SUMMARY
Conserved, multi-tasking DNA helicases mediate diverse DNA transactions and are relevant for human disease pathogenesis. These helicases and their regulation help maintain genome stability during DNA replication and repair. We show that the structural maintenance of chromosome complex Smc5-Smc6 restrains the replication fork regression activity of Mph1 helicase, but not its D-loop disruptive activity. This regulatory mechanism enables flexibility in replication fork repair without interfering with DNA break repair. In vitro studies find that Smc5-Smc6 binds to a Mph1 region required for efficient fork regression, preventing assembly of Mph1 oligomers at the junction of DNA forks. In vivo impairment of this regulatory mechanism compensates for the inactivation of another fork regression helicase and increases reliance on joint DNA structure removal or avoidance. Our findings provide molecular insights into replication fork repair regulation and uncover a role of Smc5-Smc6 in directing Mph1 activity towards a specific biochemical outcome.
INTRODUCTION
Successful completion of DNA replication requires pathways that protect, repair, or restart replication forks. These pathways and their regulation are indispensable for genome maintenance, and their dysregulation underlies the pathogenesis of cancer-prone diseases (Cox et al., 2000; Weinert et al., 2009). While many protein factors relevant to these pathways have been identified, how they act or regulate one another remains poorly understood. Insights into this question are crucial for understanding how the integrity of our genome is maintained.
Current evidence suggests that stalled or impaired replication forks can be processed in multiple ways that lead to different outcomes. One of the least understood routes is the regression of replication forks. Specialized DNA motor proteins can generate regressed DNA replication forks in vitro and in cells (e.g. Betous et al., 2012; Blastyak et al., 2007; Gari et al., 2008; Hu et al., 2012; Ralf et al., 2006; Sun et al., 2008; Zheng et al., 2011). While replication fork regression can facilitate continuity of replication when encountering an obstacle, such as by enabling lesion bypass or removal upon template strand reannealing, it can also lead to undesirable consequences. For instance, the four-way DNA junction stemming from fork regression can be processed by nucleases and/or break repair mechanism to result in illegitimate ligation or toxic intermediates (Cotta-Ramusino et al., 2005; Couch et al., 2013; Doe et al., 2002; Hu et al., 2012; Sun et al., 2008; Yeeles et al., 2013). Additionally, replication fork regression likely entails replisome disassembly, which presents a challenge to the resumption of DNA synthesis. For these reasons, alternative fork rescue mechanisms, such as translesion DNA synthesis, are thought to provide a safer means for lesion tolerance or removal while preserving replisome integrity. Conceptually, these other pathways would be facilitated by down-regulation of fork regression activity. Indeed, DNA damage checkpoint kinases can mitigate fork regression (Couch et al., 2013; Hu et al., 2012). Currently, little is known about fork regression regulatory mechanisms. To tackle this issue, we have examined the regulation of the budding yeast Mph1 helicase by the Smc5-Smc6 complex.
Mph1 and its orthologs, such as human FANCM, which is mutated in the cancer prone syndrome Fanconi anemia, and fission yeast Fml1, are multifunctional DNA motor proteins (Whitby, 2010). Fork regression by these enzymes is relevant for replication fork repair but can lead to the generation of recombination intermediates that are difficult to resolve (Chavez et al., 2011; Chen et al., 2009; Choi et al., 2010; Gari et al., 2008; Sun et al., 2008; Whitby, 2010; Zheng et al., 2011). These motor proteins are also capable of dissociating D-loops made by the Rad51 recombinase, leading to a non-crossover outcome in chromosomal break repair by homologous recombination (e. g. Crismani et al., 2012; Lorenz et al., 2012; Prakash et al., 2009; Sun et al., 2008). In mitotic cells, the D-loop dissociative activity of these proteins is favored since it minimizes the risk of the loss of heterozygosity, whereas replication fork regression catalyzed by them must be properly restrained to avoid the accumulation of toxic DNA joint molecules. How the replication fork regression activity of these proteins is specifically restrained in mitotic cells remains an open question. Interestingly, genetic and two-dimensional gel analyses have suggested a role for Smc5-Smc6, a conserved structural maintenance of chromosome complex, in Mph1 regulation (Chavez et al., 2011; Chen et al., 2009; Choi et al., 2010; Sun et al., 2008). Notably, mutations in Smc5-Smc6 lead to growth impairment, hypersensitivity to replicative stress, and the accumulation of joint DNA structures during replication (Chavez et al., 2011; Chen et al., 2009; Choi et al., 2010; Sollier et al., 2009; Torres-Rosell et al., 2007) Deletion of MPH1 or inactivation of Mph1 helicase activity alleviates these phenotypes in Smc5-Smc6 mutants, whereas overexpression of Mph1 exacerbates them (Chavez et al., 2011; Chen et al., 2009; Choi et al., 2010).
In this study, we elucidate the molecular mechanism by which Smc5-Smc6 regulates Mph1. Our results show that, via Smc5, Smc5-Smc6 interacts with and attenuates the replication fork regression activity of Mph1 without negatively impacting upon D-loop disruption catalyzed by it. Intriguingly, while the effect of Smc5 is reliant on Mph1 interaction, its DNA binding activity is dispensable for this functional attribute. By biochemical analyses, atomic force microscopy (AFM), and electron microscopy (EM), we furnish evidence that Smc5 interferes with Mph1 oligomer formation at the junction of a DNA fork structure. Our results not only offer mechanistic insights into how a conserved replication regression motor protein engages DNA forks and is regulated to minimize the generation of toxic DNA intermediates, but also reveal a novel strategy that SMC employs to channel an impaired replication fork into alternate pathways of repair and restart.
RESULTS
Regulation of Mph1 fork regression activity by Smc5-Smc6
To elucidate the action mechanism of Smc5-Smc6, we performed a battery of biochemical experiments using highly purified proteins. His6-Flag-tagged Mph1 expressed in insect cells, His6- or MBP-tagged Smc5 expressed in E. coli, and Strep-tagged Smc5, Smc6, and Smc5-Smc6 expressed in yeast were purified to greater than 85% homogeneity (Figures 1A and S1A, Supplemental Experimental Procedures). By affinity pulldown, we showed that Smc5 interacts directly with Mph1, confirming a previous finding (Figure S1B) (Chen et al., 2009). In addition, we found that Smc5-Smc6 retains this interaction while Smc6 does not bind Mph1 (Figure S1B). Thus, Smc5-Smc6 interacts with Mph1 through Smc5.
