Abstract
The post-translational palmitoylation of WNT morphogens is critical for proper signaling during embryogenesis and adult homeostasis. The addition of palmitoyl groups to WNT proteins is catalyzed by Porcupine (PORCN). However, the Wnt amino acid residues required for recognition and palmitoylation by PORCN have not been fully characterized. We show that WNT1 residues 214–234 are sufficient for PORCN-dependent palmitoylation of Ser224. Substitution of Ser224 with Thr, but not Cys, is tolerated in palmitoylation and biological assays. Our data highlight the importance of palmitoylation for WNT1 activity and establish PORCN as an O-acyl transferase for WNT1.
Keywords: WNT1, Porcupine, palmitoylation, chick, spinal cord, MBOAT
BACKGROUND
Wnt morphogens are concentration-dependent paracrine signaling proteins that play key roles in embryonic development and adult homeostasis [1-5]. Wnts undergo several post-translational modifications including cleavage of the signal peptide [6, 7], sulfation [8], glycosylation [7, 9-17], and fatty acylation [9, 11, 18-23]. Whereas the sulfation of Wnts on tyrosine residues controls their assembly into dimers (or higher order complexes) [8], the type and extent of N-linked glycosylation influence Wnt secretion [7, 9-17]. The addition of mono-unsaturated palmitic acid (palmitoylation) is required for proper gradient formation and biological activity [9, 11, 18-23]. Though the exact mechanism by which palmitoylation promotes gradient formation is not known, palmitoylation-deficient Wnts are primarily retained in the endoplasmic reticulum and are secreted to a much lesser extent compared to their wild-type counterparts [19, 21, 23, 24]. This defect in secretion might be explained by the observation that palmitoylation of Wnt is required for recognition by Wntless (WLS), a cargo protein thought to be involved in transporting Wnt through the secretory pathway [25-29]. Moreover, palmitoylation promotes Wnt-receptor interactions and signaling at the cell surface [10, 30]. The recent X-ray crystal structure for Xenopus WNT8 (XWNT8) in complex with its receptor, mouse Frizzled 8 (mFZD8) revealed that the palmitate group on Wnt makes a significant contact along a hydrophobic groove on mFZD8; this interaction enhances the affinity of XWNT8 for mouse mFZD8 [30]. The importance of palmitoylation for Wnt secretion and signaling prompted us to explore the biochemistry of this modification.
Bioinformatic studies in combination with experimental evidence suggest that Porcupine (PORCN), a member of the membrane-bound O-acyl transferase (MBOAT) superfamily, is the enzyme responsible for palmitoylating Wnts [9, 18, 19, 21, 23, 31, 32]. Consistent with the requirement of PORCN for Wnt signaling, mice lacking PORCN or WNT3, the earliest acting Wnt family member, die during gastrulation [31, 33-37]. The importance of PORCN is further demonstrated by the finding that mutations in human PORCN cause Focal Dermal Hypoplasia (FDH), a rare X-linked dominant syndrome that affects tissues of mesodermal and ectodermal origin [38, 39]. Multiple functionally important PORCN residues have already been identified in patients with FDH [40].
The investigation of PORCN specificity for fatty acyl substrates [9, 19, 30] revealed a preference for mono-unsaturated fatty acids ranging in length from thirteen to sixteen carbons [9, 19, 20, 30]. However, little is known about the protein substrate specificity of PORCN (i.e., Wnt protein) and the Wnt residues required for PORCN-dependent palmitoylation of Wnt. Mass spectrometry, X-ray crystallography and biochemical studies established that a conserved serine residue is the site of palmitoylation for WNT3A (S209) and WNT8 (S187) [9, 21, 30], yet it is not known if PORCN exclusively acylates serine residues. The contribution of neighboring amino acids to the palmitoylation site also remains unclear. In this study, we investigate the WNT1 residues required for efficient palmitoylation by PORCN and assess the biological activity of WNT1 S224A, S224C and S224T variants in cell culture and in vivo.
