Abstract
Hybrid sons between Drosophila melanogaster females and D. simulans males die as 3rd instar larvae. Two genes, D. melanogaster Hybrid male rescue (Hmr) on the X chromosome, and D. simulans Lethal hybrid rescue (Lhr) on chromosome II, interact to cause this lethality. Loss-of-function mutations in either gene suppress lethality, but several pieces of evidence suggest that additional factors are required for hybrid lethality. Here we screen the D. melanogaster autosomal genome by using the Bloomington Stock Center Deficiency kit to search for additional regions that can rescue hybrid male lethality. Our screen is designed to identify putative hybrid incompatibility (HI) genes similar to Hmr and Lhr which, when removed, are dominant suppressors of lethality. After screening 89% of the autosomal genome, we found no regions that rescue males to the adult stage. We did, however, identify several regions that rescue up to 13% of males to the pharate adult stage. This weak rescue suggests the presence of multiple minor-effect HI loci, but we were unable to map these loci to high resolution, presumably because weak rescue can be masked by genetic background effects. We attempted to test one candidate, the dosage compensation gene male specific lethal-3 (msl-3), by using RNA interference with short hairpin microRNA constructs targeted specifically against D. simulans msl-3 but failed to achieve knockdown, in part due to off-target effects. We conclude that the D. melanogaster autosomal genome likely does not contain additional major-effect HI loci. We also show that Hmr is insufficient to fully account for the lethality associated with the D. melanogaster X chromosome, suggesting that additional X-linked genes contribute to hybrid lethality.
Keywords: hybrid incompatibility, shmIRs, speciation
Speciation requires the evolution of reproductive isolating barriers that prevent the production of viable and fertile offspring between groups of individuals. Several isolating mechanisms maintain these barriers and are classified as either premating or postmating. Hybrid incompatibility (HI), the lethality and sterility of interspecific hybrid progeny, is an example of the latter. The Dobzhansky-Muller (D-M) model posits that HI is an indirect consequence of lineage-specific evolution and arises from negative epistatic interactions among alleles in the hybrid background (Maheshwari and Barbash 2011). The simplest form of the D-M model invokes two loci. For example, the ancestral genotype a1a1b1b1, where a1 and b1 are the ancestral alleles of genes a and b, diverges between two independently evolving lineages, whereby each lineage fixes a new allele, giving rise to two derived genotypes, a2a2b1b1 and a1a1b2b2. The incompatible interaction arises between the ‘a2’- and the ‘b2’-derived alleles in the hybrid. A fundamental question arising from this model is whether HI is caused by a simple interaction between 2 loci, as illustrated by this general example of a D-M interaction, or rather by complex multilocus interactions.
Matings between Drosophila melanogaster females and D. simulans males produce sterile hybrid females and invariantly lethal hybrid sons, which die as 3rd instar larvae (Barbash 2010b). Brideau et al. (2006) showed that hybrid lethality in this cross is due in part to the epistatic interaction between the genes Hybrid male rescue (Hmr), on the D. melanogaster X chromosome, and Lethal hybrid rescue (Lhr), on the D. simulans second chromosome, in a manner consistent with the D-M model (Brideau et al. 2006). Hmr and Lhr are characterized as major-effect HI genes because loss-of-function mutations in D. melanogaster Hmr or D. simulans Lhr suppress hybrid male lethality (Hutter and Ashburner 1987; Watanabe 1979). Both Hmr and Lhr are evolving rapidly due to positive selection in both the D. melanogaster and D. simulans lineages, suggesting functional divergence of the orthologs (Brideau et al. 2006; Maheshwari et al. 2008). Rescue by Lhr is asymmetric; only elimination of D. simulans Lhr rescues lethality to produce viable adult males, suggesting functional divergence of the Lhr coding sequence with respect to hybrid lethal activity (Brideau et al. 2006).
However, functional divergence of Lhr is more complex than originally proposed based on its asymmetry of rescue. Transgenic lines of D. melanogaster expressing either D. melanogaster or D. simulans Lhr transgenes were generated, and the hybrid lethal activity of each ortholog was assayed by testing for complementation (i.e., suppression) of the D. simulans Lhr1 hybrid rescue mutation (Maheshwari and Barbash 2012). Despite their extensive sequence divergence, both transgenes suppressed rescue, indicating that hybrid lethal activity is an ancestral function of Lhr (Maheshwari and Barbash 2012). Further experiments showed that D. melanogaster Lhr is expressed at a lower level in hybrids compared with D. simulans Lhr, suggesting that D. simulans Lhr may have greater hybrid lethal activity because it is expressed at a greater level in hybrids. Consistent with this interpretation, two D. melanogaster Lhr– deletions produced weak rescue to the pharate male stage (7–21% of total deficiency-carrying progeny) (Maheshwari and Barbash 2012).
Although Hmr and Lhr are major-effect hybrid lethality genes, additional factors likely contribute to lethality. Experiments performed by Muller and Pontecorvo nearly 70 years ago (1940, 1943) suggest that F1 hybrid male lethality involves interactions between loci on the D. melanogaster X (Hmr), the D. simulans 2nd chromosome (Lhr), and the D. simulans 3rd chromosome (Muller and Pontecorvo 1940; Pontecorvo 1943). More recently, Brideau et al. (2006) found that expression of D. simulans Lhr in a D. melanogaster background is not lethal, demonstrating that the interaction between D. melanogaster Hmr and D. simulans Lhr is insufficient to cause lethality (Brideau et al. 2006). Taken together, these studies strongly suggest that additional factors contribute to lethality.
Several screens of the D. melanogaster genome have searched for additional HI genes in D. melanogaster/D. simulans hybrids (Coyne et al. 1998; Presgraves 2003; Matute et al. 2010). Coyne et al. (1998) crossed D. melanogaster stocks containing deficiencies (deletions) to D. simulans males and assayed F1 hybrid female viability. This screen was designed to identify genes that cause lethality when hemizygous in a hybrid background. These regions could potentially contain genes that are haploinsufficient or recessive lethal in hybrids. The screen covered just less than 50% of the D. simulans genome and did not find any regions that caused unconditional lethality. Matute et al. (2010) repeated this screen with coverage increased to 79.4% of the genome and identified 10 regions that cause lethality when hemizygous in hybrid females (Matute et al. 2010). Presgraves (2003) performed a more sensitive screen by crossing D. melanogaster females with D. simulans males carrying the Lhr hybrid rescue mutation and assaying the viability of rescued F1 hybrid males (Presgraves 2003). This screen, unlike that of Coyne et al. (1998), can identify recessive-recessive interactions between the X chromosome and the autosomes. He screened ~70% of the D. simulans genome and found 40 nonoverlapping regions (20 lethal, 20 semilethal), that when hemizygous in hybrids, cause lethality in rescued males, concluding that recessive-recessive HI is the most common type of interaction. The genes mapped within these regions include two nucleoporins, Nup96 (Presgraves 2003) and Nup160 (Tang and Presgraves 2009).
