Abstract
Parents encountering stress environments can influence the phenotype of their offspring in a form of transgenerational phenotypic plasticity that has the potential to be adaptive if offspring are thereby better able deal with future stressors. Here, we test for the existence of anticipatory parental effects in the heat stress response in the highly polymorphic nematode Caenorhabditis remanei. Rather providing an anticipatory response, parents subject to a prior heat stress actually produce offspring that are less able to survive a severe heat shock. Selection on heat shock resistance within the larvae via experimental evolution leads to a loss of sensitivity (robustness) to environmental variation during both the parental and larval periods. Whole genome transcriptional analysis of both ancestor and selected lines shows that there is weak correspondence between genetic pathways induced via temperature shifts during parental and larval periods. Parental effects can evolve very rapidly via selection acting directly on offspring.
Keywords: experimental evolution, heat shock, phenotypic plasticity, stress resistance, transgenerational effects
1. Introduction
Experimental evolution is a powerful means of testing the effects of new environments on organismal traits. For example, when E. coli are moved from one rearing temperature to another, they adapt to the new environmental conditions within a few thousand generations [1,2]. Such experimental approaches are particularly effective at testing evolutionary hypotheses, such as the existence of potential adaptive tradeoffs in performance among environments (e.g., [1,3]). While large population sizes and fast generation times have made experimental evolution studies using microbes particularly successful (see, for example, the many examples in this volume), extending inferences from these studies to many other organisms may be somewhat limited, particularly for plants and animals. For example, most experimental evolution studies in microbes are initiated using a single clonal individual, whereas most plant and animal populations harbor substantial levels of standing genetic variation, which undoubtedly plays an important role in the response to selection [4,5]. Perhaps the most obvious difference, however, is the fact plants and animals undergo complex developmental processes during the transition from single celled egg/seed to a multicellular reproductive adult. One consequence of the dramatic difference in size and complexity between parent and ovule is the opportunity for differential provisioning of offspring with parentally-derived resources such as RNA, yolk, and endosperm. While single-celled organisms can definitely differentially provision their daughter cells [6], the vast difference in scale between parent and offspring within multicellular organisms prima facie means that the opportunity for resource partitioning is likely to be much greater.
The potential role of transgenerational effects in shaping environmental responses and evolutionary outcomes has been increasingly incorporated in biological thinking over the last few decades [7]. In addition to the direct provisioning of resources described above, other parental effects, including parentally-derived intercellular signals and epigenetic imprinting of the genome, have been found to shape a wide variety of phenotypic responses [8]. From an evolutionary point of view, such effects can be important because they create a direct correlation in phenotypes across environments that is not mediated via genetic inheritance [9,10]. For example, positively correlated maternal effects across environments (e.g., when larger mothers have resources that allow them to produce larger offspring) can lead to an acceleration of the evolutionary response at the initiation of selection, as well as a “phenotypic overshoot” when selection ceases [9]. In general, although it is clear that parental effects are ubiquitous across both plants and animals [11,12] and therefore have the potential to influence the response to selection, the extent to which these effects shape the dynamics of the evolutionary response within any given population is much less well known.
Because parental effects allow the environment experienced by the parents to shape the phenotype of the offspring, they can be thought of as a form of transgenerational phenotypic plasticity [13]. Perhaps the most interesting outcome of this type of plasticity is an “anticipatory parental effect” in which individuals use the environment that they are currently experiencing as a cue to either (1) differentially provision their offspring so that they are better equipped to respond to that environment or (2) deposit signaling compounds (i.e., mRNA or proteins) that alter gene expression within the offspring. For example, mothers experiencing a stressful environment might provide signals in their eggs that lead to increased production of stress-response proteins within the offspring [14]. In the case of stress responses, anticipatory parental effects could be characterized as “anticipatory hormesis,” in which sub-lethal exposure to a stress by parents increases the ability of their offspring to resist later exposures to that same stressor (sensu [15]). However, evidence that such anticipatory effects are common is somewhat weak [16], and parents experiencing stress may show negative effects on offspring as frequently as positive effects [17]. In particular, parental effects may reflect specific adaptations within the parents regardless of whether they increase the fitness of the offspring [18].
Experimental evolution provides a particularly powerful platform for investigating the evolution of phenotypic plasticity because alterations of individual norms of reaction (the functional relationship between environment and phenotype) can be observed directly by measuring the response in both ancestors and descendants (e.g., [19]). For example, individuals of the nematode Caenorhabditis remanei that have recently been collected from nature display much higher survival when exposed to a heat shock of 36.8°C after having been raised at 30°C than those raised at 20°C [20]. After ten generations of selection for increased heat shock resistance, however, individuals raised at 20°C and 30°C perform equally well and are significantly more resistant than their wild-caught ancestors. In other words, the importance of pre-exposure to high temperatures as an environmental cue rapidly diminished under selection.
Nematodes provide an especially appealing system for the study of experimental evolution in animals [21]. While nowhere close to microbes in terms of total population size or replication rate, the ability to raise hundreds of thousands of individuals within single replicates and their 3-4 day generation time stands out relative to other animal model systems. Most importantly, like most microbial systems, many nematodes (including C. elegans and its relatives) can be revived after being frozen, which allows ancestors and descendants to be measured contemporaneously. This ability to revive frozen samples is especially critical when looking at phenotypic sensitivity to environmental influences because it is formally impossible to distinguish temporal effects from environmental effects when separate generations are not tested at the same time.
Here, we examine the influence of the independent effects of both parental and offspring exposure to heat stress on later offspring resistance to a lethal heat shock in the highly polymorphic nematode C. remanei, both in ancestral populations that were recently collected from nature and descendent populations that had experienced ten generations of selection for heat shock resistance [20]. In addition to the evolution of the phenotypic response to temperature variation, we also examined the underlying genetic basis of the response by measuring whole-genome transcriptional differences in all possible combinations of parental and offspring environmental exposures. These analyses allow us to (1) test for the existence of anticipatory parental effects of the heat stress response, (2) ask how these parental effects evolve over time in correspondence with direct selection on the offspring phenotype, and (3) determine whether the evolution of the stress response system is constrained to particular genetic pathways that suggest either parental or offspring mediated effects.