MATERIALS and METHODS
We thank Randy Moon for supplying Super8xTopflash and Super8xFopflash [41]. We thank Andy McMahon for supplying pCIG [42], Elena Frovola for partial chick Wnt1 cDNA, Tatsuhiko Kadowaki for mPORCND [43].
Materials
Materials and their respective vendors are as follows: sucrose, fast green (Fisher); Sodium Azide (VWR); DMEM, 200mM L-Glutamine, 100x Penicillin/Streptomycin, PBS, Trypsin EDTA (Cellgro); Fetal Bovine Serum (Hyclone or Atlanta Biologicals); TX-100, PMSF (Roche); diamino 2-phenyl indole (DAPI), Tween-20, Tyrode’s solution, leupeptin, aprotinin, carboxymethylcellulose sodium salt, DMSO, fatty acid free BSA#A8806, TCEP Tris(2-carboxyethyl)phosphine hydrochloride, TBTA Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine, Copper Sulfate (Sigma); OCT (Tissue tek); pcDNA3.1(-)A, slowfade gold light anti-fade kit, dialyzed serum #26400-036, Biotin Azide (Invitrogen); paraformaldehyde (Electron Microscopy Sciences); Fugene HD Transfection Reagent, Dual Luciferase Reporter Assay System (Promega); Fertile eggs (Rhode Island Red; Petaluma Farms); HEK293T (DSHB); 3H-palmitate (MP Biomedicals); Protein A/G agarose beads, Halt protease and phosphatase inhibitor cocktail (Pierce); Immobilon-FL, Immobilon-P (Millipore); En3Hance (Perkin Elmer).
Constructs
The full-length sequence for chick WNT1 has not been reported. Therefore, we fused sequences encoding the mouse WNT1 signal peptide to the available chick WNT1 sequence [18, 44]. As the mature proteins should be entirely chick derived, we refer to this construct as chick WNT1.
Single nucleotide point mutations (resulting in S224A, S224C, and S224T substitutions) were introduced into full length WNT1 by overlap extension using PCR [45]. All sequences were confirmed by sequencing. Wild-type and mutant WNT1 cDNAs were subcloned into the pCIG expression vector. This vector drives the expression of a single bi-cistronic transcript, which encodes the protein of choice (in this case, WNT1) and a nuclear variant of Green fluorescent protein (GFP) [42]. These cDNAs were also subcloned into pCDNA 3.1 for expression in HEK293T cells.
We also generated a cDNA encoding a fusion protein, which was comprised of WNT1 (residues 1-34) at the N-terminus fused in frame to full-length GFP and WNT1 (residues 209-239). The C-terminus was comprised of the Fc region of human IgG. Shorter versions of this construct included sequences encoding WNT1 residues 214-234 and 219-229 in place of residues 209-239. Mutations encoding S224A, S224C, and S224T variants were generated as above. These fusion proteins were subcloned into pCDNA 3.1 for expression in HEK293T cells.
Antibodies
Mouse anti-chick WNT1 5F1-G11-D1 was used for western blots (1/10) while mouse anti-chick WNT1 7B3-A10-F9 was used for immunoprecipitations (1/4). Both antibodies were made in the Burrus lab (Galli et al., 2007). Mouse anti-GFP JL-8 (Clontech) (immunoprecipitation 1/750, colorimetric Westernblot 1/1500, fluorescent western blot 1/15000); Rabbit anti-Phospho Histone H3 (ser10) (Millipore) (1:1,000 dilution); IRDye800 conjugated streptavidin (Licor) (1/5000); Alexa Fluor® 680 Goat Anti-Mouse (Invitrogen) (1/4000); Goat anti mouse Cy2, Goat anti mouse Cy3, Goat anti-Rabbit DyLight649 (Jackson Immunoresearch Labs) (1/200); mouse anti-Islet1 conditioned media (Developmental Studies Hybridoma Bank) (1:20).