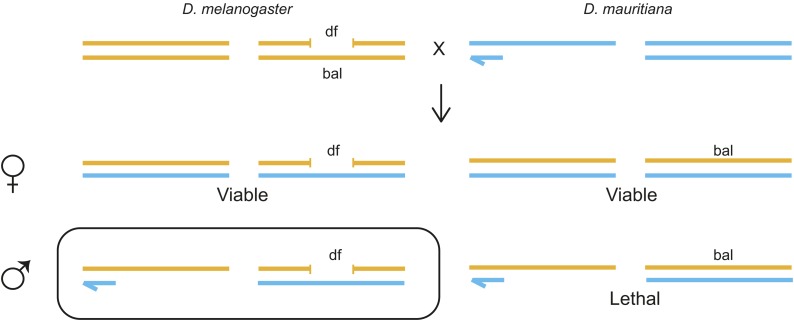
However, the genes identified in the aforementioned screens act recessively and are therefore not expected to affect F1 hybrid male viability. In contrast, both Hmr and Lhr are dominant and their presence causes lethality in the F1 generation. Here we use the Bloomington Deficiency Kit to systematically screen the vast majority of the D. melanogaster autosomal genome for genes with effects similar to Hmr and Lhr by crossing deficiency-carrying females to D. mauritiana males (Figure 1). Hmr mutations rescue hybrid males with all three sibling species of D. melanogaster but rescue best with D. mauritiana compared with D. simulans and D. sechellia (Hutter and Ashburner 1987). Because D. mauritiana and D. simulans are very closely related, having diverged within the past 250,000 years (Kliman et al. 2000; McDermott and Kliman 2008), we expect that most hybrid lethality genes are shared between the two species, but it is possible that lineage-specific HIs have evolved. For example, Nup96-dependent lethality is specific to D. simulans and D. sechellia (Barbash 2007). To further increase sensitivity, we screened for rescue to the pharate adult stage, because D. melanogaster deletions of Lhr rescue to this stage (Maheshwari and Barbash 2012). The difference in strength of rescue when deleting D. melanogaster vs. D. simulans Lhr appears to be due to a greater expression level of D. simulans Lhr in hybrids (Maheshwari and Barbash 2012). Based on these previous findings with Lhr, we suggest that our screen also has the potential to uncover genes where the D. mauritiana (or D. simulans) allele contributes more to hybrid lethality than the D. melanogaster allele.
Figure 1.
Screen design. D. melanogaster females from the Bloomington Deficiency Kit were crossed to D. mauritiana males. Female progeny inheriting either the deficiency chromosome (df) or the balancer chromosome (bal) are viable, though sterile. Males inheriting the balancer chromosome are invariably lethal. However, if the deficiency deletes a hybrid lethality gene we expect to observe rescue of this class of males (circled). We consider rescue to be survival to the pharate adult stage or beyond.
Materials and Methods
Fly stocks and crosses
D. melanogaster female flies from the Bloomington Deficiency Kit were crossed to at least two different lines of D. mauritiana (Figure 1). The D. mauritiana lines included w1f2, W139, and two different isofemale lines (105 and 207). Crosses were set up at 18° with ~20 females and ~25 males, flipped every 3−4 d for 2 wk, and progeny scored until the last fly eclosed. Pharates were then dissected to determine sex and, where possible, genotype. Rescue was calculated by dividing the number of rescued male pharates by the number of females carrying the deficiency chromosome. A D. melanogaster Lhr deletion stock (Df(2R)k08901, which we will refer to as Df(2R)Lhr–), was used as a positive control for pharate rescue (Maheshwari and Barbash 2012).
Genome coverage
The proportion of the genome covered by the screen was calculated based on either the known or estimated molecular breakpoints of the deficiencies. Approximately 40% of the deficiencies are not mapped molecularly. For these, we estimated cytological breakpoints using GBrowse on FlyBase and D. melanogaster Gene Models/Evidence (R5.48) to convert to molecular estimates. When there was uncertainty in the cytological location of a breakpoint, we took the average of the extremes of the described range. We then determined regions of overlap among deficiencies and counted each region (i.e., base pair) only once to arrive at the total number of base-pairs screened. This was done for each chromosome arm separately and then divided by the total number of base-pairs in the chromosome to determine the percentage of the chromosome arm covered.
RNAi construct design and knockdown
We used the short hairpin microRNA (shmiR) system of RNA interference (RNAi) with the Valium20 vector to attempt to knock down gene expression in hybrids (Haley et al. 2008; Ni et al. 2011). We initially designed 21-bp siRNAs, either with a single mismatch at position 2 (thought to ensure that only the antisense siRNA is loaded into the slicer complex), or without any mismatches as in (Ni et al. 2011) (Table 1). After three constructs failed to knockdown D. simulans msl-3 (sim-msl-3), we made several design modifications. First, we increased the small interfering RNA (siRNA) length to 22 bp because most short hairpin RNAs (shRNAs) made from three different shRNA constructs expressed in S2 cells were 22 bp in length (Ni et al. 2011), and most microRNAs from the endogenous miR1 locus are also 22 bp (Ruby et al. 2007). Additionally, 22 bp siRNAs were found to have better silencing than shorter siRNAs (Wu et al. 2011). Second, mismatches between the guide and passenger strands were included at positions 2 and 11 (Haley et al. 2008) to mimic the endogenous structure of miR1. Note, however, that sequencing data suggest that mismatches are not necessary to achieve preferential accumulation of the guide vs. passenger strands (Ni et al. 2011). A, C and G were mismatched with the same nucleotide; U was mismatched with C (Wu et al. 2011).
Table 1. ShmIR constructs and summary of results.
| Construct | Length | Mismatches | AS Sequence | Knockdown Assayed by RT-PCR?; Result |
|---|---|---|---|---|
| sim-Lhr-shRNA-577 | 21 | Mismatch at bp 2 | TAGATTCATTGCTAACACCAT | Yes; knockdown |
| sim-msl-3-shRNA-579 | 21 | Mismatch at bp 2 | TATTGTGATAGAAGGTCTCGG | Yes; no knockdown |
| sim-msl-3-shRNA-599 | 21 | None | TAACATAGTTCTCCCTGTCGA | N/A; lethal to both sexes in D. melanogaster |
| sim-msl-3-shRNA-600 | 21 | None | TAGTACCTTGACCATATTCCG | Yes; lethal to D. melanogaster males |
| sim-msl-3-shRNA-601 | 21 | None | TAGCGCCGTCATCACTTGCAG | Yes; no knockdown |
| sim-msl-3-shRNA-633 | 22 | Mismatches at bp 2 and 11 | TGAATGGGACCAAGTTAGTCAC | N/A; lethal to both sexes in D. melanogaster |
| sim-msl-3-shRNA-634 | 22 | Mismatches at bp 2 and 11 | TCTCCCGTGTGGAGTGGATCCA | Yes; no knockdown. |
| sim-msl-3-shRNA-635 | 22 | Mismatches at bp 2 and 11 | TCGCACATGGGCATCGACCGAT | N/A; semilethal to both sexes in D. melanogaster |
ShmIR, short hairpin microRNA; AS, anti-sense; RT-PCR, reverse-transcription polymerase chain reaction; N/A, not applicable.