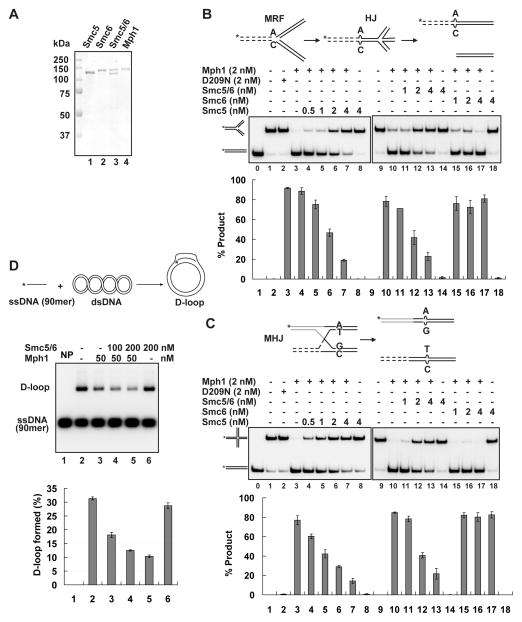
Figure 1. Inhibition of Mph1-catalyzed DNA fork regression and branch migration by Smc5-Smc6.
(A) Purified Smc5, Smc6, Smc5-Smc6, and Mph1. Coomassie Blue stain of the proteins after SDS-PAGE is shown. (B–C) The influence of Smc5, Smc6, and Smc5-Smc6 on Mph1-catalyzed regression of the movable replication fork (MRF, B) and branch migration of the movable Holliday junction (MHJ, C). DNA substrates were at 5 nM and reactions were incubated at 30 °C for 4 min. Schematics of the MRF, Holliday junction (HJ) intermediate, and final products (B) and the MHJ and final products (C) are shown on top. (D) Smc5-Smc6 does not affect Mph1-catalyzed D-loop dissociation. Rad51-mediated D-loop formation is shown on top. The D-loop dissociative activity of Mph1 was examined with and without Smc5-Smc6 after a 4-min incubation at 30 °C. NP, no protein control. Average of triplicates ± SD are graphed at the bottom in (B–D). See also Figure S1.
We first investigated how Smc5, Smc6, and Smc5-Smc6 influence the replication fork regression activity of Mph1. For this, we used a radiolabeled, oligonucleotide-based substrate resembling a replication fork, in which homology in the two duplex arms permits regression to occur (i.e. mobile replication fork structure or MRF). MRF regression mediated by Mph1 and its orthologs yields a Holliday junction (HJ) intermediate that is branch migrated to form two duplex molecules, one of which harbors the radiolabel (Figure 1B) (Gari et al., 2008; Sun et al., 2008; Zheng et al., 2011). In the presence of ATP, Mph1 catalyzes fork regression efficiently, such that 2 nM of the protein could convert ~ 90% of the substrate (5 nM) into product after 4 min of incubation (Figure 1B). Importantly, the addition of a stoichiometric quantity of Smc5 or Smc5-Smc6 (0.5–4 nM) led to increasingly strong inhibition of product formation (Figure 1B). For instance, 4 nM Smc5 or Smc5-Smc6, inhibited this reaction > 4 fold, while the same concentration of Smc6 had no effect. Smc proteins and the Smc complex are devoid of MRF-processing ability (Figure 1B), and, as expected (Zheng et al., 2011) and the helicase-defective mph1-D209N mutant protein is ineffective (Figure 1B).
We next asked whether Smc5, Smc6 and their complex would affect Mph1-dependent processing of the HJ, a structure derived from fork regression (Figure 1B). We employed a radiolabeled mobile HJ (MHJ), branch migration of which yields two duplexes with one harboring the radiolabel (Figure 1C) (Gari et al., 2008; Zheng et al., 2011). As for the MRF, stoichiometric amounts of Smc5 and Smc5-Smc6, but not Smc6, were able to strongly inhibit branch migration of the MHJ by Mph1 (Figure 1C). Neither the Smc species nor mph1-D209N dissociated the MHJ (Figure 1C). These results demonstrate that Smc5-Smc6 does not process MRF or MHJ structures but effectively restrains Mph1 from doing so, and that Smc5 is key to mediating this effect.
Specificity of Smc5-Smc6 action
To determine the specificity of the inhibitory action of Smc5 and Smc5-Smc6, we first tested how they affected FANCM and Rad54 that exhibit similar activities as Mph1 (Bugreev et al., 2006; Gari et al., 2008). Neither Smc5-Smc6 (Figures S1C–S1D) nor Smc5 (Figure S1E–S1F) had a significant effect on the processing of MRF and MHJ by FANCM or by Rad54. These results indicate that the inhibitory effect of Smc5-Smc6 and Smc5 on Mph1-mediated reactions does not stem from a nonspecific steric effect on substrate accessibility.
We then tested whether Smc5-Smc6 also affects the D-loop dissociation activity of Mph1 using oligonucleotide-based or Rad51-generated D-loops. By itself, Smc5-Smc6 could not disrupt these substrates and even appeared to enhance Mph1-mediated dissociation (Figure 1D and data not shown). This stands in contrast to the complex’s strong inhibitory effect on Mph1-mediated MRF and MHJ processing. Lastly, we found that Smc5-Smc6 exerts no effect on the 3′-5′ helicase activity of Mph1 and has no helicase activity on its own (data not shown).