Cell Culture
HEK293T cells were grown in standard medium (DMEM with 10% fetal bovine serum, 4mM L-glutamine, and 1x penicillin/streptomycin) on 100mm plates in humidified incubators set to 10% CO2.
Palmitoylation assays
Two different palmitoylation assays were employed in these studies. The differences in these assays simply reflect the timing of their development. For the first, HEK293T cells were transiently transfected with WNT1, spGFP:WNT1 (209-239), or spGFP:WNT1 (209-239) S224A using the FUGENE HD transfection kit (Promega) according to standard protocol. Cells were metabolically labeled with 3H-palmitate for 5-8 hrs. Antibodies against WNT1 or GFP were used to immunoprecipitated proteins from cell lysates as previously described (Galli2011). After separation by SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE), gels were electroblotted onto PVDF, treated with En3Hance and subjected to autoradiography at −80°C with an intensifying screen for 3-4 months.
For the second assay, HEK293T cells were seeded in a 6-well plate, incubated overnight, and co-transfected with 1.65μg of pCDNA 3.1 encoding wild-type and mutant spGFP:WNT1(209-239):Fc fusions along with 1.65μg of pCDNA 3.1 encoding eGFP (as filler) or mPORCND using the FUGENE HD transfection kit (Promega) according to standard protocol. ω-Alkyne palmitic acid (Alk-C16) was synthesized as previously described [46] and used to metabolically label HEK293T cells [9, 47]. After metabolic labeling of cells with Alk-C16 for 20-24 hrs [48], Fc containing proteins were immunoprecipitated with Protein A/G beads [49]. Proteins retained on the beads were subjected to click chemistry with biotin azide before separation by SDS-PAGE and analysis by Western blot [9, 47]. Western blots were probed with IRDye800 conjugated streptavidin, and anti-GFP or anti-WNT1 antibodies followed by goat Alexa Fluor® 680-conjugated anti-mouse secondary antibody.
8xSuperTopFlash Wnt signaling assay
HEK293T cells were transfected with pCDNA3.1 expression constructs encoding wild-type and variant WNT1 proteins [19]. DNA quantities used in transfections are as follows: pcDNA3.1 GFP, pcDNA3.1 cWNT1, pcDNA3.1 cWNT1S224A, pcDNA3.1 cWNT1S224C, and pcDNA3.1 cWNT1S224T at 0.25μg/well; Super8xTopFlash or Super8xFopFlash at 0.01μg/well; RL-CMV at 0.01ng/well. Cells were incubated overnight, lysed and measured as per Promega Dual-Luciferase Reporter Assay protocol. Luciferase measurements were carried out in a TD-20/20 luminometer.
In ovo electroporation of chick embryos
Fertile eggs (Petaluma Farms) were incubated at 37°C between 48-52hrs to HH12-14 chick embryos. Before injecting the posterior spinal cord with a mixture of methylcellulose, fast green, and DNA, a thin strip of parafilm was placed on both sides of the embryos, leaving only the neural tube exposed for electroporation. The electrodes were placed on top of the parafilm to lessen damage to the embryo. Electroporations were performed with nail polish-coated platinum electrodes placed at either side of the neural tube, and four 50ms pulses at 45 volts were applied to transfect DNA. DNA was transfected into cells on one side of the neural tube as a result. Successfully electroporated embryos were incubated to 48hrs later to observe gene expression, then harvested at HH20-22 and fixed in PBS containing 4% paraformaldehyde.
Immunostaining
Fixed embryos were prepared for embedding by immersion in sucrose gradient solutions containing sodium azide. Fixed embryos were embedded using Tissue Tek OCT and transverse sections (10μm thick) were acquired using a Leica Cryostat. Sections were subjected to immunostaining with the anti-phospho histone H3 antibody to monitor proliferation and the Islet1 antibody to assess the specification of DI3 interneurons and motor neurons. Sections were stained with DAPI to visualize nuclei.