2. Results
2.1 Parental environment alters plasticity for heat stress resistance
We analyzed three different sets of C. remanei populations for resistance to a severe heat shock of 36.8°C following the four possible combinations of parents and offspring being raised at 20°C or 30°C. As described in [20], one population was derived from a set of 26 natural isolates (isofemale lines) a few generations removed from nature, yielding a highly polymorphic base population (“ancestor”). The next population was an experimental population that was selected for heat shock resistance to 36.8-37.2°C (“heat”) for 10 generations. Each replicate population comprised 1000-2000 mating individuals. Exposure to acute stress occurred either every second generation or when the population produced ≥24,000 eggs, whichever occurred later (~30 generations total). The final population was subject to the same handling as the heat-selected lines, but without an actual heat shock (“control”), yielding a total of 30 generations of adaptation to general laboratory conditions. Two replicates of the selected lines and four replicates of the control lines were maintained [20]. Polymorphism within C. remanei populations ~5% [22-24], so the response to selection observed here should be based almost entirely on segregating variation.
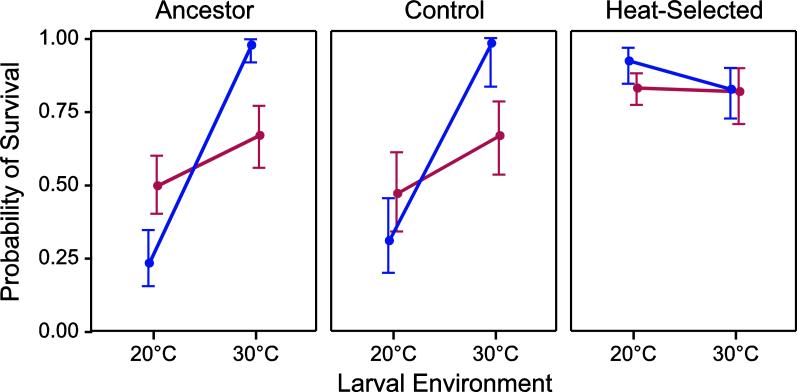
For individuals from the recently isolated ancestral population, if parents are raised in permissive conditions at 20°C, then exposure to a mild heat stress during larval development improved resistance to subsequent heat shock (Fig. 1; F1,21 = 54.15, P < 0.001), which is consistent with previous observations [20]. In fact, the plastic response induced by the larval worms under stress is sufficient to ensure almost complete survival during subsequent heat shock, before any selection for increased stress resistance has even occurred. However, the protective effect of larval exposure to 30°C was mediated by an interaction between the environment experienced by the parents and embryos (Fig. 1; F1,20 = 32.03, P < 0.001). Specifically, if the parents were previously exposed to heat stress at 30°C, then larval survival significantly improved when the larvae themselves were raised under non-stressful conditions at 20°C (t20 = 2.92, P = 0.009). However, when parents were raised at 30°C, the heat shock resistance of larvae reared at 30°C was greatly reduced compared to when parents were raised at 20°C. Pre-exposure to heat during the larval period was therefore only strongly protective in larvae whose parents were not heat stressed, while being only mildly protective for larvae whose parents were.
Fig. 1.
Phenotypic plasticity for heat stress resistance in ancestral and selected populations. Interaction plots depict the effects of larval environment on the proportion of worms surviving heat shock when the parents were reared at 20°C (blue) or 30°C (red) in each of the experimental populations. Conditional means from the GLM with 95% confidence intervals are plotted.
2.2 Loss of transgenerational plasticity after experimental evolution
In lines selected for resistance to heat shock, sensitivity to both the larval and parental environments was lost relative to ancestor and a laboratory-adaptation control (Fig. 1). In this population, acclimating the larvae to 30°C prior to heat shock did not provide the protective effect observed in the other populations (F1,21 = 1.97, P = 0.175). The loss of plasticity (“genetic assimilation”; [20,25]) of the phenotype was observed regardless of the environment in which the parents were raised (F1,20 = 2.12, P = 0.161). In each treatment combination, survival was at least 80% following heat shock. The lack of environmental sensitivity was not simply because the populations were no longer affected by the heat shock, as survival had not yet reached the maximum of 100% in any treatment condition in the heat-selected line.
2.3 Gene regulation is influenced by parental environment
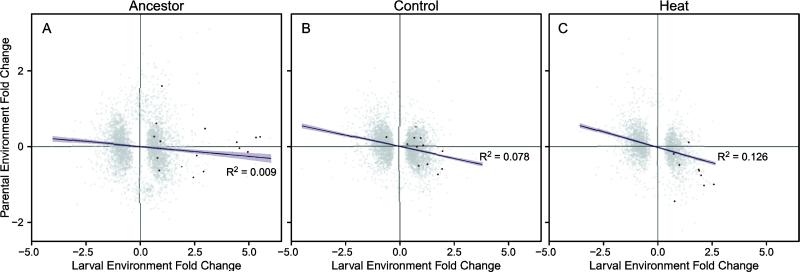
Given that the parental environment contributed to differences in heat resistance later in life, we hypothesized that the environment experienced in the previous generation would influence the regulation of genes prior to heat shock and therefore the differences in phenotype across parental environments. In the ancestral population, 480 genes had a significant change in expression related to the parental environment, while 3316 genes were differentially expressed across larval environments (Fig. 2A; determined using a general linear model with a false discovery rate of 0.05). In addition, 189 genes differed in their response across larval environments, depending on the prior conditions experienced by the parents. Similarly, in the evolved populations, the largest transcriptional effect was observed in response to changing larval environments, but somewhat surprisingly a substantial number of genes continued to show differences in expression following parental stress (Fig. 2B,C).
Fig. 2.
Comparison of parental and larval environmental effects on gene regulation in the experimentally evolved populations. (A) Ancestral populations, (B) laboratory adaptation controls, and (C) populations selected for heat shock resistance during the larval period. Only genes that were differentially expressed (FDR < 5%) across parental or larval environments are shown (gray). Heat shock proteins that were significantly differentially expressed are highlighted in red. Lines in each panel indicate the linear fit from the regression model (±95% CI). All correlations are P < 0.0001.