Dependence of Smc5 action on Mph1 interaction
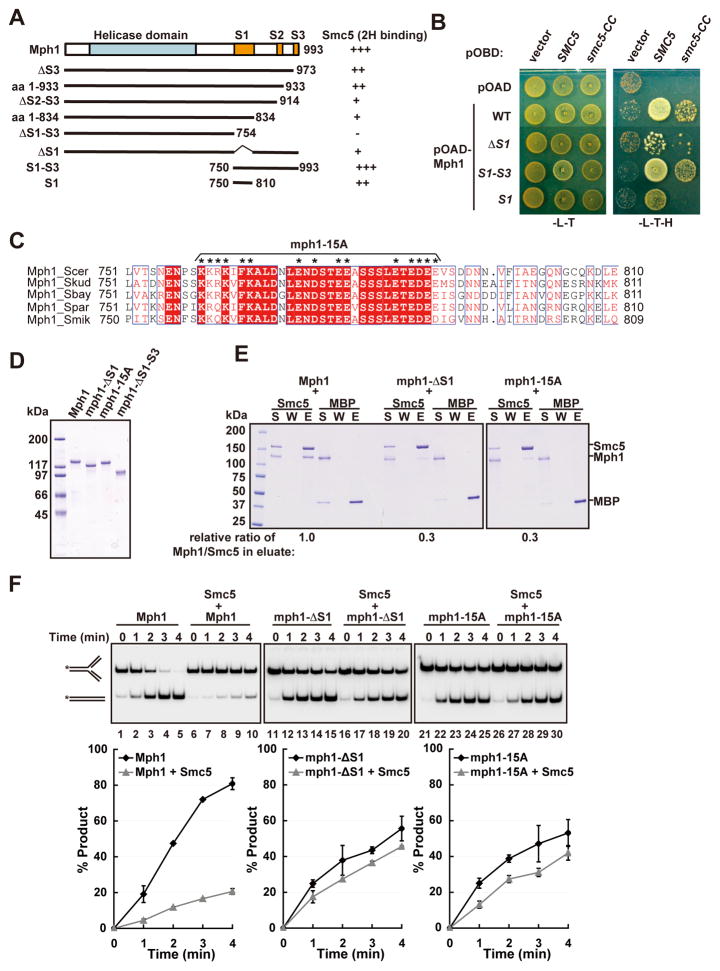
We strived to understand the mechanism by which Smc5-Smc6 negatively regulates Mph1 in fork regression and HJ branch migration. First, we asked if this regulation relies on physical interaction between Mph1 and Smc5. As summarized in Figure 2A and exemplified in Figure 2B, yeast two-hybrid analysis found that the region spanning amino acid residues 754-993 in Mph1 is both necessary and sufficient for binding Smc5 and a minimal Smc5 region critical for Mph1 interaction (smc5-CC, see below). Additional deletion analysis identified three segments - S1 (residues 750-810), S2 (residues 914-933), and S3 (residues 973-993) - involved in Smc5 interaction, as deletion of each segment reduced Smc5 binding, with S1 conferring partial interaction with Smc5 (Figures 2A and 2B).
Figure 2. Smc5 regulation of Mph1 is contingent upon their interaction.
(A) Yeast two hybrid result that delimit the Smc5 interaction domain in Mph1. The relative strength of the Mph1 species for Smc5 interaction is indicated. (B) Examples of yeast two hybrid assays involving Mph1 fragments with Smc5 or the coiled coil domain of Smc5 (smc5-CC). (C) Alignment of Mph1 S1 region from Saccharomyces species. Fifteen conserved residues changed to alanine in mph1-15A are indicated by asterisks. (D) Purified Mph1 and its three mutant variants. Coomassie Blue stain of the proteins after SDS-PAGE is shown. (E) Mutant mph1 proteins exhibit reduced Smc5 binding. Affinity pulldown results are shown and the relative ratio of Mph1 or its mutants to Smc5 in the eluate (E) is indicated. S: supernatant. W: wash. (F) Deleting or mutating the S1 region of Mph1 largely abolishes Smc5 inhibitory effect in Mph1-catalyzed MRF regression. Mph1 or its mutants (2 nM) with or without Smc5 (4 nM) were incubated with MRF (5 nM) at 30 °C for indicated time. Average of triplicates ± SD are graphed at the bottom. See also Figure S2 and Figure S3.
To ascertain the relevance of the Mph1-Smc5 interaction, we generated ΔS1 and ΔS1-S3 deletion mutants. In addition, as the S1 segment is conserved among Mph1 orthologues from Saccharomyces species particularly at hydrophobic and charged residues, some of these residues were replaced by alanine to yield the mph1-15A mutant (Figure 2C). Affinity pulldown using the three purified mph1 mutant proteins (Figure 2D) showed that mph1-ΔS1 and mph1-15A are significantly impaired for Smc5 binding (Figure 2E), and mph1-ΔS1-S3 is completely defective in this regard (Figure S2A).
We found that none of the above mutations affects the ATPase activity of Mph1 (Figure S2B), its D-loop dissociative activity (Figure S2C), or MRF binding (Figures S2D–S2E, and data not shown). In addition, they show very similar profiles as the wild-type protein when analyzed by circular dichroism (Figures S2G–S2H). These analyses demonstrate that the mutant mph1 proteins are properly folded. We did find that the mutants are moderately impaired for MRF regression activity (Figures 2F and S2I). Thus, the S1-S3 region appears to be required for maximal fork regression by Mph1 but not for DNA binding, ATPase, or D-loop disruption. As the S1-S3 region does not bind MRF (Figure S2F), its effect on replication fork regression is not due to DNA association. Importantly, replication fork regression by mph1-ΔS1 and mph1-15A is only slightly affected by Smc5 (Figure 2F), and mph1-ΔS1-S3 is unresponsive to Smc5 (Figure S2I). Similarly, MHJ branch migration mediated by these mutant proteins is either unresponsive or largely unaffected by Smc5 (Figure S2J–S2K). Thus, regulation of Mph1-mediated fork regression or branch migration is contingent upon physical interaction with Smc5 in a region of Mph1 required for maximal enzyme activity.
Like other members of the SMC family, Smc5 harbors three domains: a “hinge”, an extended anti-parallel coiled coil, and a globular “head” composed of the N- and C-termini (Figure S3A). Smc5 coiled coil fragments, but not the hinge or head region, interact with Mph1 in yeast two hybrid analysis (Figure S3A), and the larger coiled coil region is as proficient as full-length Smc5 in binding Mph1 or S1-S3 (Figures S3A and 2B). While this Smc5 fragment is refractory to purification, the coiled coil and hinge fragment (smc5-ΔNC) can be purified, and it associates with Mph1 with the same affinity as Smc5 (Figure S3B). Importantly, smc5-ΔNC reduces Mph1 fork regression activity as much as Smc5 (Figure S3C). Since smc5-ΔNC is devoid of any MRF binding ability while Smc5 has only a modest affinity for MRF (Figure S3D), inhibition of Mph1 is independent of DNA binding by Smc5. This conclusion is also consistent with the lack of interference by Smc5 in FANCM- and Rad54- mediated DNA processing (Figure S1E–S1F).