Confocal microscopy
Single-slice or stacked images of tissue sections were obtained using a Zeiss LSM710 confocal microscope, using a 20x objective at 0.9 × digital zoom to obtain representative spinal cord images, and using a 40X oil objective at 0.6 × digital zoom to obtain cell count images. GFP was visualized using an argon 488-nm line, Cy3 was visualized using a HeNe 543-nm line, Dy649 was visualized using a HeNe 633-nm line, and DAPI was visualized using a diode 405-nm line. Final images were processed in Adobe Photoshop.
Quantification and statistical analysis
Single-slice images were used for quantification of proliferative cells, DI3 interneurons, and MNs. The Automatic Nuclei Counter plug-in for ImageJ was extended and used to automate the process of counting cells. Images were processed in Adobe Photoshop, Adobe Illustrator, and ImageJ to enable cell counting by the ImageJ software. Data were analyzed using a two-tailed unpaired Student’s t-test.
Cross-species alignment of Wnt sequences surrounding the palmitoylation site
Wnt sequences were aligned using Clustal Omega (version 1.2.1) [50, 51]. The accession numbers for sequences used in our alignment are as follows: chick WNT1 (AY753286), mouse WNT1 (NM_021279), mouse WNT3A (NM_009522), Xenopus Laevis WNT8 (NM_001088168), zebrafish WNT1 (NM_001201398), Drosophila melanogaster WG (NM_078778), Caenorhabditis elegans EGL-20 (NM_069353), Amphimedon queenslandia (sponge) WNT (EU285557), Aurelia aurita (moon jelly) WNT1 (KC341730), Sea Urchin WNT1 (NM_001123500), Amphioxus WNT1 (AF061974), Leech WNT1 (GU289716), Sea squirt WNT (AB210617), and Arctic lamprey (KF318011). Because the chick WNT1 sequence is a partial sequence, the numbering shown in the alignment does not reflect the actual position of the amino acid in the full-length protein (S181 in the alignment = S224 in the presumed full length protein). Further, our sequencing tells us that the amino acid at position 227 in the full-length protein (184 in the alignment) is a cysteine instead of an arginine residue. Thus, we made this substitution in our alignment.
RESULTS
Generation of a fusion protein that is palmitoylated in a PORCN-dependent manner
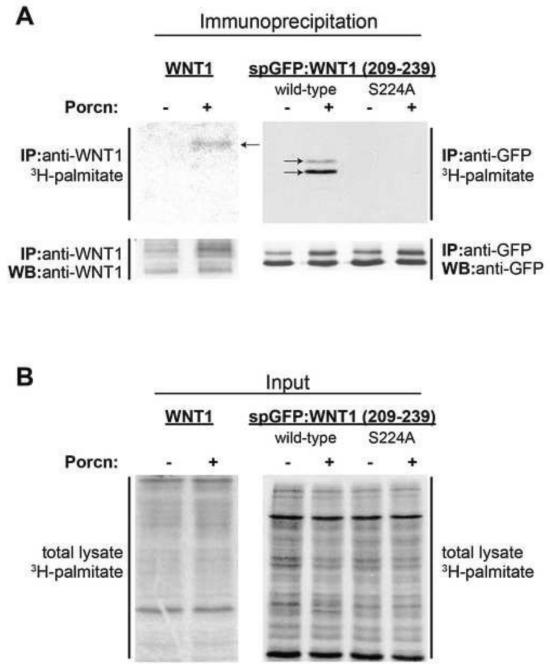
Previous studies from our lab demonstrated that PORCN promotes the hydrophobic modification of a secreted GFP variant fused to a 31 amino acid WNT1 sequence, which flanks the serine residue (S224) known to be palmitoylated in full length Wnts (Fig 1, [19]). To confirm that the increased hydrophobicity of this fusion protein was indeed caused by an increase in palmitoylation, we co-transfected HEK293T cells with full length WNT1 (as a positive control) or the spGFP:WNT1 (209-239) fusion protein along with PORCN or empty vector (control). After labeling the cells with 3H-palmitate, WNT1 and spGFP:WNT1(209-239) were immunoprecipitated, separated via SDS-PAGE and then detected by autoradiography (Fig 2). As expected, PORCN promoted the palmitoylation of full-length WNT1 (Fig 2A). Though the observed signal is weak, it is consistent with the relatively low levels of WNT1 expression (Fig 2A).