Reverse-transcription polymerase chain reaction (RT-PCR):
cDNA synthesis was performed as in Maheshwari and Barbash (2012). RT-PCR for Lhr used the primers GTAGCTTTCTCTTGGCGCTCTT and GTAAGTGAACTGAAGCTGCGTTGG, which span a fixed indel between D. melanogaster and D. simulans Lhr, amplifying products of 278 bp and 326 bp from D. melanogaster and D. simulans, respectively. RT-PCR for D. melanogaster msl-3 (mel-msl-3) used the primers AGGAAAAACCCCCGTCCGGA and GCGTGCTGTTTGCCTAGTACCTT. RT-PCR for sim-msl-3 used the primers AGGAGAAACCCCCGCCACCC and GCGTGCTGTTTGCCTAGTACCTT.
Molecular breakpoint determination
Df(3L)BSC27 (6867) was outcrossed to a sequenced wild-type DGRP strain (NC486). Df(3L)BSC33 (6964) is balanced over TM2, and we did not outcross this stock to NC486 because Ubx was not sufficiently expressed to accurately genotype the progeny. DNA was isolated from 3-day-old females (n = 10) using the QIAGEN DNeasy Blood and Tissue Kit according to the supplemental insect protocol. The Epigenomics Core Facility at Weill Cornell Medical Center constructed the libraries and performed paired-end sequencing.
Raw reads (50 bp) were aligned separately to the reference genome (Dmel r5) using the default settings of BWA 0.6.2. Three different methods were used to analyze the sequences: split-read mapping, paired-end mapping, and depth of coverage. The split-read mapping was performed by Pindel (Ye et al. 2009), which identifies paired reads in which only one read maps to the reference genome. This mapped read serves as an anchor point for the detection of the other read. Depending upon the value of the expected sizes of copy number variants (CNVs), Pindel then splits the unmapped read into two or three fragments and maps them separately. Paired-end analysis was performed using Hydra and Delly (Quinlan et al. 2010; Rausch et al. 2012), which identify read-pairs that do not map to the reference genome with the expected size and orientation and then clusters them based on which CNV they support. This approach successfully identified the deletion in Df(3L)BSC27. Finally, depth of coverage was assessed using BEDTools to calculate the coverage per 50-bp interval and plotted in R. Breakpoints can be determined to within 500 bp with this method, which confirmed the predicted breakpoints in Df(3L)BSC33. Illumina sequence data are available from the NCBI website under the study accession SRP044233.
Results
D. melanogaster Lhr rescues F1 hybrid male lethality
We recently reported that D. melanogaster Lhr (mel-Lhr) has weak hybrid lethal activity (Maheshwari and Barbash 2012). Two different Lhr– deletions produced ~7–21% rescue of males to the pharate adult stage in crosses to D. mauritiana (Maheshwari and Barbash 2012). We therefore used Df(2R)Lhr– as a positive control for hybrid male rescue in our screen and observed rescue with two different stocks of D. mauritiana, averaging 7–8% at 18° (Table 2).
Table 2. Genome coverage of screen.
| Arm | All Stocks Screened | Sensitivity |
|
|---|---|---|---|
| ≥50 Df-Carrying Females Obtained | ≥100 Df-Carrying Females Obtained | ||
| 2L | 92.4% | 52.8% | 21.9% |
| 2R | 81.1% | 56.3% | 23.9% |
| 3L | 86.5% | 83.8% | 61.9% |
| 3R | 94.0% | 91.2% | 91.0% |
| Total of chromosomes 2 and 3 | 88.9% | 72.5% | 52.4% |
However, as reported in Maheshwari and Barbash (2012), we also found that other Lhr– deletions (Df(2R)BSC44 and Df(2R)BSC161), did not rescue males to the pharate stage. We conclude that our screen is vulnerable to false negatives due to background effects in the genome that affect our ability to detect weak rescue. This finding complicates mapping efforts because stocks that do not rescue are uninformative.
Lack of evidence for additional major-effect HI loci in the D. melanogaster genome
We screened 278 stocks from the Bloomington Deficiency Kit, covering approximately 89% of the autosomal genome (Supporting Information, Table S1). The average number of female progeny per cross was 238, with a slight bias for progeny carrying the deficiency chromosome (t-test, P < 0.001). However, the numbers varied widely among crosses due to variable mating efficiency. A total of 72.5% and 52.4% of crosses produced a minimum of 50 and 100 deficiency-carrying females, respectively, summed over replicates. The percentage of each chromosome arm covered is shown in Table 2.
We did not find any regions in the D. melanogaster genome, which, when removed, produce viable hybrid males (a single live male was found in one replicate with Df(3L)XDI98 but not in three other replicates). Therefore, within the regions of the genome that we screened, there are likely no additional major-effect D. melanogaster hybrid lethality genes. However, we found two adjacent deficiencies on 3L that give rescue comparable to positive controls with Lhr– deficiencies, Df(3L)BSC27 (65D4-65E6) and Df(3L)BSC33 (65E10-65F6) (Table 3). We also found weak rescue with deficiencies spanning 75A6-76D5 (Table 3). Further characterization of these two regions is described in the following sections. Three additional regions, two on 3L and one on 3R, also produced pharate males but were not pursued for further study due to low or variable rescue and/or the paucity of additional deficiency stocks (Table 3). For example, Df(3L)emc-E12 produced 6.7% rescue with D. mauritiana W139 but no rescue with strain iso 105. Thus, we conclude that rescue to the pharate stage is a rare event and is vulnerable to background effects.