Next, we compared the gene expression changes resulting from the differences between parental and larval environments to determine whether the expression differences set up in the parental environment would predict the response in the larvae directly. Given that the environmental stimulus is temperature for both the parents and the larvae, we predicted that we would observe a positive correlation in the regulation of genes across parental and larval environments. Surprisingly, the changes in gene expression attributed to parental vs. larval effects were not strongly correlated in any of the populations (Fig. 2), and in all populations the correlation was slightly negative, rather than positive. If the same genes responded similarly to increases in temperature in both parents and offspring, then we would expect a strong positive correlation between these responses rather than a weak negative correlation.
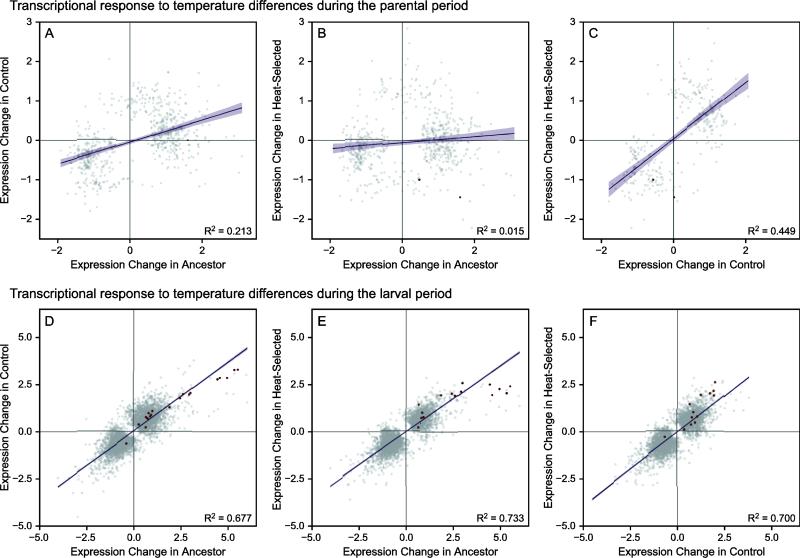
3.4 Degradation of parental response through selection
Next, we asked if gene regulation due to each environment was strongly correlated among populations. Between the ancestral population and control population, we found that the differences in expression due to parental environment in each population were only weakly correlated (Fig. 3A-C; R2 = 0.213, P < 0.0001). Between the ancestral population and the heat-selected population, the correlation in the transcriptional response was extremely weak (R2 = 0.015, P < 0.0001), although all of these correlations are highly statistically significant because of the large number of genes involved. Surprisingly, however, the control and heat-selected populations did show a moderate positive correlation in gene expression changes (R2 = 0.449, P < 0.0001), possibly highlighting a role for laboratory adaptation in driving the transcriptional changes among populations.
Fig. 3.
Differences in environmental effects on gene expression across populations. Population differences in parental environment effects (panels A-C) on gene expression and larval environment effects (D-F) on expression are shown for each pairwise combination of experimental populations (ancestor, control, and selection for heat-shock resistance). Only genes that were differentially expressed (FDR < 5%) for each comparison are shown (gray), with heat shock proteins highlighted in red. Lines in each panel indicate the linear fit from the regression model (±95% CI). All correlations are P < 0.0001.
In contrast to the transgenerational influences, the influence of larval environment on transcription was strongly positively correlated among all pairs of populations (Fig. 3D-F). This pattern confirms the results that we have previously reported for these populations [20]. We also examined the changes in expression for several families of heat-shock proteins (HSPs), many of which show strong responses to larval heat stress in our populations. The HSPs also conformed to the general pattern observed among pairs of populations, and these genes tended to be differentially regulated across larval environments in a similar fashion in all populations.
3.5 Experimental evolution of regulatory pathways affected by heat stress
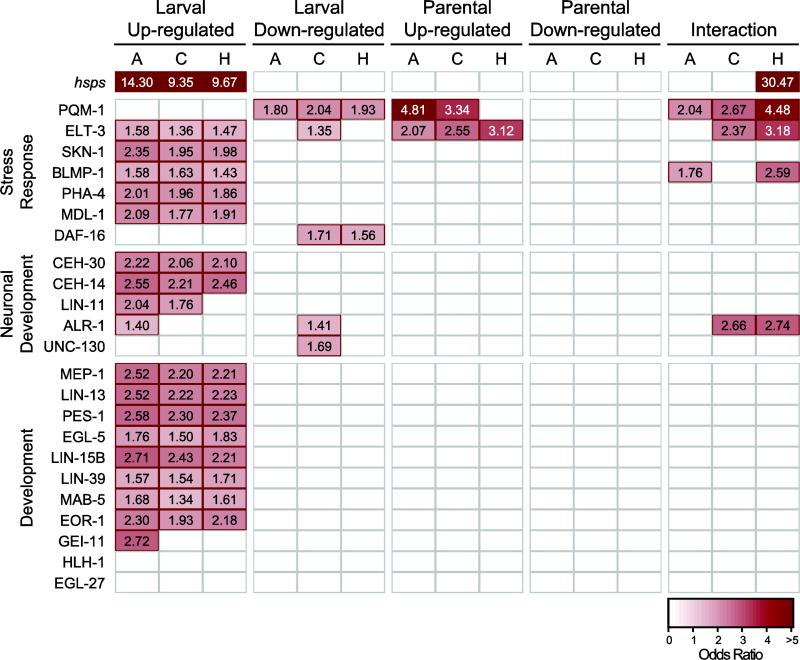
In C. elegans, several transcription factors are known to be critical regulators of cellular responses to stress [26,27]. However, these regulators may not be differentially expressed in response to stress themselves, but rather undergo protein modifications to activate them under certain conditions. For example, the FOXO transcription factor DAF-16 is a major target of the insulin/insulin-like growth factor signaling (IIS) pathway in worms, and is responsible for mediating responses to heat and oxidative stress, among others [28,29]. DAF-16 is normally localized in the cytoplasm, but in stress conditions, DAF-16 is activated and transported to the nucleus, where it regulates transcription of many target genes [30,31]. We identified C. remanei homologs of known binding targets of 23 transcription factors with known roles in development and homeostasis [32], and tested for significant enrichment among the up- and down-regulated gene sets from each population. We also tested for enrichment of the heat shock proteins, as this set of genes exhibits a well-characterized response to increases in temperature in C. elegans and other model organisms.