Interference of Mph1-substrate engagement by Smc5
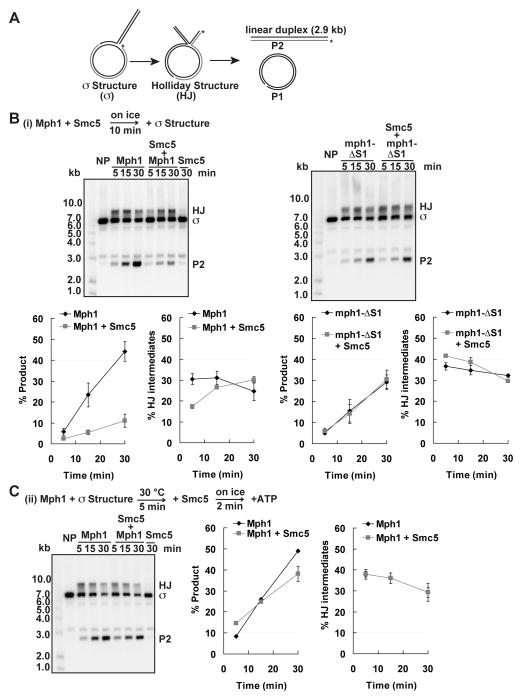
To obtain additional mechanistic insights, we examined a plasmid-based sigma-shaped structure that more closely resembles a replication fork (Blastyak et al., 2007; Ralf et al., 2006). With this substrate, fork regression yields a HJ, which can branch migrate over 2.9 kb to generate a radiolabeled linear product and an unlabeled circular one (Figure 3A). This large substrate is ideal for order-of-addition experiments that would allow us to distinguish between whether Smc5 inhibits Mph1 engagement with the substrate or interferes with Mph1 function at a later step. Pre-incubation of Mph1 with Smc5 led to a strong inhibition of product formation (Figure 3B, left). This inhibition again depends on Smc5-Mph1 interaction, as Smc5 had little effect on product formation by mph1-ΔS1 (Figure 3B, right). In contrast, Mph1 pre-bound to the sigma structure is unresponsive to Smc5 (Figure 3C). These observations suggest that Smc5 blocks the productive engagement of Mph1 with its substrate rather than antagonizing substrate-bound Mph1.
Figure 3. Inhibition of Mph1-catalyzed sigma structure processing by Smc5.
(A) Schematic of the plasmid-based sigma structure and its processing. Fork regression converts the sigma structure (σ) into a HJ, and complete branch migration yields a nicked circular duplex (P1) and radiolabeled linear duplex (P2). (B) Smc5 inhibits processing of the sigma structure by Mph1 but has no effect on the equivalent reaction mediated by mph1-ΔS1. Mph1 or mph1-ΔS1 (4 nM) were preincubated with Smc5 (40 nM) before adding 0.6 nM of the sigma substrate and incubation at 30 °C for 5, 15 and 30 min. (C) Pre-incubation of sigma structure substrate with Mph1 alleviates inhibition by Smc5. NP, no protein control. The values plotted are the averages of triplicates ± SD (B–C).
Prevention of Mph1 oligomerization at DNA junctions by Smc5
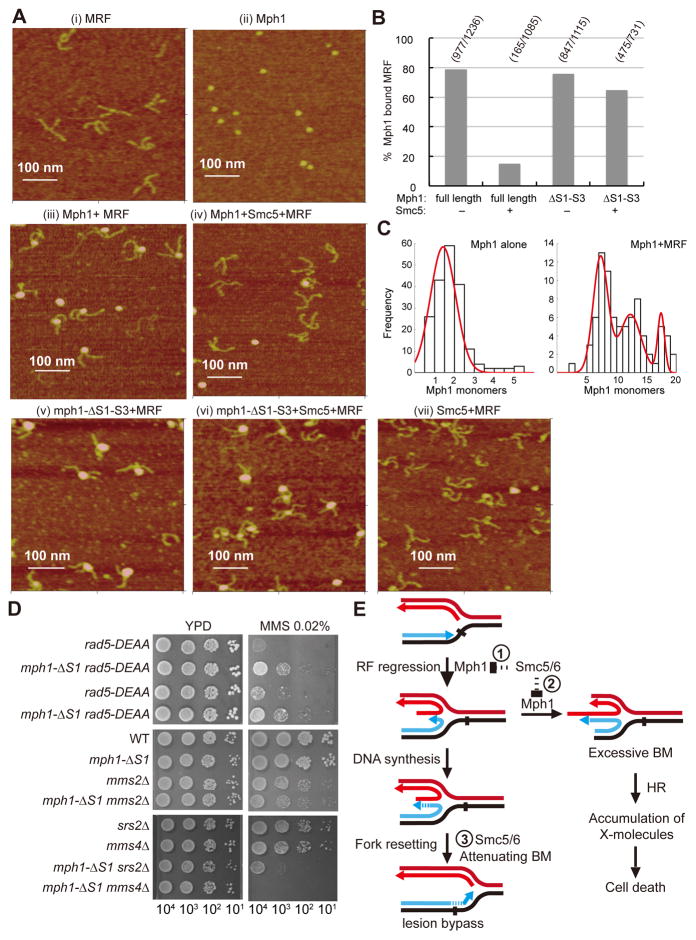
AFM allows direct imaging of DNA, proteins, and their complexes at the single molecule level (Hansma et al., 2004). MRF with two arms of 170 base pairs and one arm of 150 base pairs and monomeric Mph1 are clearly discernable by AFM (Figures 4A (i–ii) and S4A–S4B). When Mph1 was incubated with this substrate, ~80% of the DNA molecules became Mph1-bound at the junction point within the substrate (Figures 4A (iii) and 4B). Junction-bound Mph1 was larger than Mph1 alone (compare Figure 4A (ii) vs. (iii)). Volume analysis from AFM images indicated that Mph1 protein consists of a mixture of monomers and dimers (Figure 4C). In contrast, the predominant DNA junction bound Mph1 harbored 6–8 protomers with additional populations being equivalent to ~12 and ~18 protomers (Figure 4C). These analyses thus showed that Mph1 associates with the DNA junction of a MRF as defined multimeric complexes.
Figure 4. AFM analysis of the effect of Smc5 on Mph1 engagement with MRF and genetic tests of mph1-ΔS1.
(A) Representative AFM images are shown for the following: MRF alone (i), Mph1 alone (ii), Mph1 incubated with MRF (iii), Mph1 and Smc5 incubated with MRF (iv), mph1-ΔS1-S3 incubated with MRF (v), mph1-ΔS1-S3 and Smc5 incubated with MRF (vi), and Smc5 incubated with MRF (vii). (B) Quantification of the AFM data. The numbers ofMph1-MRF complexes over the total number of MRF molecules sampled are indicated. For example, among 1236 MRF molecules examined, 977 were bounded by Mph1, whereas when Mph1 was pre-incubated with Smc5, only 165 were bounded by Mph1 among 1085 MRF molecules examined. (C) Distribution analysis of protein volume from AFM images of Mph1 protein alone and Mph1 bound to MRF DNA, expressed as Mph1 monomer equivalents. For Mph1 protein alone the fitted red line, R2=0.98, has a mean=1.4 ± 0.9 (S.D.) (n=167). The distribution of particle volume for Mph1 complexes bound to MRF DNA plotted in a histogram was best fitted to a Gaussian distribution with three populations (red line, R2=0.95, mean of the first peak=7.3 ± 1.7 (S.D.), mean of the second peak=12 ± 2.5 (S.D.), mean of the third peak=17.4 ± 0.8 (S.D.), (n=82). (D) mph1-ΔS1 rescues the MMS sensitivity of the rad5 helicase dead allele (rad5-DEAA) but not that of mms2Δ, and is sensitized by srs2Δ and mms4Δ. The number of cells spotted is indicated for each column. (E) Model depicting the roles of Mph1 and Smc5-Smc6 in replication fork repair. See main text for details. See also Figure S4 and Table S2.