Table 3. Regions that rescue hybrid males to the pharate adult stage.
| Region | Deficiency (% Rescue) | Molecular Breakpointsa |
Inferred Molecular Breakpointsb |
||
|---|---|---|---|---|---|
| Lhr | Df(2R)BSC49 (21.4%) | 12738807 | 13290649 | ||
| Df(2R)k08901 (7.21%/8.31%) | 13309963 | 13340212 | |||
| Df(2R)BSC44 | 13166788 | 13309036 | |||
| Df(2R)BSC161 | 13192288 | 13372333 | |||
| 61A-62E5 | Df(3L)emc-E12 (6.7%) | 206780 | 885293 | ||
| Df(3L)R-G7 (3.0%) | 1863545 | 2541764 | |||
| Df(3L)Ar14-8 | 641337 | 1615040 | |||
| Df(3L)BSC181 | 1688724 | 1841694 | |||
| 65D-66C5 | Df(3L)BSC27 (5.6%/10.8%) | 6935985 | 7149104 | ||
| Df(3L)RM5-2c | 6999777 | 7879617 | |||
| Df(3L)BSC33 (12.8%) | 7271620 | 7319021 | |||
| Df(3L)GN24 | 3922651 | 5203390 | |||
| Df(3L)XDI98 | 3967594 | 4134155 | |||
| Df(3L)ZN47 | 5096316 | 6696471 | |||
| Df(3L)W5.4 | 5919622 | 7029849 | |||
| Df(3L)BSC411 | 5969060 | 6618726 | |||
| Df(3L)Exel6109 | 6736213 | 6936639 | |||
| Df(3L)BSC224 | 6957557 | 7150109 | |||
| Df(3L)BSC374 | 6957558 | 7032145 | |||
| Df(3L)RM5-1 | 6999777 | 7287396 | |||
| Df(3L)Exel6110 | 7087906 | 7149284 | |||
| Df(3L)BSC117 | 7242575 | 7328086 | |||
| Df(3L)pbl-X1 | 7349893 | 8129687 | |||
| Df(3L)Exel8104 | 7353086 | 7522363 | |||
| Df(3L)ZP1 | 7889239 | 8254722 | |||
| 75A6-76D5 | Df(3L)W10 (1.9%) | 17867203 | 18202039 | ||
| Df(3L)fz2 (1.7%) | 19148197 | 19226562 | |||
| Df(3L)BSC20 (0.8%) | 19360266 | 19492579 | |||
| Df(3L)kto2 (3.5%) | 19380732 | 19924632 | |||
| Df(3L)BSC8 | 17656096 | 18009745 | |||
| Df(3L)Cat | 18056276 | 18834273 | |||
| Df(3L)ED4782 | 18988994 | 19163802 | |||
| Df(3L)XS533 | 19481010 | 20314886 | |||
| 83B7-83D1 | Df(3R)BSC47 (3.9%/1.7%) | 1509535 | 1756808 | ||
| Df(3R)BSC464 | 1474083 | 2037668 | |||
In cases in which rescue occurred with both strains, percent rescue with D. mauritiana W139 is presented first, followed by percent rescue with D. mauritiana iso 105. Overlapping deficiencies that do not rescue also are listed.
Breakpoints molecularly mapped.
Breakpoints inferred based on cytology.
Df(3L)RM5-2 produced pharate hybrid males in initial crosses, but further testing failed to reproduce this result. See Table S1.
Characterization of 75A-76D region
The initial cross with Df(3L)W10 produced 7.5% hybrid pharate males with D. mauritiana iso 105, but additional replicates failed to produce rescue. Summing across replicates yields a final rescue of 1.9%. Two deficiencies, Df(3L)BSC8 and Df(3L)Cat, which when combined encompass all of Df(3L)W10, did not rescue. However, other deficiencies adjacent to Df(3L)W10 did rescue. Df(3L)fz2, which is ~80 kb and predicted to be ~1 Mb distal to Df(3L)W10 rescued males at a similar low level (1.7%). Two additional deficiencies, Df(3L)BSC20 and Df(3L)kto2, ~130 kb distal to Df(3L)fz2 gave weak rescue (0.8% and 3.5%, respectively). These results suggest the possibility of multiple minor-effect genes in the 75A-76D region, but the low and variable level of rescue precluded further mapping.
Characterization of the 65D-E region
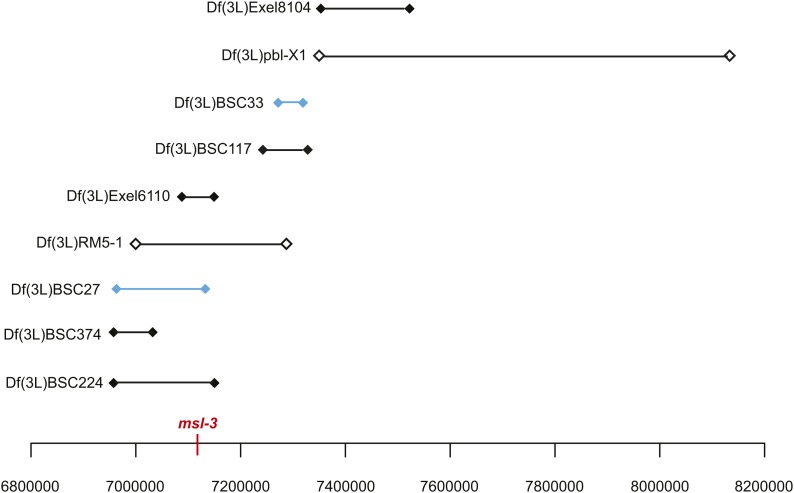
The Df(3L)BSC27 and Df(3L)BSC33 deletions both rescued pharate males greater than 10%. They are predicted to be 213 kb and 96 kb, respectively, and to be separated by 122 kb (Figure 2). When crossed together, we found trans-heterozygotes are viable, which is consistent with nonoverlapping deficiencies. These results suggest either that there are two hybrid lethality loci in the region, or that they affect a single gene that is between them.
Figure 2.
Deficiencies screened spanning 64C-66A. Df(3L)BSC27 and Df(3L)BSC33 spanning 65D4-65F6 produced rescued hybrid male pharates when crossed to D. mauritiana (blue bars). Seven deficiencies spanning this region that did not rescue are also shown. Filled endpoints denote molecularly defined deletions, whereas open endpoints indicate estimated breakpoints. Deficiencies Df(3L)Exel6110 and Df(3L)BSC27 were tested for complementation with msl-31; neither complemented msl-31, consistent with their molecularly mapped breakpoints. Complementation results are presented in Table S2.
None of 13 additional deficiencies in this region produced pharate males (Figure 2 and Table 3). The nonrescuing deficiencies include Df(3L)BSC224, which deletes all but 22 kb of Df(3L)BSC27, and Df(3L)BSC117, which encompasses the rescuing Df(3L)BSC33. One possibility is that the rescuing deletion stocks contain second-site mutations that are responsible for the rescue. However, considering our aforementioned results showing variable rescue with Lhr– deletions, we conclude that Df(3L)BSC27 and Df(3L)BSC33 are identifying two candidate regions but that our power to further map the hybrid rescue gene(s) is severely limited by false negatives.