Not surprisingly, we observed strong enrichment (Fisher's Exact Test, P < 0.002) of HSPs that are up-regulated under larval stress (Fig. 4). In addition, the genes up-regulated under larval stress were also highly enriched for regulatory targets of most of the transcription factors for which binding data was available (Fig. 4), indicating that larval stress had broad effects on developmental programs as well as stress response pathways. These effects were observed in the evolved populations as well as in the ancestor. The genes significantly down-regulated under larval stress tended to be enriched for targets of two transcription factors with important roles in mediating stress response: DAF-16 and PQM-1. PQM-1 is a C2H2 zinc finger and leucine zipper-containing protein [33]. In C. elegans, PQM-1 is responsive to certain types of oxidative stress [33] and, together with DAF-16, is a key regulatory target of the insulin-like signaling pathway [34].
Fig. 4.
Enrichment of regulatory targets of key transcription factors among differentially expressed genes. Red boxes indicate significant enrichment (one-tailed Fisher's exact test; P < 0.002) within each set of up- or down-regulated genes, and odds ratios for each significantly enriched set are given. Transcription factors with described roles in mediating stress response, dauer formation, or longevity are classified as potential stress response factors, but may also have other important roles in development. Proteins that influence the specification and development of sensory neurons are classified here as neuronal development factors. A, ancestor; C, control evolved population; H, heat-selected population.
In contrast, the genes with increased expression across parental environments only showed significant enrichment of targets of two transcription factors. PQM-1 targets were enriched in this group, as were targets of ELT-3, a GATA transcription factor that functions during hypodermal development in C. elegans [35], which may also function downstream of IIS to influence longevity [36], pathogen resistance [37], and osmotic stress response [38]. Interestingly, although there was relatively strong enrichment for PQM-1 targets in the ancestor and control populations (odds ratio of 4.81 and 3.34, respectively), the heat-selected population showed no significant enrichment of this group, consistent with a loss of transgenerational effects within these populations.
Genes with significant parent-offspring interaction effects were also significantly enriched for targets of several of the transcription factors. Targets of PQM-1 and ELT-3 were also enriched among this group. In the lab-adapted populations, we also observed enrichment of ALR-1 targets, an ARX homolog involved in the development of touch receptor neurons in C. elegans [39,40]. BLMP-1 targets were also enriched among the genes with significant interactions. While BLMP-1 is an important regulator of developmental timing in C. elegans [41,42], it also is expressed in the sensory neurons in the amphids and phasmids [43], and acts downstream of the IIS and TGF-β pathways to trigger the formation of stress-resistant dauer larvae and influence longevity [41].
3. Discussion
Organisms are often faced with variable environments and have evolved adaptive strategies, such as behavioral or phenotypic plasticity, to cope with changing conditions. In many cases not only does the present generation respond to the changing environment, but previous generations—parents, grandparents, or even great-grandparents—can influence their descendants’ response to future stress [8]. If the parents’ experience accurately predicts the conditions their offspring will face, then such transgenerational plasticity can benefit the offspring and thereby lead to adaptive evolution [13,18].
While some evidence exists for the transmission of anticipatory plasticity from parent to offspring, the general pattern across plants and animals appears to be quite weak [16]. Our results highlight at least one reason why this might be the case. It makes intuitive sense that parental environment could provide an important cue for offspring environment. Equally important, however, the environment experienced by parents has the potential to directly influence the parental phenotype, which in turn can influence offspring quality [17,18]. Here, we find a complex interaction between parental environment and offspring outcome. Parents raised at high temperatures give rise to offspring that fare more poorly when reared in high temperatures, whereas the converse is true for parents and offspring raised at more permissive temperatures (Fig. 1). In other words, parental exposure to heat stress drastically degrades the normally robust hormetic response of larvae that increases resistance to heat shock after initial exposure to a mild heat stress. One reasonable explanation for this finding is that parents raised at high temperatures experience levels of stress that are sufficiently high to not only up-regulate appropriate stress pathways, but also to generate significant damage within the parents themselves, leading to the net effect of lowering offspring quality in stressful environments. While it is temping to focus specifically on maternal effects in this context, it has recently been shown that C. elegans males can transfer mRNA to eggs during fertilization [44], providing at least one plausible alternative mechanism. However, more likely are direct maternal influences on offspring performance. This could be something as straightforward as heat-stressed mothers providing less or lower quality yolk to her eggs or heat damage to maternally provided proteins [45], to something more complex such as dis-regulation of maternal mRNA provisioning to the eggs [46,47]. A more careful analysis of the phenotypes of eggs and early stage larvae from stressed parents may provide more direct insights into these effects.
Interestingly, the negative effects of stress exposure during the parental generation are lost during the response to selection to continued high heat stress during the early larval period (Fig. 1). Importantly, at no time during the experimental evolution did the parents experience high temperature conditions, as all individuals were reared at 20°C except during the brief periods of larval heat shock. Thus the observed change in transgenerational plasticity results from a correlated response to direct selection on stress resistance in the offspring. A correlated response of this sort could arise from a genetic correlation between traits associated with the parental effects and the offspring trait under selection [9]. This overall effect is very similar to the pattern of genetic assimilation for plasticity to temperature shifts in the offspring environment (Fig. 1; [20]). One characterization of the larval heat shock response is that it became more robust (less variable) to variation in both parental and larval environments over the course of selection for increased resistance. Thus, transgenerational parental effects have also become “assimilated”, although in the opposite direction of larval phenotypic plasticity. Although we have emphasized the negative effects of parents exposed to high heat when the larvae also experience high heat, larvae raised at lower temperatures whose parents have been raised at high temperatures do in fact perform better than worms with no history of exposure to heat. Thus there is some potential for additional protection from parental signaling, and consistent regulation of these effects may be the source of the assimilated parental response.