Figure 1. Schematic of the base constructs used in our studies.
For these studies we made constructs encoding full-length WNT1 as shown (as per the methods section, mouse sequences encoding residues 1-63 were appended to chick sequencing encoding residues 64-370). After cleavage of the signal peptide between residues 27 and 28 (demarcated by a white line), residues 29-63 are from mouse WNT1 while the residues 64-370 are from chick WNT1. Consensus glycosylation sequences are shown by Y’s. The position of the palmitoylated serine residue (S224) is indicated by a black vertical line. This residue was altered in additional constructs encoding variants with S224A, S224C, and S224T mutations (not shown). We also generated an spGFP:WNT1 (209-239) fusion protein, which consists of amino acids 1-34 of mouse WNT1 at the N-terminus. This sequence encodes the signal peptide, the signal peptide cleavage site, and one glycosylation site (N29). The middle of the fusion protein consists of GFP while the C-terminal ends consists of a short chick WNT1 sequence (209-239) surrounding the palmitoylated serine residue. A parallel fusion protein with the S224A mutation was also made. Lastly, to facilitate our studies in which we used click chemistry to detect WNT1 palmitoylation, we generated a fusion protein in which spGFP:WNT1(209-239) was fused to the Fc region of human IgG. Again, variants of this protein include S224A, S224C, and S224T substitutions (not shown). We also generated a series of fusions encoding shorter WNT1 sequences surrounding S224 (not shown).
Figure 2. spGFP:WNT1 (209-239) is palmitoylated in a PORCN-dependent manner.
HEK293T cells were co-transfected with WNT1 or spGFP:WNT1 (209-239) along with GFP (control) or PORCN. Cells expressing WNT1 (full length or the spGFP:Wnt1(209-239) fusion protein) were metabolically labeled with 3H-palmitate. Full length WNT1 was immunoprecipitated with anti-WNT1 antibodies while the fusion protein was immunoprecipitated with anti-GFP antibodies. Proteins were separated via SDS-PAGE, electroblotted onto a PVDF membrane, treated with En3Hance reagent, and subjected to autoradiography for 3 months. (A) PORCN promotes the incorporation of 3H-palmitate into full length WNT1 and spGFP:WNT1(209-239). 3H-palmitate is incorporated into monoglycosylated (upper band) and non-glycosylated (lower band) spGFP:WNT1(209-239). No 3H palmitate was incorporated into the S224A variant of the spGFP:WNT1 fusion protein. The Western blot of immunoprecipitated WNT1 and spGFP:WNT1 shows that the levels of these proteins are relatively constant in the presence and absence of co-transfected PORCN. (B) The amount of 3H-palmitate incorporated into total cell lysates was equivalent. Due to the lengthy exposures required, this experiment was carried out only twice.
PORCN also promoted the palmitoylation of the spGFP:WNT1 (209-239) fusion protein, indicating that this WNT1-derived short polypeptide serves as a substrate for PORCN-dependent palmitoylation (Fig 2A). The lack of incorporation of 3H-palmitate into spGFP:WNT1 (209-239) S224A in the absence or presence of PORCN co-expression is consistent with the hypothesis that S224 is the site of palmitoylation. As a control, we show consistent levels of palmitoylation in the total cell lysates for all samples (Fig 2B).