We also used metal shadowing EM to show that Mph1 specifically recognizes the junction point in the MRF (Figure S4C). Moreover, compared with Mph1 alone (Figure S4C (iii), (v) and (vi), circled) and Mph1 associated with a 3′ ssDNA overhang (Figure S4C (ii), yellow arrows), Mph1 species bound to the MRF junction (Figure S4C (iii)–(vi), white arrows) are clearly larger, again indicative of protein oligomerization. The EM experiments thus provide further support for Mph1 oligomerization at DNA junctions.
Importantly, AFM analysis showed that Smc5, added in slight excess over Mph1, reduces the frequency of Mph1-MRF complexes about 5 fold (Figures 4A (iv) and 4B). Like Mph1, mph1-ΔS1-S3 also forms oligomers at the MRF junction, but these are resistant to Smc5 (Figures 4A (v–vi) and 4B). No significant binding of Smc5 to the MRF was detected by this method (Figure 4A (vii)). Taken together, these results reveal that Mph1 oligomerizes on the junction point within the MRF, and that Smc5 prevents Mph1-substrate engagement in a manner that requires its interaction with Mph1.
Genetic compensation and sensitization by mph1-ΔS1
We examined mph1ΔS1 mutant cells genetically to understand the biological relevance of Mph1 regulation. This mph1 mutant is most suitable for in vivo analysis, as the S2 and S3 regions in Mph1 are involved in additional interactions, such as with the ssDNA binding protein RPA (Banerjee et al., 2008). Our biochemical data predicted that mph1-ΔS1 would confer increased fork regression ability, but this effect could be mild, as the mph1-ΔS1 protein retains residual Smc5 binding (Figures 2B and 2E). To test this prediction, we first examined whether mph1-ΔS1 would compensate for the loss of fork regression function of the Rad5 protein. Rad5 has been implicated in replication fork rescue, and mutation of residues D681 and E682 to alanine (rad5-DEAA) in helicase motif II abolishes its fork regression activity and results in sensitivity to the fork blocking agent MMS (Blastyak et al., 2007; Gangavarapu et al., 2006; Minca and Kowalski, 2010). We found that mph1-ΔS1 indeed partially suppresses rad5-DEAA MMS sensitivity (Figure 4D). As Rad5 also participates in lesion bypass along with the Mms2-Ubc13 ubiquitin enzyme complex, we examined whether mph1-ΔS1 would alleviate the MMS sensitivity of mms2Δ cells but found no suppression (Figure 4D). Thus, mph1-ΔS1 appears to specifically compensate for the rad5 mutant’s fork regression defect.
We reasoned that increased fork regression by mph1-ΔS1 would also increase reliance on enzymes that resolve structures stemming from regressed replication forks. We found that although mph1-ΔS1 by itself shows only mild sensitivity to MMS, its survival in the presence of MMS requires Srs2, which prevents the formation of recombination intermediates, and Mms4, which resolves these intermediates (Figure 4D and S4D). Specifically, a strong synergism of mph1-ΔS1 with the mms4Δ or srs2Δ mutation was seen (Figure 4D). Together, the genetic interactions documented herein complement our biochemical results in showing that impaired Smc5-mediated regulation of Mph1 activity renders cells reliant on enzymes for joint DNA molecule removal or avoidance, yet compensates for the defect in another DNA motor protein.
DISCUSSION
Our results show that Smc5-Smc6 restrains the replication fork regression and DNA branch migration activities of Mph1, but not the latter’s D-loop dissociative and DNA unwinding functions. We have also presented several lines of evidence to support the conclusion that Smc5 regulation of Mph1 does not require Smc5 DNA binding, but rather stems from its direct interaction with Mph1. These results challenge the canonical view of SMC proteins as a DNA-tethering entity whose coiled coil domain serves only a structural role, and suggest that SMC complexes employ versatile and dynamic protein interactions as an additional means of regulating DNA metabolism/transactions. AFM and EM analyses have provided evidence that Mph1 functions as an oligomer to mediate fork regression. To our knowledge, this is the first molecular description of how a replication fork regression enzyme engages its substrate and is consistent with previous accounts of oligomerization of DNA branch migrating enzymes (Petukhova et al., 1999; Ristic et al., 2001; West, 1997). Our results suggest that Smc5 regulates Mph1 by preventing the ability of the latter to oligomerize at fork junctions by targeting a region needed for optimal enzymatic activity.
Based on our findings and the literature, we present the following model for the relationship between Smc5-Smc6 and Mph1 in DNA replication (Figure 4E). Smc5-Smc6, shown to localize in vivo to stalled forks (Ampatzidou et al., 2006; Lindroos et al., 2006), restrains the fork regression and branch migration activities of Mph1 (➀ and ➁). While some degree of regression can be beneficial for fork restart (Michel et al., 2001; Whitby, 2010), studies presented here and elsewhere indicate that it must be tightly controlled (Couch et al., 2013; Hu et al., 2012) to allow translesion DNA synthesis-mediated fork bypass and prevent the formation of excessive joint DNA molecules (Chavez et al., 2011; Chen et al., 2009). We speculate that after DNA synthesis templated from the sister strand, Smc5-Smc6 may favor the reversal of fork regression by restraining Mph1 activity (➃). Our results shed light on a novel function of Smc5-Smc6 and should provide a molecular context for understanding how fork regression is regulated in other eukaryotic cells. As Smc5 alone is sufficient for Mph1 inhibition, other subunits of the complex must perform additional functions integral to the regulatory mechanisms documented for this complex, such as protein modifications by SUMO and ubiquitin as well as chromosome loading (Chiolo et al., 2011; Doyle et al., 2010; Duan et al., 2009; Kegel et al., 2010; Zhao and Blobel, 2005). Future work to elucidate how this SMC complex integrates multiple roles will provide further insights into genome protection mechanisms.
EXPERIMENTAL PROCEDURES
Preparation of proteins
Due to space limitation, the purification of recombinant proteins (Mph1 and its mutants, Smc5 and its fragments, Smc6, and Smc5-Smc6) is described in detail in the Supplemental Experimental Procedures.