What is the underlying functional basis of these responses? The effects could be mediated through transcriptional differences or alternative effects. For example, in C. elegans maternally controlled sugar deposition and homeostasis in the egg can tradeoff against an offspring's capacity to respond to anoxic stress [48] (see also [49]). A parental effect of this sort does not necessarily require a transcriptional response of the offspring. Here, examination of larval transcriptomes resulting from all possible combinations of parental and larval environments for both ancestral and evolved populations yields results that are largely compatible with our phenotypic observations. First, as previously observed, there was a large transcriptional response caused by the change in larval environment (Fig. 3; [20]). This response remains largely consistent even after selection for increased heat shock resistance had shifted the pattern of plasticity observed at the whole-phenotype level.
In contrast to the direct effects of the larval rearing environment, a much smaller fraction of the genome appears to be involved in the response to differences in parental environment. Perhaps most interesting, there is little correlation between the transcriptional response observed by shifting the parental environment and that observed by an equivalent shift in the larval environment (Fig. 2). This lack of correspondence becomes even more marked after ten generations of experimental evolution. Thus the parental and larval responses are largely decoupled from one another, which is consistent with our observation of a lack of anticipatory phenotypic plasticity for heat shock resistance in the ancestor and the latter disappearance of any parental influence in the selection lines. Interestingly, both the control and heat shock selected lines actually evolve to become more similar to one another in terms of their responses to changes in the parental environment. The cause of this effect is currently unclear, but must involve novel adaptation to the laboratory environment.
Analysis of specific genetic pathways that might be involved in these responses yields a similar picture. In C. elegans, the heat stress response is known to involve a number of interacting neuroendocrine signaling systems [26,27]. One would therefore expect differential regulation of stress response pathways in the face of increased temperature stress, which we do indeed observe (Fig. 4). Strikingly, however, nearly every biological system responds strongly to increases in temperature during the larval period, so this is a highly generalized response. In contrast, only a few specific stress response systems appear to be induced via transgenerational influences (e.g., PGM-1; [34]). These same systems also tend to be implicated in the observed negative interaction between parental and larval temperature effects (Fig. 4). Because the first phases of development are initiated before the egg is even laid, it is actually impossible for us to strictly separate parental effects from larval effects very early in development. However, the lack of correspondence between the parental and larval responses, which are generated just a few hours later in development, strongly suggests that these are unlikely to be solely larval responses.
4. Conclusion
While it seems obvious to assume that parents would want to signal to their offspring so as to prepare them for the potential of coming stresses, in reality the evolution of this capacity depends on the predictability of environmental cues over time and the balance of interests between parental and offspring fitness [50]. For natural populations of the nematode C. remanei, this balance appears to have swung toward the parents, such that parents that experience stressful environments produce offspring that are less capable of responding to changes in their environment than parents reared in a benign environment. This lack of anticipatory signal is reflected both in the phenotypic response of heat shock resistance and in the transcriptional response of the offspring. Selection for increased larval stress resistance essentially wipes out any signal of parental influence, as offspring evolve a robust resistance to elevated temperatures that is independent of parental influences. Transgenerational effects are likely to be important for a wide variety of biological systems. It is clear from this work that (1) they should not be assumed to be strictly adaptive and (2) they can be rapidly labile to evolutionary change. Experimental evolution coupled with comprehensive genomic analyses should be a powerful tool for examining these questions in greater depth.
5. Methods
5.1 Experimental evolution
All populations of C. remanei described were maintained on Nematode Growth Medium-lite (NGM-lite, U.S. Biological) seeded with Escherichia coli strain OP50 [51]. The ancestral population used in this study was created as described previously [20]. Briefly, 26 isofemale strains were derived from natural isolates collected in Ontario, Canada. These strains were crossed together in a controlled fashion to promote equal genetic contributions from each strain, resulting in a genetically heterogeneous cohort that was representative of the natural genetic variation within the natural population. The resulting population (C. remanei strain PX443) was cryogenically preserved after it was created. Frozen stocks of this ancestral population were later thawed for use in experimental evolution and phenotyping assays. Experimentally evolved populations were subjected to ten generations of heat shock at an average initial temperature of 36.8°C, whereas control populations were maintained in a similar fashion, but without being subjected to a heat shock (described in [20]).
5.2 Heat stress response phenotype and parental effects
We used the ancestor, one representative control population, and one heat-selected population to test for parental effects on heat stress resistance. Frozen stocks of worms that had undergone 10 generations of acute selection were thawed at 20°C and allowed to recover for two generations prior to phenotyping to minimize effects attributable to freezing. In the third generation post-thaw, each population was divided into two different parental environments. One subset was maintained in the standard lab environment at 20°C, while the remaining worms were transferred to a mildly stressful heat environment at 30°C. When a population had produced eggs, worms were stage-synchronized by treating the population with a bleach solution [52]. After bleaching, half of the eggs recovered from each parental treatment were kept in buffer at 20°C until hatching (approximately 20-24 hours), while the remainder were kept at 30°C. This constitutes the “larval environment” for each population. Note that the parental environment was also experienced by the embryonic worms (up to ~30 cells), before the eggs were laid and age synchronization occurs, and thus may have induced a combination of parental and embryonic effects on stress resistance. After hatching, worms in L1 diapause suspended in liquid buffer were exposed to an acute heat shock at an average temperature of 36.8°C as described in [20].
5.3 Statistical analysis of phenotypic results
Heat stress resistance is best interpreted as the proportion of individuals surviving following acute heat shock. We assumed that the total number of individuals in each trial was equal to the average count from the three control plates from the same line that were subjected to a mock treatment. In any case in which the number of surviving worms from the shock treatment was greater than this total, the number of survivors was assumed to be equal to the total (100% survival). We tested for differences in heat shock resistance using a generalized linear model (GLM) implemented in the lme4 package [53] in the R statistical environment [54]. Three factors—selection regime, parental environment, and larval environment—were tested in the full model, as well as their interactions. We used a logit link function and quasi-binomial error distribution, to allow for overdispersion in the response variable. In addition, we tested for interactions between the parental and larval environments within each population separately, using a similar set of reduced models.