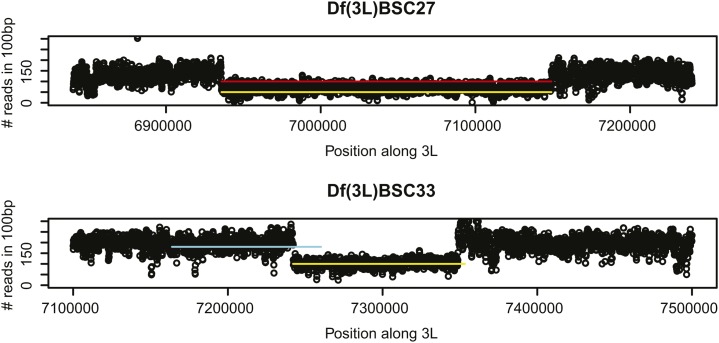
Characterization of molecular breakpoints by Next-Generation Sequencing of rescuing 3L deficiencies
We attempted to confirm the breakpoints of the rescuing deletions by short-read sequencing. We identified the breakpoints of Df(3L)BSC27 using both Hydra and Delly, which use paired-end read analysis (Quinlan et al. 2010; Rausch et al. 2012). The breakpoints were identified to be within 200 bp of those reported previously (Table 3). Pindel, which uses split-read analysis, did not identify this deletion, likely due to the 50-bp read length; however, our analysis of read-depth further supported this deletion (Figure 3). Neither paired-end nor split-read analysis determined the correct deletion in Df(3L)BSC33. Pindel identified a deletion partially overlapping the predicted region and matching the predicted size but it was not supported by read-depth analysis (Figure 3). However, read-depth analysis did identify a deletion that corresponds well to the predicted molecular breakpoints previously reported (Table 3). We conclude that the Df(3L)BSC27 and Df(3L)BSC33 deletion stocks are correct.
Figure 3.
Coverage plots of deficiency stocks based on sequencing data. Coverage analysis supports the predicted deletions for Df(3L)BSC27 and Df(3L)BSC33. The number of reads mapping within 100-bp intervals is plotted against the corresponding position on 3L for the sequenced deficiency lines. Segments in yellow represent the predicted location of deletions based on previous molecular or cytological estimates. Segments in red and cyan represent deletions predicted by paired-end approaches (Hydra and/or Delly) and Pindel, respectively.
Sixty-three genes are annotated within Df(3L)BSC27 and Df(3L)BSC33 (Table 4). We prioritized candidates based on shared characteristics with Hmr and Lhr, such as encoding proteins that are nuclear, rapidly evolving, highly expressed in ovaries, chromatin-binding, contain MADF and/or BESS domains, and are heterochromatic. Seven genes (tow, msl-3, Mis12, cdc27, bin, MED4, and mei-P22) encode nuclear proteins involved in processes including dosage compensation, mitosis, transcription, and meiotic recombination. The msl-3 protein contains a chromatin-binding chromo domain (Koonin et al. 1995) as well as an MRG domain that is implicated in chromatin remodeling (Bertram and Pereira-Smith 2001), whereas bin has a forkhead DNA-binding domain (Pérez Sánchez et al. 2002). Mis12 localizes specifically to the kinetochore and functions in mitotic spindle formation (Goshima et al. 2007). CG9948 is largely uncharacterized but is of interest because it contains a MADF domain, similar to Hmr. Five of the candidate genes encode proteins highly expressed in the ovaries (RpL18, CG9953, Cdc27, Galphai, and sgl) with functions that include translation, proteolysis, mitosis, receptor binding, and oxidoreductase activity.
Table 4. Genes mapped within rescuing deficiencies.
| Deficiency | No. Genes (High-Priority Candidates) | Genesa |
|---|---|---|
| Df(3L)BSC27 | 44 (9) | tow, msl-3, Cpr65Ec, CG17744, CG10077, Surf1, corn, form3, Cpr65Eb, mp, Mis12, melt, Galphai, CG9953, CG9948, CR32385, CG10063, CG34030, CG43439, bin, CG14823, CG10075, CG32391, CG8629, Cpr65Ea, CG15829, CG8641, CG8628, CG32388, BBS1, Dbi, CG10064, sgl, Prat2, ms(3)04202, mp, Me, Vn, CS3-1, rip, dv, E(Ubx)3L, jv |
| Df(3L)BSC33 | 21 (4) | MED4, unc-13-4A, mRpL50, CG14830, Neos, CG14829, CR43470, mei-P22, Dscam2, RpL18, CG14826, BHD, Cdc27, ms(3)04202, CS3-1, anon-65Ea, CG8628, Dscam2, corn, E(Ubx)3L, form3 |
High-priority candidates are indicated in bold, using criteria described in the section Results.
We focused on the gene male specific lethal-3 (msl-3) in part because, like Hmr and Lhr, it encodes an adaptively evolving chromatin-binding protein (Rodriguez et al. 2007). The msl-3 protein is part of the dosage compensation complex (DCC) that binds to the X chromosome in males to mediate hypertranscription (Gorman et al. 1995). A dosage compensation defect does not appear to be the direct cause of hybrid lethality because X chromosome transcripts are not preferentially affected in lethal hybrids (Wei et al. 2014). However, several observations suggest that dosage compensation genes may interact with hybrid lethality. Components of the DCC fail to localize to the X chromosome and H4K16Ac is not enriched on the X chromosome in hybrid males compared with pure species males (Pal Bhadra et al. 2006), although a later study did detect Msl-2 protein on the X (Thomae et al. 2013). Additionally, mutations in D. melanogaster DCC genes, including msl-3, mildly enhance hybrid male viability when partially rescued by Lhr alleles (Barbash 2010a).