Replication fork regression and branch migration assays
The substrates (MRF, MHJ, and sigma structure) were prepared as described (Blastyak et al., 2007; Gari et al., 2008; Ralf et al., 2006). The replication fork regression and branch migration reactions followed protocols used previously to characterize Mph1/FANCM activities (Gari et al., 2008; Singh et al., 2010; Zheng et al., 2011), as detailed in the Supplemental Experimental Procedures. We note that previous studies have shown that all the Mph1-catalyzed reactions, including DNA fork regression, DNA branch migration, DNA unwinding, and D-loop disruption, require ATP hydrolysis (Gari et al., 2008; Prakash et al., 2009; Sun et al., 2008; Zheng et al., 2011). Additional experimental details for the replication fork regression and DNA branch migration assays, affinity pulldown assay, ATPase assay, yeast two hybrid assay, AFM analysis, metal shadowing electron microscopy analysis, DNA mobility shift assay and D-loop formation and disruption assays are provided in the Supplemental Experimental Procedures. The yeast strains used in this study are summarized in Table S1.
Supplementary Material
Figure S1. Physical interaction of Smc5-Smc6 with Mph1 and specific inhibition of Mph1-catalyzed DNA fork regression and branch migration by Smc5-Smc6, related to Figure 1.
(A) Silver stain of purified His6-Smc5, MBP-Smc5, Strep-Smc5, Strep-Smc6, Myc-Smc5-Strep-Smc6, and His6-Flag-Mph1 after SDS-PAGE.
(B) Affinity pulldown to examine the interaction of Smc5, Smc6, or Smc5-Smc6 with Mph1. Strep-tagged Smc5 (lane 1–3), Smc6 (lane 4–6), and Smc5-Smc6 (lane 7–9) were mixed with purified His6-Flag-Mph1 and incubated with Strep tactin resin. The supernatant (S), wash (W) and eluate (E) fractions were analyzed by SDS-PAGE and immunoblotted with anti-FLAG or anti-Strep antibodies. As Smc5, Smc6 and Mph1 migrate at similar positions on SDS-PAGE, immunoblot was used to distinguish these proteins.
(C–D) Time course experiments showing that Smc5-Smc6 (4 nM) inhibits MRF (5 nM) regression (C) and MHJ (5 nM) branch migration (D) catalyzed by Mph1 (2 nM) but has no effect on the equivalent reactions catalyzed by FANCM (2 nM) or Rad54 (2 nM).
(E–F) Time course experiments showing that Smc5 (4 nM) inhibits MRF (5 nM) regression (E) and MHJ (5 nM) branch migration (F) catalyzed by Mph1 (2 nM) but has no effect on the equivalent reactions catalyzed by FANCM (2 nM) and Rad54 (2 nM). Experiments were performed as in Figure 1B and 1C and the results were quantified and plotted. The error bars represent standard deviation from three independent experiments.
Figure S2. Mph1 mutants diminishing Smc5 interaction are folded properly and refractory to Smc5 inhibition, related to Figure 2.
(A) The mph1-ΔS1-S3 protein fails to interact with Smc5. Affinity pulldown results are shown and the relative ratio of Mph1 or mph1-ΔS1-S3 to Smc5 in the eluate (E) is indicated. Experiments were performed as in Figure 2E. S: supernatant and W: wash.
(B) mph1 mutants are proficient for ATPase activity. The ssDNA-dependent ATPase activity of Mph1 (WT) and mutant mph1 proteins was determined and quantified as described Experimental Procedure.
(C) The mph1 mutants are proficient in D-loop dissociation. The ability of Mph1 and mutant mph1 proteins (each at 100 nM) to dissociate Rad51-made D-loops was determined. Experiments were performed as in Figure 1D. NP, no protein control. The results were quantified and graphed. The values are the average of triplicates ± SD.
(D–F) Binding of the MRF structure by Mph1 (D), mph1-ΔS1-S3 (E), and mph1-S1-S3 (F). Radiolabeled MRF (5 nM) were incubated with Mph1 (5–80 nM), mph1-ΔS1-S3 (10–60 nM) or the mph1-S1-S3 fragment (10–60 nM), and the mix was examined by agarose gel. Interaction between Mph1 or mph1-ΔS1-S3 with MRF DNA were indicated by the shifted bands representing DNA-protein complexes. The results were quantified and plotted.
(G–H) Circular Dichroism spectrum (G) and secondary structure prediction (H) of Mph1 and mutants. Secondary structure prediction based on the Circular Dichroism spectra is derived by CDNN 2.1 (Applied Photophysics Ltd). Note that that the small changes seen for the mph1-ΔS1-S3 can be attributed to the truncation of 240 residues in this mutant.
(I–J) mph1-ΔS1-S3 is refractory to Smc5 inhibition of MRF regression (I) and MHJ branch migration (J). The indicated concentration of Mph1, mph1-ΔS1-S3 and Smc5 were incubated with 5 nM substrates at 30 °C for 4 min. Experiments were performed as in Figure 1B–1C.
(K) Deleting or mutating the S1 region of Mph1 largely abolishes Smc5 inhibitory effect in Mph1-catalyzed MHJ branch migration. Mph1 or its mutant (2 nM) with or without Smc5 (4 nM) was incubated with MHJ (5 nM) at 30 °C for the indicated time. Averaged values from triplicates ± SD are graphed at the bottom.
Figure S3. The Smc5 domain that interacts with Mph1 but is devoid of DNA binding activity is sufficient for Mph1 inhibition, related to Figure3.
(A) Summary of yeast two-hybrid results for the interaction between Smc5 and its fragments with Mph1. The Smc5 species were fused to the Gal4 activation domain in the pOAD vector and tested against Mph1 fused to the Gal4 DNA binding domain in the pOBD vector. Selection was conducted on -Leu-Trp-His (-L-T-H) medium, and strong or moderate interaction strength is designated +++ or +, respectively.
(B) MBP-tagged Smc5 (WT) and smc5-ΔNC (residues 149-944) were tested for interaction with Mph1 by affinity pulldown.
(C) smc5-ΔNC is proficient in inhibiting Mph1-catalyzed MRF regression. The indicated concentration of Mph1, Smc5 and smc5-ΔNC were incubated with 5 nM MRF substrate at 30 °C for 4 min. Experiments were performed as in Figure 1B.
(D) Smc5, but not smc5-ΔNC, interacts with MRF. Smc5 and smc5-ΔNC (10–80 nM) were tested for MRF (5 nM) binding and the results were quantified and plotted. The error bars in panels C and D represent standard deviation from three independent experiments.
