landes Bioscience, Georgetown. [Google Scholar]
- Ayme-Southgate A., Southgate R., Saide J., Benian G. M., Pardue M. L., 1995. Both synchronous and asynchronous muscle isoforms of projectin (the Drosophila bent locus product) contain functional kinase domains. J. Cell Biol. 128: 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Lopez L. A., Alexandre C., Mitchell A., Pasakarnis L., Vincent J. P., 2013. Accelerated homologous recombination and subsequent genome modification in Drosophila. Development 140: 4818–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P., Liu J.-L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports 4: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K. J., Carroll D., 2014. Targeted genome engineering techniques in Drosophila. Methods 68: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer, K. J., J. K. Trautman, M. Christian, T. J. Dahlem, C. M. Lake et al., 2013a Comparing zinc finger nucleases and transcription activator-like effector nucleases for gene targeting in Drosophila. G3 (Bethesda) 3: 1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer, K. J., J. K. Trautman, K. Mukherjee, and D. Carroll, 2013b Donor DNA utilization during gene targeting with zinc-finger nucleases. G3 (Bethesda) 3: 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher R., Hollmann M., Merk K., Nitschko V., Obermaier C., et al. , 2014. Efficient chromosomal gene modification with CRISPR/cas9 and PCR-based homologous recombination donors in cultured Drosophila cells. Nucleic Acids Res. 42: e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., et al. , 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyrberg C. C., Labeit S., Bullard B., Leonard K., Fyrberg E., 1992. Drosophila projectin: relatedness to titin and twitchin and correlation with lethal(4) 102 CDa and bent-dominant mutants. Proc. Biol. Sci. 249: 33–40. [DOI] [PubMed] [Google Scholar]
- Gokcezade J., Sienski G., Duchek P., 2014. Efficient CRISPR/Cas9 plasmids for rapid and versatile genome editing in Drosophila. G3 (Bethesda) 4: 2279–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013a Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Wildonger J., Harrison M. M., O’Connor-Giles K. M., 2013b CRISPR/Cas9-mediated genome engineering and the promise of designer flies on demand. Fly (Austin) 7: 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., et al. , 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjieconomou D., Rotkopf S., Alexandre C., Bell D. M., Dickson B. J., et al. , 2011. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nat. Methods 8: 260–266. [DOI] [PubMed] [Google Scholar]
- Hsu P. D., Lander E. S., Zhang F., 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J. K., Sander J. D., 2013. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Ueda R., 2013. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195: 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnlein R. P., Frommer G., Friedrich M., Gonzalez-Gaitan M., Weber A., et al. , 1994. spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J. 13: 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer C. C. H., Ejsmont R. K., Schönbauer C., Schnorrer F., Tomancak P., 2010. In vivo RNAi rescue in Drosophila melanogaster with genomic transgenes from Drosophila pseudoobscura. PLoS ONE 5: e8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Radut D., Sekelsky J. J., 2004. End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics 168: 2067–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H.-M., Lee T., Bullock S. L., 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111: E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Sun J., Housden B. E., Hu Y., Roesel C., et al. , 2013. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. USA 110: 19012–19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, X., Z. Yang, D. Mao, Z. Chang, H.-H. Qiao et al., 2014 Performance of the Cas9 nickase system in Drosophila melanogaster. G3 (Bethesda). 4: 1955–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Jato S., Busturia A., Herr W., 2011. Drosophila melanogaster dHCF interacts with both PcG and TrxG epigenetic regulators. PLoS ONE 6: e27479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D., Joung J. K., 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M., Murray J. I., Schanze K., Pozniakovski A., Niu W., et al. , 2012. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer F., Schönbauer C., Langer C. C. H., Dietzl G., Novatchkova M., et al. , 2010. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature 464: 287–291. [DOI] [PubMed] [Google Scholar]
- Schönbauer C., Distler J., Jährling N., Radolf M., Dodt H.-U., et al. , 2011. Spalt mediates an evolutionarily conserved switch to fibrillar muscle fate in insects. Nature 479: 406–409. [DOI] [PubMed] [Google Scholar]
- Sebo Z. L., Lee H. B., Peng Y., Guo Y., 2014. A simplified and efficient germline-specific CRISPR/Cas9 system for Drosophila genomic engineering. Fly (Austin) 8: 52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., Schulze K. L., Haelterman N. A., Pan H., He Y., et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitkunat M., Schnorrer F., 2014. A guide to study Drosophila muscle biology. Methods 68: 2–14. [DOI] [PubMed] [Google Scholar]
- Xue, Z., M. Ren, M. Wu, J. Dai, Y. S. Rong et al., 2014 Efficient gene knock-out and knock-in with transgenic Cas9 in Drosophila. G3 (Bethesda) 4: 925–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Ren M., Wang Z., Zhang B., Rong Y. S., et al. , 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 195: 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Chen H., Liu J., Zhang H., Yan Y., et al. , 2014. Various applications of TALEN- and CRISPR/Cas9-mediated homologous recombination to modify the Drosophila genome. Biol. Open 3: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ferreira I. R. S., Schnorrer F., 2014. A simple TALEN-based protocol for efficient genome-editing in Drosophila. Methods 69: 32–37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.