Glycosylation promotes, but is not required for, the palmitoylation of WNT1
The migration of spGFP:WNT1 (209-239) as two bands is consistent with previous studies demonstrating that this fusion protein is glycosylated on N29 (Fig 2, UPPER PANEL) [19]. Here, we observe that the non-glycosylated (lower molecular weight) and the mono-glycosylated (higher molecular weight) WNT1-derived fusion peptides both incorporated 3H-palmitate, consistent with recent studies on WNT3A showing that glycosylation is not required for the protein’s palmitoylation [9].
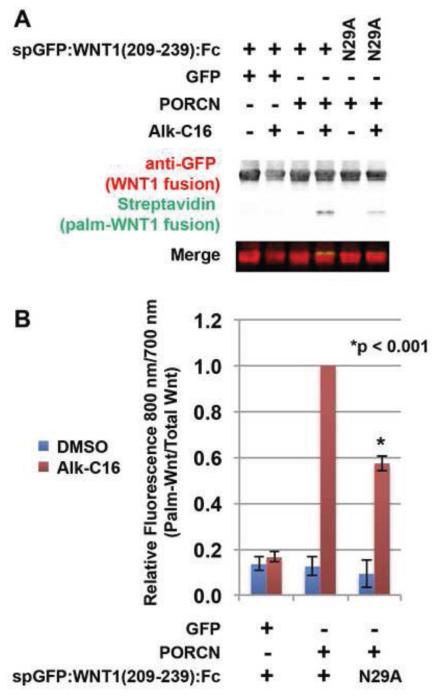
To further assess if glycosylation plays a role in PORCN-dependent Wnt palmitoylation, we used click chemistry to compare the degree of palmitoylation of a spGFP:WNT1(209-239):Fc fusion with a variant in which the glycosylated asparagine was substituted with an alanine (N29A). Cells transfected with PORCN and spGFP:WNT1(209-239):Fc or the spGFP:WNT1 (209-239):Fc N29A mutant were labeled with DMSO (carrier) or ω-alkyne-palmitic acid (Alk-C16) before lysis. Fusion proteins precipitated using Protein A/G beads were then subjected to a bio-orthogonal click chemistry reaction with biotin-azide. This reaction generates a covalent linkage between the alkyne and azide moiety. Proteins were then analyzed by western blot with an anti-GFP antibody (red) and streptavidin-fluorophore (green). In contrast to the studies above, the non-glycosylated and monoglycosylated forms of the Fc fusion constructs appear as a single band in this experiment (Fig 3A). This was not unexpected as the MW of the fusion protein with the Fc tag used here is significantly larger than that of the fusion protein (lacking Fc tag) used above in Figure 2. This difference in size likely contributed to the lack of resolution for the two differentially glycosylated forms of the fusion peptide. Our analysis shows that the glycosylation-deficient variant (N29A) displays roughly half of the level of palmitoylation observed in the glycosylated spGFP:WNT1(209-239):Fc version (Fig 3B). Thus, we conclude that even though glycosylation of distal sequences is not required for the palmitoylation of WNT1, it seems to promote palmitoylation.
Figure 3. Glycosylation promotes, but is not absolutely required for Wnt1 palmitoylation.
HEK293T cells were co-transfected with Fc fusion constructs encoding wild-type spGFP:WNT1 (209-239) or mutant spGFP:WNT1 (209-239) bearing a N29A mutation, along with GFP (control) or PORCN. The N29A mutation in the Fc fusion protein generates a non-glycosylated variant [19]. Cells were metabolically labeled with DMSO carrier (− Alk-C16) or Alkyne-palmitate (+ Alk-C16) for 24hrs, lysed, and precipitated with protein A/G beads [49]. The beads were then subjected to click chemistry with biotin-azide and then separated by SDS-PAGE and analysis by Western Blot [48]. Western Blots were probed with an anti-GFP antibody followed by Alex680-conjugated secondary (red) and IRDye800-conjugated streptavidin (green). The blot was scanned using a Licor Odyssey CLX scanner. (A) In the presence of PORCN, the glycosylation deficient variant N29A is palmitoylated, albeit at reduced levels. (B) Quantification of relative amounts of palmitate labeled-spGFP:Wnt1:Fc and total spGFP:Wnt1:Fc allows us to conclude that while glycosylation does promote palmitoylation, it is not an absolute prerequisite. Data shown are the average of two (N29A) or four (wild-type) independent replicates. Error bars represent +/− standard error. A Student’s t-test was used to assess statistical significance.