Figure S4. AFM and EM visualization of Mph1 oligomerization on fork junctions, and mph1ΔS1 genetic tests, related to Figure 4.
(A) Schematic of the replication fork substrate for AFM.
(B) Processing of the replication fork by Mph1 to form dsDNA products. The reactions were resolved in 1% agarose gel and stained with SYBR Gold (Invitrogen).
(C) Mph1 oligomerization on movable replication forks visualized after tungsten rotary metal shadow casting. Top, schematic of the 3′ DNA overhang and the movable replication fork substrates for EM analysis. Bottom, representative EM micrographs are shown for the following: MRF alone (i), Mph1 incubated with the 3′-overhang (ii), Mph1 incubated with the MRF (iii), (iv), (v), (vi). The unbound Mph1 molecules are circled. MRF- or 3′-overhang-bound Mph1 oligomers are indicated by the white or yellow arrows, respectively. Scale bars represent 100 nm.
(D) mph1-ΔS1 showed mild sensitivity at 0.05% MMS.
Table S1. Yeast two-hybrid plasmids and strains used in the study, related to Experimental Procedures.
Table S2. Oligonucleotides used for constructing the movable replication forks for AFM analyses, related to Figure 4.
Acknowledgments
We apologize to colleagues whose work is not cited due to space limitation. This study was supported by US National Institutes of Health grants GM057814, ES007061, ES015632, and GM080670, American Cancer Society grant RSG-12-013-01-CCG, Marie Curie Reintegration Grant (FP7-276898), The Dutch Technology Foundation STW NanoNextNL consortium and a Leukemia and Lymphoma Society Scholar Award. We thank members of the Sung and Zhao laboratories, particularly Xiao Peng, for helpful comments on the manuscript, Walter J Chazin for assistance in CD analysis, Jack Griffith for the use of his EM facility, and Damien D’Amours for providing the Smc5 and Smc6 expression plasmids.
Footnotes
Supplemental Information includes Experimental Procedures, four figures, and two tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ampatzidou E, Irmisch A, O’Connell MJ, Murray JM. Smc5/6 is required for repair at collapsed replication forks. Mol Cell Biol. 2006;26:9387–9401. doi: 10.1128/MCB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Smith S, Oum JH, Liaw HJ, Hwang JY, Sikdar N, Motegi A, Lee SE, Myung K. Mph1p promotes gross chromosomal rearrangement through partial inhibition of homologous recombination. J Cell Biol. 2008;181:1083–1093. doi: 10.1083/jcb.200711146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- Chavez A, Agrawal V, Johnson FB. Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes Smc5/6 complex. J Biol Chem. 2011;286:5119–5125. doi: 10.1074/jbc.M110.201608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Choi K, Szakal B, Arenz J, Duan X, Ye H, Branzei D, Zhao X. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc Natl Acad Sci USA. 2009;106:21252–21257. doi: 10.1073/pnas.0908258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Szakal B, Chen YH, Branzei D, Zhao X. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2306–2314. doi: 10.1091/mbc.E10-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, Sogo J, Foiani M. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Couch FB, Bansbach CE, Driscoll R, Luzwick JW, Glick GG, Betous R, Carroll CM, Jung SY, Qin J, Cimprich KA, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27:1610–1623. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R. FANCM limits meiotic crossovers. Science. 2012;336:1588–1590. doi: 10.1126/science.1220381. [DOI] [PubMed] [Google Scholar]
- Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J Biol Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Sarangi P, Liu X, Rangi GK, Zhao X, Ye H. Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell. 2009;35:657–668. doi: 10.1016/j.molcel.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangavarapu V, Haracska L, Unk I, Johnson RE, Prakash S, Prakash L. Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:7783–7790. doi: 10.1128/MCB.01260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- Hansma HG, Kasuya K, Oroudjev E. Atomic force microscopy imaging and pulling of nucleic acids. Curr Opin Struct Biol. 2004;14:380–385. doi: 10.1016/j.sbi.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Hu J, Sun L, Shen F, Chen Y, Hua Y, Liu Y, Zhang M, Hu Y, Wang Q, Xu W, et al. The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell. 2012;149:1221–1232. doi: 10.1016/j.cell.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Kegel A, Sjogren C. The Smc5/6 complex: more than repair? Cold Spring Harb. Symp Quant Biol. 2010;75:179–187. doi: 10.1101/sqb.2010.75.047. [DOI] [PubMed] [Google Scholar]
- Lindroos HB, Strom L, Itoh T, Katou Y, Shirahige K, Sjogren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Lorenz A, Osman F, Sun W, Nandi S, Steinacher R, Whitby MC. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science. 2012;336:1585–1588. doi: 10.1126/science.1220111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Flores MJ, Viguera E, Grompone G, Seigneur M, Bidnenko V. Rescue of arrested replication forks by homologous recombination. Proc Natl Acad Sci USA. 2001;98:8181–8188. doi: 10.1073/pnas.111008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minca EC, Kowalski D. Multiple Rad5 activities mediate sister chromatid recombination to bypass DNA damage at stalled replication forks. Mol Cell. 2010;38:649–661. doi: 10.1016/j.molcel.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- Ristic D, Wyman C, Paulusma C, Kanaar R. The architecture of the human Rad54-DNA complex provides evidence for protein translocation along DNA. Proc Natl Acad Sci USA. 2001;98:8454–8460. doi: 10.1073/pnas.151056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Driscoll R, Castellucci F, Foiani M, Jackson SP, Branzei D. The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair. Mol Biol Cell. 2009;20:1671–1682. doi: 10.1091/mbc.E08-08-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A, Machin F, Pasero P, Lisby M, Haber JE, Aragon L. Anaphase onset before complete DNA replication with intact checkpoint responses. Science. 2007;315:1411–1415. doi: 10.1126/science.1134025. [DOI] [PubMed] [Google Scholar]
- Weinert T, Kaochar S, Jones H, Paek A, Clark AJ. The replication fork’s five degrees of freedom, their failure and genome rearrangements. Curr Opin Cell Biol. 2009;21:778–784. doi: 10.1016/j.ceb.2009.10.004. [DOI] [PubMed] [Google Scholar]
- West SC. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair. 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol. 2013;5:a012815. doi: 10.1101/cshperspect.a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XF, Prakash R, Saro D, Longerich S, Niu H, Sung P. Processing of DNA structures via DNA unwinding and branch migration by the S. cerevisiae Mph1 protein. DNA Repair. 2011;10:1034–1043. doi: 10.1016/j.dnarep.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Physical interaction of Smc5-Smc6 with Mph1 and specific inhibition of Mph1-catalyzed DNA fork regression and branch migration by Smc5-Smc6, related to Figure 1.