siRNA was ineffective in silencing in D. simulans msl-3
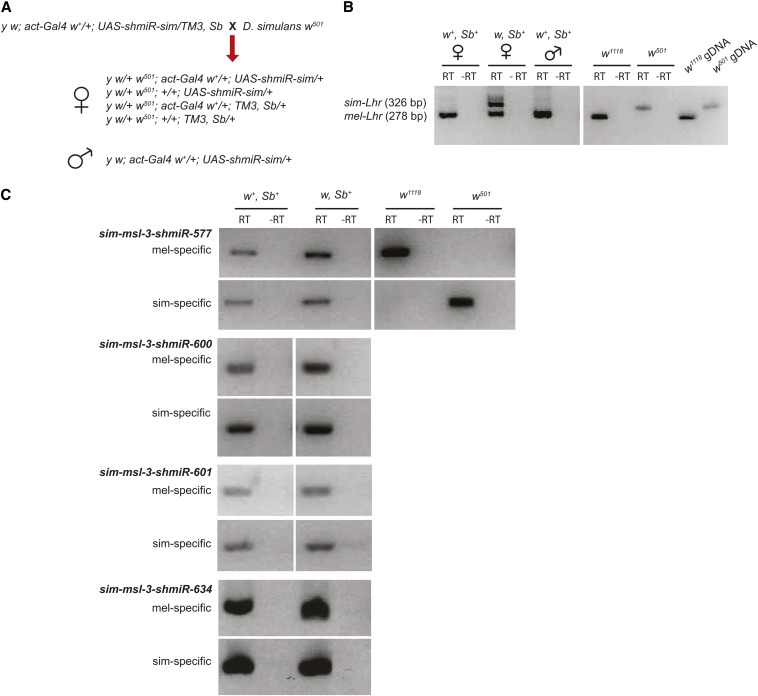
We first tested D. melanogaster msl-3 as a candidate responsible for rescue by crossing msl-31 females to D. mauritiana. No live or pharate hybrid males were observed (270 and 84 msl-31/+ hybrid females recovered in crosses with D. mauritiana iso 105 and W139, respectively). We next attempted to test sim-msl-3 by knocking down its expression in hybrids using RNAi. The experimental challenge was to design an RNAi construct that would target the D. simulans ortholog but not the D. melanogaster ortholog, because removing both copies of msl-3 would result in male lethality. The shmiR method seemed ideal because it can achieve potent knockdown by expressing a single 21-bp siRNA from a modified miRNA-based vector (Haley et al. 2008). We used the Valium 20 transformation vector, which expresses under the control of upstream activation sequence (UAS) sequences when crossed to a strain expressing the Gal4 activator protein (Ni et al. 2011). Our strategy was to transform these RNAi constructs into the attP2 site on D. melanogaster chromosome 3 and then cross transformed stocks to a strain containing actin-Gal4 on chromosome 2. These flies will express ubiquitously the shmiR, and when females are crossed to D. simulans males, one-quarter of the F1 hybrid progeny will inherit both the Gal4 driver and the UAS-driven shmiR.
As a positive control, we first designed a vector that targets an insertion that is specific to D. simulans Lhr (sim-Lhr) (Table 1 and Table 5). Expression of this construct in D. melanogaster produced no phenotype. All four expected genotypes of F1 hybrid females were recovered. RT-PCR analysis showed that only mel-Lhr is expressed in act-Gal4/+; UAS-shmIR-sim-Lhr/+ whereas both orthologs are expressed in control hybrid females (+/+; UAS-shmIR-sim-Lhr/+), demonstrating that the construct specifically knocks down sim-Lhr expression (Figure 4B). Interestingly, at 25° females expressing the shmiR against sim-Lhr had the greatest viability, which is consistent with sim-Lhr having a dominant effect on hybrid female viability at greater temperatures (Barbash et al. 2000). As predicted, knockdown of sim-Lhr rescued hybrid males, and RT-PCR analysis again demonstrated specific knockdown of the sim-Lhr otholog (Figure 4).
Table 5. Suppression of hybrid lethality by UAS-shmIR-sim-Lhr.
| Temperature | Sex of Progeny | GAL4/+; UAS/+ (w+ Sb+) | GAL4/+; +/+ (w+ Sb) | +/+; UAS/+ (w Sb+) | +/+; +/+ (w Sb) |
|---|---|---|---|---|---|
| 25° | Female | 217 | 32 | 165 | 47 |
| Male | 144 | 0 | 0 | 0 | |
| 18° | Female | 62 | 79 | 58 | 57 |
| Male | 58 | 0 | 1 | 0 |
y w; P{w[+mC]=Act5C-GAL4}25FO1 /+; φ{UAS-shmIR-sim-Lhr}attP2, v+ y+/TM3,Sb D. melanogaster females were crossed to w501 D. simulans males. The transgenes are abbreviated as “GAL4” and “UAS” in the table headings. Number of progeny of the indicated genotype (phenotype) are listed.
Figure 4.
RT-PCR tests of shmiR knock-down of D. simulans Lhr and msl-3. (A) D. melanogaster females expressing either Lhr or msl-3 siRNAs targeting the D. simulans orthologs (abbreviated as UAS-shmiR-sim) were crossed to D. simulans males. w+, Sb+ hybrid progeny (1/4 of the total) inherit both the Gal4 driver and UAS-shmiR. The males will survive if the UAS-shmiR construct knocks down expression of a hybrid lethality gene. (B and C) RT-PCR tests of knockdown. (B) Hybrid male and female progeny carrying both act-Gal4 and UAS-shmiR-sim-Lhr (w+, Sb+) express D. melanogaster mel-Lhr (278 bp) but not sim-Lhr (326 bp), assayed using a single primer pair that detects an insertion in sim-Lhr. Hybrid females carrying only UAS-shmiR-sim-Lhr (w, Sb+) were used as a control and express both orthologs. Right panel is RT-PCR and genomic DNA (gDNA) controls from D. melanogaster w1118 and D. simulans w501. (C) None of four tested sim-msl3-shmiR constructs silence sim-msl-3 expression in hybrid female progeny. Separate PCRs were performed using primer pairs specific to either mel-msl-3 or sim-msl-3, as confirmed using controls as described previously. Hybrid females carrying both act-Gal4 and UAS-shmiR-sim-msl-3 (w+, Sb+) expressed both msl-3 orthologs. As a control, progeny only inheriting UAS-shmiR-sim-msl-3 (w, Sb+) were also assayed (except for sim-msl-3-shmiR-577 where both w, Sb+ and w, Sb animals were pooled), and expressed both orthologs as expected. RT-PCR, reverse-transcription polymerase chain reaction.
We designed seven different shmiRs against sim-msl-3 that each had mismatches to mel-msl-3 (Table 1). Some were designed with modifications from published schemes (see the section Materials and Methods). Three of the constructs were lethal or semilethal to both sexes when expressed in D. melanogaster (Table 1), which must be due to off-target effects because msl-3 is only required in males. A fourth construct (msl-3-shRNA-600) was lethal only to D. melanogaster males. Because msl-3 is expressed in females, we could assay the effect of this shmiR, and found by RT-PCR that it does not reduce msl-3 expression (Figure 4C). Therefore, this construct likely has a post-transcriptional effect on mel-msl-3.
The remaining three constructs were viable within D. melanogaster and thus could be crossed to D. simulans as we did for the Lhr control. None produced any hybrid males, but RT-PCR demonstrated that none silenced sim-msl-3 expression. We were thus unable to test whether sim-msl-3 affects hybrid male viability.
mel-Hmr does not cause lethality to Xsim hybrid males
Our screen was limited to the autosomes, but we wished to test whether D. melanogaster (mel-Hmr) can account entirely for the lethal effect of Xmel in hybrid males. Xsim (and Xmau) hybrid males are viable (Sturtevant 1920; Hutter et al. 1990), and a simple prediction is that the presence of mel-Hmr will kill Xsim hybrid males if mel-Hmr is the sole X-linked difference between these species involved in hybrid lethality. Hybrid sons carrying the paternal X chromosome can be generated by crossing D. simulans males to compound-X D. melanogaster females. We tested the role of mel-Hmr by using a mel-Hmr-HA transgene (Satyaki et al. 2014) and found that it had no effect on Xsim hybrid male viability (Table 6). We conclude that additional genes and/or sequences on Xmel are required for the fully penetrant lethality of Xmel hybrid males.