5.4 Transcriptional profiling of populations
Tissue samples for transcriptional profiling were obtained from each of the populations that were used for phenotyping. Frozen stocks of worms from each population were thawed at 20°C, and were maintained at that temperature until the population size was at least 200,000 individuals. At that time, a subset of the population was transferred to a 30°C incubator, while the remaining worms were kept at 20°C (the parental environment). Worms were then allowed to lay eggs, which were stage-synchronized as previously described by incubating for 20 hours in liquid medium without food. Half of the embryos from each parental environment developed at 20°C during this stage-synchronization period, whereas the remainder developed at 30°C. We defined the larval environment as the temperature experienced by the worms during the 20-hour stage-synchronization. After synchronization, larval worms were passed through a 20-μm Nitex screen to filter out any unhatched eggs and dead adults. Approximately 15 μl of L1 tissue (~100,000 individuals) was flash-frozen in TRIzol (Ambion). Total RNA was isolated from tissue samples and Illumina sequencing libraries were prepared as described in [20]. For the 20°C parental condition, we collected 6 replicate samples per larval treatment from at least two independently thawed vials for each line. These samples were the same as those previously analyzed in [20]. In the 30°C parental condition, 2 replicate samples were collected for each larval treatment from one of the thaws used for the 20°C parental treatment. All samples were sequenced in five lanes on an Illumina HiSeq 2000 at the University of Oregon Genomics Core Facility.
5.5 Differential expression analysis
Quality filtering of raw sequence reads was performed using the process_shortreads component of the Stacks software package [55,56]. Reads were discarded if they failed the Illumina chastity filter, had uncalled bases, or if the sample identity could not be determined because the barcode was unidentified. Reads with ambiguous barcodes were rescued if they had no more than one mismatch from a known barcode. We aligned all reads that passed the quality filters against a new C. remanei reference genome assembled from C. remanei strain PX356 (Fierst et al, in preparation) using GSNAP [57]. We used annotated gene models for this assembly to guide the alignment of sequence reads across the exon boundaries, but allowed GSNAP to also identify novel splice sites. We used the htseq-count tool from the Python package HTSeq (http://www-huber.embl.de/users/anders/HTSeq/) to count reads that aligned to annotated protein-coding genes. Reads were counted using the “union” mode in htseq-count, which only counts reads if they overlap a single gene model unambiguously.
We performed independent filtering of the dataset prior to model fitting [58] to exclude the 40% of genes with the lowest variance across all samples from all subsequent analyses. This filtering strategy improves power by removing the genes—and therefore unnecessary multiple tests—that were undetected in all samples and unlikely to be statistically different across treatments. Differential expression analysis was conducted in R, using the DESeq2 package [59,60], which uses a negative binomial error distribution to test for differential gene expression. Library size for each replicate sample was normalized using DESeq based on the entire dataset, however each population was analyzed separately. The effects of the parental environment, larval environment, and their interaction were included in the full model. Significance values for differential expression were adjusted for multiple comparisons using the Benjamini-Hochberg method [61], and model effects were deemed significant if they reached a 5% false discovery rate (FDR).
To compare transcriptional plasticity among different treatments and different populations, we fit an ordinary least squares (OLS) linear model to the log2 transformed fold change in expression of the significantly differentially expressed genes in each pairwise comparison of interest. Within each population, we compared the expression change attributable to the larval environment against the expression change due to parental environment in this way. We also compared the fold change attributed to each treatment (larval or parental environment) between each pair of populations using a similar analysis.
5.6 Enrichment of transcription factor targets
To understand how parental or larval stress affects major developmental pathways, we looked for enrichment of gene targets for several transcription factors that have critical roles in development or stress response. In each set of differentially expressed genes for each population, we tested for enrichment of known regulatory targets of 23 transcription factors for which binding data is available for the related nematode C. elegans. Binding targets for all transcription factors except for the FOXO transcription factor DAF-16 were obtained from the C. elegans modENCODE project [32]. These targets were all identified from chromatin immunoprecipitation sequencing (ChIP-seq). Putative target genes bound by DAF-16 have been previously identified using two different approaches: ChIP [62] and DNA adenine methyltransferase identification (DamID) [63]. We included all genes bound by DAF-16 within their promoter region from both studies as possible DAF-16 targets. Putative transcription factor target genes could be included multiple gene sets, if they are bound by more than one factor.
C. remanei homologs for each of the C. elegans transcription factor targets were determined based on the annotations that have been curated in the WS220 release of WormBase [64]. Homologous genes identified by any method were included as possible transcription factor targets in C. remanei. In cases where multiple C. remanei genes were matched to a single gene in C. elegans, all possible homologous genes were included in the gene set, since no information was available to determine whether transcription factor binding was preserved preferentially in either possible homolog.
Modules were tested for significant enrichment of target genes bound by each transcription factor using a one-tailed Fisher's exact test. In addition, we tested for enrichment of the C. remanei heat shock proteins examined previously [20]. Since a total of 24 gene sets were tested, we determined that there was significant enrichment by using a Bonferroni corrected α-value of P < 0.05 / 24 (<0.002).
Acknowledgments
We thank the many people who have assisted with maintaining the selection lines, and to Janna Fierst for support in the use of her preliminary reassembly of the C. remanei genome.
Funding
This work was supported by a GRFP and DDIG (1210922) from the National Science Foundation to KLS, a Ruth L. Kirschstein NRSA Postdoctoral Fellowship to RMR (AG032900), National Institutes of Health grants to PCP (AG022500, AG043988, GM096008) and WAC (RR032670), and the Ellison Medical Foundation fellowship to PCP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Database Links
RNA-seq data are deposited in the NCBI Gene Expression Omnibus (GEO) database as part of series GSE56510 with accession numbers GSM1362987–1363022.