PORCN is an O-acyl transferase with specificity to serine and threonine, but not cysteine, residues
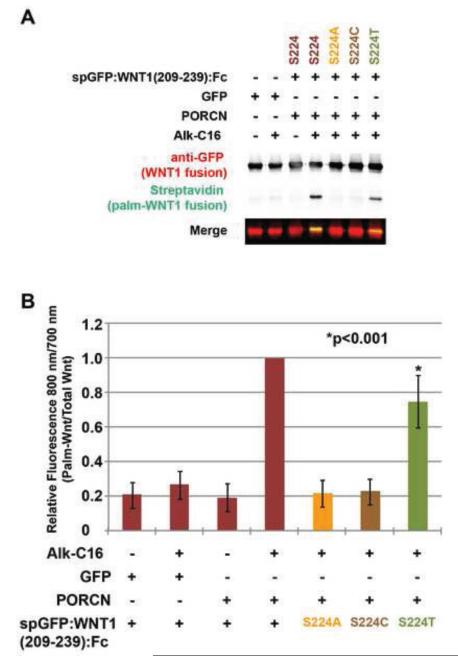
Palmitoyl acyltransferases are known to acylate cysteine, serine, and threonine residues [52]. To test whether PORCN fatty acylates these residues, we co-transfected HEK293T cells with constructs encoding PORCN and spGFP:WNT1(209-239):Fc fusions bearing S224A, S224C or S224T substitutions. Consistent with previous results [19], quantification of the amount of palmitate labeled spGFP(209-239):WNT1:Fc as compared to total spGFP(209-239):WNT1:Fc revealed that PORCN did not promote the palmitoylation of the S224A nor the S224C variants (Fig 4A, B). Interestingly, PORCN-dependent lipid modification was observed in the S224T variant, albeit to lower levels compared to the wild-type protein (Fig 4A, B), indicating that PORCN catalyzes the transfer of fatty acids onto threonine residues. Overall, our data are consistent with a model in which PORCN acts as an O-acyl transferase and not an S-acyl transferase.
Figure 4. PORCN promotes the palmitoylation of serine, threonine, but not cysteine residues.
HEK293T cells were co-transfected with constructs encoding wild-type and variant spGFP:WNT1(209-239) fusion proteins. The variants possessed S224A, S224C, or S224T substitutions. Cells were metabolically labeled with DMSO carrier (− Alk-C16) or Alkyne-palmitate (+ Alk-C16) for 24hrs. After lysis, Fc containing proteins were precipitated with protein A/G beads. Proteins adhered to the beads were then subjected to bio-orthogonal click chemistry with biotin-azide and then separated by SDS-PAGE and analysis by Western Blot. Western blots were probed with anti-GFP followed by Alex680-conjugated secondary antibody (red) and IRDye800-conjugated streptavidin (green). Blots were scanned using a Licor Odyssey CLX scanner. (A) Overexpression of PORCN in the presence of Alk-C16 promotes the incorporation of Alkyne-palmitate into the wild-type and S224T versions of spGFP:WNT1(209-239). The S224A and S224C variants do not incorporate Alkyne palmitate. (B) Quantitation of relative amounts of incorporated palmitate (800 nm) and GFP (700 nm) shows that PORCN can indeed promote the lipid modification of threonine, but to a lesser degree than for the wild-type protein. Data shown are the average of three independent replicates. Error bars represent +/− standard error. A Student’s t-test was used to assess statistical significance.