Table 6. A mel-Hmr transgene does not reduce Xsim hybrid male viability.
| Hybrid Females |
Hybrid Males |
|||
|---|---|---|---|---|
| C(1)DX; +/+ | C(1)DX; ø{mel-Hmr-HA}/+ | w/Y; +/+ | w/Y; ø{mel-Hmr-HA}/+ | Relative Viability |
| 0 | 0 | 132 | 108 | 81.8%a |
C(1)DX, y w f/Y; ø{mel-Hmr-HA}/+ D. melanogaster females were crossed to w501/Y D. simulans males at 22-23°.
Not significant by χ2 test (P > 0.05).
Discussion
Major-effect vs. minor-effect HI genes
We used the Bloomington Deficiency Kit to screen for dominant suppressors of lethality in interspecific hybrids between D. melanogaster and D. mauritiana. Our screen is different from previous screens (Coyne et al. 1998; Presgraves 2003; Matute et al. 2010) in that it is designed to identify putative HI loci that cause dominant lethality, like Hmr and Lhr. The removal of either Hmr or Lhr suppresses hybrid male lethality, which classifies them as major-effect HI genes. Because we only screened for hybrid rescue with D. mauritiana, our screen also is predicated on the assumption that additional HI loci will be similar to Hmr and cause lethality with all three sibling species of D. melanogaster.
Our failure to observe any adult males suggests that the D. melanogaster genome does not harbor additional major-effect HI loci within the regions screened, but this does not exclude the possibility that there are additional factors of minor effect that contribute to hybrid lethality. We identified four regions, in addition to the region on 2R containing Lhr, which when deleted rescue hybrid male lethality to the pharate stage, suggesting the presence of minor-effect HI loci. However, the weak nature of these rescuing effects makes them susceptible to suppression by background effects. Without the ability to confirm them with multiple overlapping deficiencies, we conclude that the rescuing deficiencies identify regions that can be tentatively considered to contain minor-effect hybrid lethality loci.
The regions deleted by Df(3L)BSC27 and Df(3L)BSC33 gave the strongest level of rescue. Surprisingly, these two deletions are distinct and do not overlap, as predicted by estimated cytological breakpoints and confirmed by complementation crosses and sequencing. It is possible, however, that both deletions affect a single gene. There are ~93 kb between Df(3L)BSC27 and Df(3L)BSC33. Most enhancers in Drosophila are within 10 kb of their target sequence, but longer-range interactions are known (Berman et al. 2004). For example, the cut gene is regulated by an enhancer 80 kb upstream of its promoter (Jack and Delotto 1995). A recent genome-wide study estimated that ~28% of enhancers are >20 kb from their targets and can be more than 100 kb away (Kvon et al. 2014).
Pontecorvo’s experiments (1943) suggest that gene(s) on the D. simulans 3rd chromosome contribute to hybrid lethality. Based on previous findings that D. melanogaster Lhr has a weak effect on hybrid lethality, we reasoned that our screen has the ability to identify major-effect genes in D. simulans by detecting weak effects of deleting the D. melanogaster ortholog. We attempted to test msl-3 as one such candidate by knocking down expression of sim-msl-3 using shmiRs but unfortunately failed to do so.
Challenges with using shmiRs to knockdown gene expression
The shmiR system is attractive because the expression of a single siRNA allows the design of siRNAs that target only one of the two orthologs in hybrids. We were successful in specifically targeting sim-Lhr by designing a shmiR targeting a species-specific insertion in the sim-Lhr coding sequence. However, we were unable to achieve specific knockdown of sim-msl-3. Three constructs caused lethality to both sexes within D. melanogaster (Table 1), which must be off-target effects as msl-3 is only required in males. One shmiR (msl-3-sim-shmiR-600) was lethal in D. melanogaster males, even though it has a mismatch to the D. melanogaster msl-3 ortholog near its center. We found that msl-3 mRNA expression is not reduced in the lethal males, suggesting that msl-3 translation is likely being blocked via a microRNA-like effect of the seed sequence. Because as little as 6 bp of sequence homology near the 5′ end of a microRNA can be sufficient for its activity (Lewis et al. 2005), this hypothesis would explain why the shmiR designed to target sim-msl-3 would be able to target the D. melanogaster ortholog. This type of off-target effect will be challenging to predict because it requires limited homology and suggests the need for caution when expressing shmiRs at a high level. The clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) system may prove an effective alternative in the future for targeting mutations to D. simulans candidate HI genes.
The potential role of X-chromosome genes
The X chromosome contains 15% of D. melanogaster genes (Adams 2000), but we could not screen it because deletions of the X are lethal in males. We showed here that mel-Hmr is insufficient to explain the X-linked portion of hybrid male lethality, because a mel-Hmr-HA transgene does not induce lethality of Xsim hybrid males (Table 6). This result is consistent with previous findings that a similar transgene significantly reduces viability of Xmel/Xsim hybrid females but does not induce full lethality (Barbash et al. 2003), in contrast to the invariant lethality of Xmel/Xmel hybrid females (Hutter et al. 1990).
Df(1)307-1-2 identifies a candidate region in 9D that is adjacent to but nonoverlapping with Hmr (Barbash et al. 2003), but the causal gene remains unidentified. A recent screen examined Y-linked duplications of Xmel regions for lethality in Xsim hybrid males and identified 2 candidate regions (Matute and Gavin-Smyth 2014). One of these regions (9C-10B) is not a new discovery, as Dp(1;2)v+75d covering 9A2-10C2 was previously shown to reduce Xmau, Xsec, and Xsim hybrid male viability (Barbash et al. 2000; Orr and Irving 2000). The duplication also significantly reduces Xmel/Xmau and Xmel/Xsim hybrid female viability (Barbash et al. 2000), presumably due at least in part to the aforementioned effect of Hmr. This effect in females demonstrates that the 9C-10B duplication causes dominant lethality by interacting with D. simulans and D. mauritiana genes that are also dominantly acting and could be autosomal or X-linked, in contrast to the conclusion that it represents a dominant-recessive interaction between the X chromosomes (Matute and Gavin-Smyth 2014).
We have shown that Hmr cannot be the sole cause of the duplication-induced hybrid male lethality (Table 6), although it may be contributing. We suggest that lethality may result from the cumulative dosage increase of multiple duplicated genes, because well-characterized HI genes act as gain-of-function alleles in a hybrid background (Maheshwari and Barbash 2011). These putative dosage effects can only be tested for in a hybrid background. We reiterate here that testing fitness effects of chromosome aberrations within D. melanogaster is a useful general control but has no bearing on whether or not duplications or deficiencies are responsible for dosage effects in hybrids, because HI genes by definition have distinct (and often opposite) properties in hybrid vs. pure species backgrounds (Maheshwari and Barbash 2011).