(A) Silver stain of purified His6-Smc5, MBP-Smc5, Strep-Smc5, Strep-Smc6, Myc-Smc5-Strep-Smc6, and His6-Flag-Mph1 after SDS-PAGE.
(B) Affinity pulldown to examine the interaction of Smc5, Smc6, or Smc5-Smc6 with Mph1. Strep-tagged Smc5 (lane 1–3), Smc6 (lane 4–6), and Smc5-Smc6 (lane 7–9) were mixed with purified His6-Flag-Mph1 and incubated with Strep tactin resin. The supernatant (S), wash (W) and eluate (E) fractions were analyzed by SDS-PAGE and immunoblotted with anti-FLAG or anti-Strep antibodies. As Smc5, Smc6 and Mph1 migrate at similar positions on SDS-PAGE, immunoblot was used to distinguish these proteins.
(C–D) Time course experiments showing that Smc5-Smc6 (4 nM) inhibits MRF (5 nM) regression (C) and MHJ (5 nM) branch migration (D) catalyzed by Mph1 (2 nM) but has no effect on the equivalent reactions catalyzed by FANCM (2 nM) or Rad54 (2 nM).
(E–F) Time course experiments showing that Smc5 (4 nM) inhibits MRF (5 nM) regression (E) and MHJ (5 nM) branch migration (F) catalyzed by Mph1 (2 nM) but has no effect on the equivalent reactions catalyzed by FANCM (2 nM) and Rad54 (2 nM). Experiments were performed as in Figure 1B and 1C and the results were quantified and plotted. The error bars represent standard deviation from three independent experiments.
Figure S2. Mph1 mutants diminishing Smc5 interaction are folded properly and refractory to Smc5 inhibition, related to Figure 2.
(A) The mph1-ΔS1-S3 protein fails to interact with Smc5. Affinity pulldown results are shown and the relative ratio of Mph1 or mph1-ΔS1-S3 to Smc5 in the eluate (E) is indicated. Experiments were performed as in Figure 2E. S: supernatant and W: wash.
(B) mph1 mutants are proficient for ATPase activity. The ssDNA-dependent ATPase activity of Mph1 (WT) and mutant mph1 proteins was determined and quantified as described Experimental Procedure.
(C) The mph1 mutants are proficient in D-loop dissociation. The ability of Mph1 and mutant mph1 proteins (each at 100 nM) to dissociate Rad51-made D-loops was determined. Experiments were performed as in Figure 1D. NP, no protein control. The results were quantified and graphed. The values are the average of triplicates ± SD.
(D–F) Binding of the MRF structure by Mph1 (D), mph1-ΔS1-S3 (E), and mph1-S1-S3 (F). Radiolabeled MRF (5 nM) were incubated with Mph1 (5–80 nM), mph1-ΔS1-S3 (10–60 nM) or the mph1-S1-S3 fragment (10–60 nM), and the mix was examined by agarose gel. Interaction between Mph1 or mph1-ΔS1-S3 with MRF DNA were indicated by the shifted bands representing DNA-protein complexes. The results were quantified and plotted.
(G–H) Circular Dichroism spectrum (G) and secondary structure prediction (H) of Mph1 and mutants. Secondary structure prediction based on the Circular Dichroism spectra is derived by CDNN 2.1 (Applied Photophysics Ltd). Note that that the small changes seen for the mph1-ΔS1-S3 can be attributed to the truncation of 240 residues in this mutant.
(I–J) mph1-ΔS1-S3 is refractory to Smc5 inhibition of MRF regression (I) and MHJ branch migration (J). The indicated concentration of Mph1, mph1-ΔS1-S3 and Smc5 were incubated with 5 nM substrates at 30 °C for 4 min. Experiments were performed as in Figure 1B–1C.
(K) Deleting or mutating the S1 region of Mph1 largely abolishes Smc5 inhibitory effect in Mph1-catalyzed MHJ branch migration. Mph1 or its mutant (2 nM) with or without Smc5 (4 nM) was incubated with MHJ (5 nM) at 30 °C for the indicated time. Averaged values from triplicates ± SD are graphed at the bottom.
Figure S3. The Smc5 domain that interacts with Mph1 but is devoid of DNA binding activity is sufficient for Mph1 inhibition, related to Figure3.
(A) Summary of yeast two-hybrid results for the interaction between Smc5 and its fragments with Mph1. The Smc5 species were fused to the Gal4 activation domain in the pOAD vector and tested against Mph1 fused to the Gal4 DNA binding domain in the pOBD vector. Selection was conducted on -Leu-Trp-His (-L-T-H) medium, and strong or moderate interaction strength is designated +++ or +, respectively.
(B) MBP-tagged Smc5 (WT) and smc5-ΔNC (residues 149-944) were tested for interaction with Mph1 by affinity pulldown.
(C) smc5-ΔNC is proficient in inhibiting Mph1-catalyzed MRF regression. The indicated concentration of Mph1, Smc5 and smc5-ΔNC were incubated with 5 nM MRF substrate at 30 °C for 4 min. Experiments were performed as in Figure 1B.
(D) Smc5, but not smc5-ΔNC, interacts with MRF. Smc5 and smc5-ΔNC (10–80 nM) were tested for MRF (5 nM) binding and the results were quantified and plotted. The error bars in panels C and D represent standard deviation from three independent experiments.
Figure S4. AFM and EM visualization of Mph1 oligomerization on fork junctions, and mph1ΔS1 genetic tests, related to Figure 4.
(A) Schematic of the replication fork substrate for AFM.
(B) Processing of the replication fork by Mph1 to form dsDNA products. The reactions were resolved in 1% agarose gel and stained with SYBR Gold (Invitrogen).
(C) Mph1 oligomerization on movable replication forks visualized after tungsten rotary metal shadow casting. Top, schematic of the 3′ DNA overhang and the movable replication fork substrates for EM analysis. Bottom, representative EM micrographs are shown for the following: MRF alone (i), Mph1 incubated with the 3′-overhang (ii), Mph1 incubated with the MRF (iii), (iv), (v), (vi). The unbound Mph1 molecules are circled. MRF- or 3′-overhang-bound Mph1 oligomers are indicated by the white or yellow arrows, respectively. Scale bars represent 100 nm.
(D) mph1-ΔS1 showed mild sensitivity at 0.05% MMS.
Table S1. Yeast two-hybrid plasmids and strains used in the study, related to Experimental Procedures.
Table S2. Oligonucleotides used for constructing the movable replication forks for AFM analyses, related to Figure 4.