References
- 1.Bennett AF, Lenski RE. An experimental test of evolutionary trade-offs during temperature adaptation. Proc. Natl. Acad. Sci. U.S.a. 2007;104(Suppl 1):8649–8654. doi: 10.1073/pnas.0702117104. doi:10.1073/pnas.0702117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett AF, Lenski RE, Mittler JE. Evolutionary adaptation to temperature. I. Fitness responses of Escherichia coli to changes in its thermal environment. Evolution. 1992;46:16. doi: 10.1111/j.1558-5646.1992.tb01981.x. doi:10.2307/2409801. [DOI] [PubMed] [Google Scholar]
- 3.Cooper VS, Bennett AF, Lenski RE. Evolution of thermal dependence of growth rate of Escherichia coli populations during 20,000 generations in a constant environment. Evolution. 2001;55:889–896. doi: 10.1554/0014-3820(2001)055[0889:eotdog]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 4.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 2010;20:R208–15. doi: 10.1016/j.cub.2009.11.055. doi:10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Messer PW, Petrov DA. Population genomics of rapid adaptation by soft selective sweeps. Trends in Ecology & Evolution. 2013;28:659–669. doi: 10.1016/j.tree.2013.08.003. doi:10.1016/j.tree.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilbert DW, Piggot PJ. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 2004;68:234–262. doi: 10.1128/MMBR.68.2.234-262.2004. doi:10.1128/MMBR.68.2.234-262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badyaev AV, Uller T. Parental effects in ecology and evolution: mechanisms, processes and implications. Philosophical Transactions of the Royal Society B: Biological Sciences. 2009;364:1169–1177. doi: 10.1098/rstb.2008.0302. doi:10.1098/rstb.2008.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton T, Metcalfe NB. Can environmental conditions experienced in early life influence future generations? Proceedings of the Royal Society B: Biological Sciences. 2014;281:20140311–20140311. doi: 10.1098/rspb.2014.0311. doi:10.1111/j.1365-294X.2011.05402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkpatrick M, Lande R. The evolution of maternal characters. Evolution. 1989;43:485. doi: 10.1111/j.1558-5646.1989.tb04247.x. doi:10.2307/2409054. [DOI] [PubMed] [Google Scholar]
- 10.Wolf JB, E.D.B. The coadaptation of parental and offspring characters. Evolution. 1998;52:299. doi: 10.1111/j.1558-5646.1998.tb01632.x. doi:10.2307/2411068. [DOI] [PubMed] [Google Scholar]
- 11.Mousseau TA, Dingle H. Maternal effects in insect life histories. Annual Review of Entomology. 1991;36:511–534. [Google Scholar]
- 12.Roach DA, Wulff RD. Maternal effects in plants. Annual Review of Ecology and Systematics. 1987:209–235. doi:10.1152/physiolgenomics.00052.2004. [Google Scholar]
- 13.Mousseau TA, Fox CW. The adaptive significance of maternal effects. Trends in Ecology & Evolution. 1998;13:403–407. doi: 10.1016/s0169-5347(98)01472-4. [DOI] [PubMed] [Google Scholar]
- 14.Ho DH, Burggren WW. Epigenetics and transgenerational transfer: a physiological perspective. Journal of Experimental Biology. 2010;213:3–16. doi: 10.1242/jeb.019752. doi:10.1242/jeb.019752. [DOI] [PubMed] [Google Scholar]
- 15.Calabrese EJ, Baldwin LA. Chemical hormesis: its historical foundations as a biological hypothesis. Hum Exp Toxicol. 2000;19:2–31. doi: 10.1191/096032700678815585. [DOI] [PubMed] [Google Scholar]
- 16.Uller T, Nakagawa S, English S. Weak evidence for anticipatory parental effects in plants and animals. J Evolution Biol. 2013;26:2161–2170. doi: 10.1111/jeb.12212. doi:10.1111/jeb.12212. [DOI] [PubMed] [Google Scholar]
- 17.Bernardo J. The particular maternal effect of propagule size, especially egg size: patterns, models, quality of evidence and interpretations. American Zoologist. 1996;36:216–236. [Google Scholar]
- 18.Marshall DJ, Uller T. When is a maternal effect adaptive? Oikos. 2007;116:1957–1963. [Google Scholar]
- 19.Bennett AF, Lenski RE. Evolutionary adaptation to temperature II. Thermal niches of experimental lines of Escherichia coli. Evolution. 1993:1–12. doi: 10.1111/j.1558-5646.1993.tb01194.x. [DOI] [PubMed] [Google Scholar]
- 20.Sikkink KL, Reynolds RM, Ituarte CM, Cresko WA, Phillips PC. Rapid evolution of phenotypic plasticity and shifting thresholds of genetic assimilation in the nematode Caenorhabditis remanei. G3 (Bethesda) 2014;4:1103–1112. doi: 10.1534/g3.114.010553. doi:10.1534/g3.114.010553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray JC, Cutter AD. Mainstreaming Caenorhabditis elegans in experimental evolution. Proceedings of the Royal Society B: Biological Sciences. 2014;281:20133055. doi: 10.1098/rspb.2013.3055. doi:10.1098/rspb.2013.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jovelin R, Ajie BC, Phillips PC. Molecular evolution and quantitative variation for chemosensory behaviour in the nematode genus Caenorhabditis. Mol Ecol. 2003;12:1325–1337. doi: 10.1046/j.1365-294x.2003.01805.x. [DOI] [PubMed] [Google Scholar]
- 23.Cutter AD, Baird SE, Charlesworth D. High nucleotide polymorphism and rapid decay of linkage disequilibrium in wild populations of Caenorhabditis remanei. Genetics. 2006;174:901–913. doi: 10.1534/genetics.106.061879. doi:10.1534/genetics.106.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jovelin R, Dunham JP, Sung FS, Phillips PC. High nucleotide divergence in developmental regulatory genes contrasts with the structural elements of olfactory pathways in Caenorhabditis. Genetics. 2009;181:1387–1397. doi: 10.1534/genetics.107.082651. doi:10.1534/genetics.107.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waddington CH. Genetic assimilation of an acquired character. Evolution. 1953;7:118–126. doi:10.2307/2405747. [Google Scholar]
- 26.Morimoto RI. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 2011;76:91–99. doi: 10.1101/sqb.2012.76.010637. doi:10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez M, Snoek LB, De Bono M, Kammenga JE. Worms under stress: C. elegans stress response and its relevance to complex human disease and aging. Trends Genet. 2013;29:367–374. doi: 10.1016/j.tig.2013.01.010. doi:10.1016/j.tig.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- 29.Hsu A-L, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. doi:10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 30.Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Current Biology. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 31.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. doi:10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 32.Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2011;21:245–254. doi: 10.1101/gr.114587.110. doi:10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tawe WN, Eschbach ML, Walter RD, Henkle-Dührsen K. Identification of stress-responsive genes in Caenorhabditis elegans using RT-PCR differential display. Nucleic Acids Research. 1998;26:1621–1627. doi: 10.1093/nar/26.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, Bussemaker HJ. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013;154:676–690. doi: 10.1016/j.cell.2013.07.006. doi:10.1016/j.cell.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilleard JS, Shafi Y, Barry JD, McGhee JD. ELT-3: A Caenorhabditis elegans GATA factor expressed in the embryonic epidermis during morphogenesis. Developmental Biology. 1999;208:265–280. doi: 10.1006/dbio.1999.9202. doi:10.1006/dbio.1999.9202. [DOI] [PubMed] [Google Scholar]