Supplementary Material
Acknowledgments
We thank Dr. Margarida Moreira for help with identifying deficiency breakpoints; Kevin Wei, Michael McGurk, and Daniel Zinshteyn for helpful comments on the manuscript; and the Bloomington Drosophila Stock Center for the many deficiency stocks. Supported by National Institutes of Health GM074737 to D.A.B.
Footnotes
Supporting information is available online at http://www.g3journal.org/lookup/suppl/doi:10.1534/g3.114.014076/-/DC1
Communicating editor: R. Kulathinal
Literature Cited
- Adams M. D., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Barbash D. A., 2007. Nup96-dependent hybrid lethality occurs in a subset of species from the simulans clade of Drosophila. Genetics 176: 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., 2010a Genetic testing of the hypothesis that hybrid male lethality results from a failure in dosage compensation. Genetics 184: 313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., 2010b Ninety years of Drosophila melanogaster hybrids. Genetics 186: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., Roote J., Ashburner M., 2000. The Drosophila melanogaster hybrid male rescue gene causes inviability in male and female species hybrids. Genetics 154: 1747–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., Siino D. F., Tarone A. M., Roote J., 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. USA 100: 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman B. P., Pfeiffer B. D., Laverty T. R., Salzberg S. L., Rubin G. M., et al. , 2004. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 5: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram M. J., Pereira-Smith O. M., 2001. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene 266: 111–121. [DOI] [PubMed] [Google Scholar]
- Brideau N. J., Flores H. A., Wang J., Maheshwari S., Wang X., et al. , 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295. [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Simeonidis S., Rooney P., 1998. Relative paucity of genes causing inviability in hybrids between Drosophila melanogaster and D. simulans. Genetics 150: 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman M., Franke A., Baker B. S., 1995. Molecular characterization of the male-specific lethal-3 gene and investigations of the regulation of dosage compensation in Drosophila. Development 121: 463–475. [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., et al. , 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B., Hendrix D., Trang V., Levine M., 2008. A simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev. Biol. 321: 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter P., Ashburner M., 1987. Genetic rescue of inviable hybrids between Drosophila melanogaster and its sibling species. Nature 327: 331–333. [DOI] [PubMed] [Google Scholar]
- Hutter P., Roote J., Ashburner M., 1990. A genetic basis for the inviability of hybrids between sibling species of Drosophila. Genetics 124: 909–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack J., DeLotto Y., 1995. Structure and regulation of a complex locus: the cut gene of Drosophila. Genetics 139: 1689–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliman R. M., Andolfatto P., Coyne J. A., Depaulis F., Kreitman M., et al. , 2000. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156: 1913–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Zhou S., Lucchesi J. C., 1995. The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 23: 4229–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon E. Z., Kazmar T., Stampfel G., Yáñez-Cuna J. O., Pagani M., et al. , 2014. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512: 91–95. [DOI] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P., 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- Maheshwari S., Barbash D. A., 2011. The genetics of hybrid incompatibilities. Annu. Rev. Genet. 45: 331–355. [DOI] [PubMed] [Google Scholar]
- Maheshwari S., Barbash D. A., 2012. Cis-by-Trans regulatory divergence causes the asymmetric lethal effects of an ancestral hybrid incompatibility gene. PLoS Genet. 8: e1002597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S., Wang J., Barbash D. A., 2008. Recurrent positive selection of the Drosophila hybrid incompatibility gene Hmr. Mol. Biol. Evol. 25: 2421–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute D. R., Gavin-Smyth J., 2014. Fine mapping of dominant X-linked incompatibility alleles in Drosophila hybrids. PLoS Genet. 10: e1004270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute D. R., Butler I. A., Turissini D. A., Coyne J. A., 2010. A test of the snowball theory for the rate of evolution of hybrid incompatibilities. Science 329: 1518–1521. [DOI] [PubMed] [Google Scholar]
- McDermott S. R., Kliman R. M., 2008. Estimation of isolation times of the island species in the Drosophila simulans complex from multilocus DNA sequence data. PLoS ONE 3: e2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., Pontecorvo G., 1940. Recombinants between Drosophila species, the F1 hybrids of which are sterile. Nature 146: 199–200. [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., et al. , 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8: 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., Irving S., 2000. Genetic analysis of the hybrid male rescue locus of Drosophila. Genetics 155: 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal Bhadra M., Bhadra U., Birchler J. A., 2006. Misregulation of sex-lethal and disruption of male-specific lethal complex localization in Drosophila species hybrids. Genetics 174: 1151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez Sánchez C., Casas-Tintó S., Sánchez L., Rey-Campos J., Granadino B., 2002. DmFoxF, a novel Drosophila fork head factor expressed in visceral mesoderm. Mech. Dev. 111: 163–166. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G., 1943. Viability interactions between chromosomes of Drosophila melanogaster and Drosophila simulans. J. Genet. 45: 51–66. [Google Scholar]
- Presgraves D. C., 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163: 955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R., Clark R. A., Sokolova S., Leibowitz M. L., Zhang Y., et al. , 2010. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res. 20: 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T., Zichner T., Schlattl A., Stutz A. M., Benes V., et al. , 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M. A., Vermaak D., Bayes J. J., Malik H. S., 2007. Species-specific positive selection of the male-specific lethal complex that participates in dosage compensation in Drosophila. Proc. Natl. Acad. Sci. USA 104: 15412–15417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J. G., Stark A., Johnston W. K., Kellis M., Bartel D. P., et al. , 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17: 1850–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyaki P. R. V., Cuykendall T. N., Wei K. H.-C., Brideau N. J., Kwak H., et al. , 2014. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet. 10: e1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1920. Genetic Studies on drosophila simulans. I. Introduction. Hybrids with drosophila melanogaster. Genetics 5: 488–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Presgraves D. C., 2009. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323: 779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomae A. W., Schade G. O. M., Padeken J., Borath M., Vetter I., et al. , 2013. A pair of centromeric proteins mediates reproductive isolation in Drosophila species. Dev. Cell 27: 412–424. [DOI] [PubMed] [Google Scholar]
- Watanabe T. K., 1979. A gene that rescues the lethal hybrids between Drosophila melanogaster and Drosophila simulans. Jpn. J. Genet. 54: 325–331. [Google Scholar]
- Wei K. H.-C., Clark A. G., Barbash D. A., 2014. Limited gene misregulation is exacerbated by allele-specific upregulation in lethal hybrids between Drosophila melanogaster and Drosophila simulans. Mol. Biol. Evol. 31: 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Ma H., Ye C., Ramirez D., Chen S., et al. , 2011. Improved siRNA/shRNA functionality by mismatched duplex. PLoS ONE 6: e28580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K., Schulz M. H., Long Q., Apweiler R., Ning Z., 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.