- 36.Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, et al. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. doi:10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pujol N, Zugasti O, Wong D, Couillault C, Kurz CL, Schulenburg H, et al. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathogens. 2008;4:e1000105. doi: 10.1371/journal.ppat.1000105. doi:10.1371/journal.ppat.1000105.g006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohlfing A-K, Miteva Y, Hannenhalli S, Lamitina T. Genetic and physiological activation of osmosensitive gene expression mimics transcriptional signatures of pathogen infection in C. elegans. PLoS ONE. 2010;5:e9010. doi: 10.1371/journal.pone.0009010. doi:10.1371/journal.pone.0009010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melkman T, Sengupta P. Regulation of chemosensory and GABAergic motor neuron development by the C. elegans Aristaless/Arx homolog alr-1. Development. 2005;132:1935–1949. doi: 10.1242/dev.01788. doi:10.1242/dev.01788. [DOI] [PubMed] [Google Scholar]
- 40.Tucker M, Sieber M, Morphew M, Han M. The Caenorhabditis elegans aristaless orthologue, alr-1, is required for maintaining the functional and structural integrity of the amphid sensory organs. Mol. Biol. Cell. 2005;16:4695–4704. doi: 10.1091/mbc.E05-03-0205. doi:10.1091/mbc.E05-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horn M, Geisen C, Cermak L, Becker B, Nakamura S, Klein C, et al. DRE-1/FBXO11-dependent degradation of BLMP-1/BLIMP-1 governs C. elegans developmental timing and maturation. Developmental Cell. 2014;28:697–710. doi: 10.1016/j.devcel.2014.01.028. doi:10.1016/j.devcel.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang T-F, Cho C-Y, Cheng Y-T, Huang J-W, Wu Y-Z, Yeh AY-C, et al. BLMP-1/Blimp-1 regulates the spatiotemporal cell migration pattern in C. elegans. PLoS Genet. 2014;10:e1004428. doi: 10.1371/journal.pgen.1004428. doi:10.1371/journal.pgen.1004428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reece-Hoyes JS, Shingles J, Dupuy D, Grove CA, Walhout AJM, Vidal M, et al. Insight into transcription factor gene duplication from Caenorhabditis elegans promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. doi:10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoeckius M, Grun D, Kirchner M, Ayoub S, Torti F, Piano F, et al. Global characterization of the oocyte-to-embryo transition in Caenorhabditis elegans uncovers a novel mRNA clearance mechanism. The EMBO Journal. 2014;33:1751–1766. doi: 10.15252/embj.201488769. doi:10.15252/embj.201488769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimble J, Sharrock WJ. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Developmental Biology. 1983;96:189–196. doi: 10.1016/0012-1606(83)90322-6. [DOI] [PubMed] [Google Scholar]
- 46.Wood WB, Hecht R, Carr S, Vanderslice R, Wolf N, Hirsh D. Parental effects and phenotypic characterization of mutations that affect early development in Caenorhabditis elegans. Developmental Biology. 1980;74:446–469. doi: 10.1016/0012-1606(80)90445-5. [DOI] [PubMed] [Google Scholar]
- 47.Mains PE, Sulston IA, Wood WB. Dominant maternal-effect mutations causing embryonic lethality in Caenorhabditis elegans. Genetics. 1990;125:351–369. doi: 10.1093/genetics/125.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frazier HN, III, Roth MB. Adaptive sugar provisioning controls survival of C. elegans embryos in adverse environments. Current Biology. 2012:1–5. doi: 10.1016/j.cub.2009.03.066. doi:10.1016/j.cub.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harvey SC, Orbidans HE. All eggs are not equal: the maternal environment affects progeny reproduction and developmental fate in Caenorhabditis elegans. PLoS ONE. 2011;6:e25840. doi: 10.1371/journal.pone.0025840. doi:10.1371/journal.pone.0025840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burgess SC, Marshall DJ. Adaptive parental effects: the importance of estimating environmental predictability and offspring fitness appropriately. Oikos. 2014:no–no. doi:10.1111/oik.01235. [Google Scholar]
- 51.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. doi:10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bates D, Maechler M, Bolker B, Walker S. lme4: Linear mixed-effects models using Eigen and S4. 2014 Accessed Online: December. [Google Scholar]
- 54.R Development Core Team R: A language and environment for statistical computing. 2014 [Google Scholar]
- 55.Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. Stacks: building and genotyping loci de novo from short-read sequences. G3 (Bethesda) 2011;1:171–182. doi: 10.1534/g3.111.000240. doi:10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. doi:10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. doi:10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bourgon R, Gentleman R, Huber W. Independent filtering increases detection power for high-throughput experiments. Proceedings of the National Academy of Sciences. 2010;107:9546–9551. doi: 10.1073/pnas.0914005107. doi:10.1073/pnas.0914005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. doi:10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. bioRxiv. 2014 doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995:289–300. [Google Scholar]
- 62.Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. doi:10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 63.Schuster E, McElwee JJ, Tullet JMA, Doonan R, Matthijssens F, Reece-Hoyes JS, et al. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol. 2010;6:399. doi: 10.1038/msb.2010.54. doi:10.1038/msb.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Research. 2010;38:D463–7. doi: 10.1093/nar/gkp952. doi:10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]