WNT1 S224T retains biological activity in cultured cells
To test whether the palmitoylation of WNT1 is correlated with its biological activity, we compared the ability of WNT1 S224A, WNT1 S224C, and WNT1 S224T to induce signalling in HEK293T cells, as measured by 8X SuperTopFlash reporter assay, which assesses β-catenin-dependent signaling [41]. Non-palmitoylated Wnt variants (S224A and S224C) showed lack of biological activity as compared to wild-type WNT1 (Fig 5). By contrast, the WNT1 S224T variant promoted signaling via the β-catenin-dependent pathway and exhibits roughly 50% of the activity of wild-type WNT1 (Fig 5). These results highlight the importance of palmitoylation for Wnt biological activity and suggest that serine to threonine mutations could be tolerated in vivo. Interestingly, Moon Jelly Wnt contains a threonine residue instead of serine (see sequence alignment in Fig. 8A), consistent with our observations that that palmitoylation of WNT1 on a threonine residue is tolerated in cell culture and in vivo (see below).
Figure 5. Wnt1 S224T is biologically active in an 8X SuperTopFlash Assay.
HEK293T cells were co-transfected with GFP, WNT1, WNT1S224A, WNT1S224C, or WNT1S224T; Super8xTopFlash or Super8xFopFlash bearing mutated TCF binding sites; and Renilla Luciferase. Cells were incubated overnight, lysed, and assayed for the ratio of Firely Luciferase to Renilla Luciferase using a Dual-Luciferase Reporter Assay. Luciferase measurements were performed using a TD-20/20 luminometer. Over-expression of Wnt1 and Wnt1 S224T activated the TopFlash reporter, while Wnt1S224A and Wnt1S224C did not. Data shown are from 3-9 independent experiments. Each independent experiment had 5 replicates. Error bars represent +/− standard error. A Student’s t-test was used to assess statistical significance.
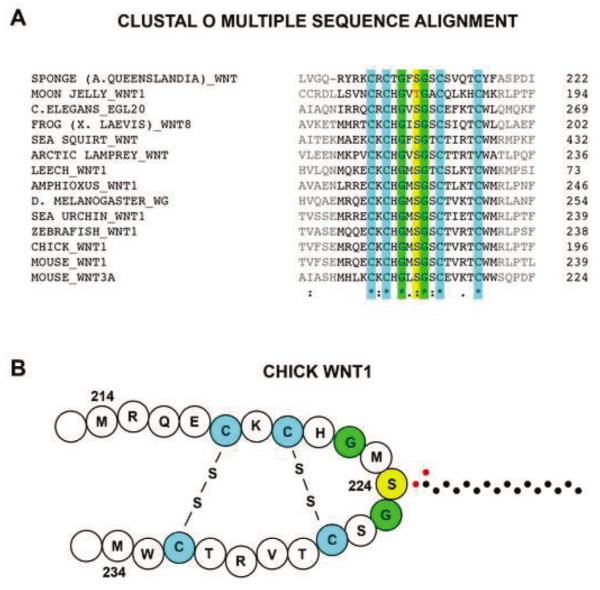
Figure 8. Cross-species alignment of Wnt1 shows a conserved serine or threonine residue involved in PORCN-dependent palmitoylation.
(A) A comparison of Wnt1 sequences of various animal species was performed using a Clustal Omega cross-species alignment. The sequences corresponding to the 31-residue fragment are shown in this alignment. The 5 amino acids on each end of this peptide, which are not required for palmitoylation, are shown in gray. This alignment shows that the moon jelly has a threonine residue in place of the serine. Furthermore, the conservation of only 6 of the 20 internal residues flanking the palmitoylated serine/threonine (yellow) suggest the importance of the four cysteine residues (green) and the two glycine residues (blue). (B) Based on the known structure of XWNT8 [30], we present a working model of the secondary structure of chick WNT1. The carbons of the palmitate group are shown as black dots while the oxygens are depicted by red dots. Symbols are defined as follows: “*” = identical; “:” = conserved substitution; “.” = semi-conserved substitution.
WNT1 S224T is biologically active in the developing spinal cord
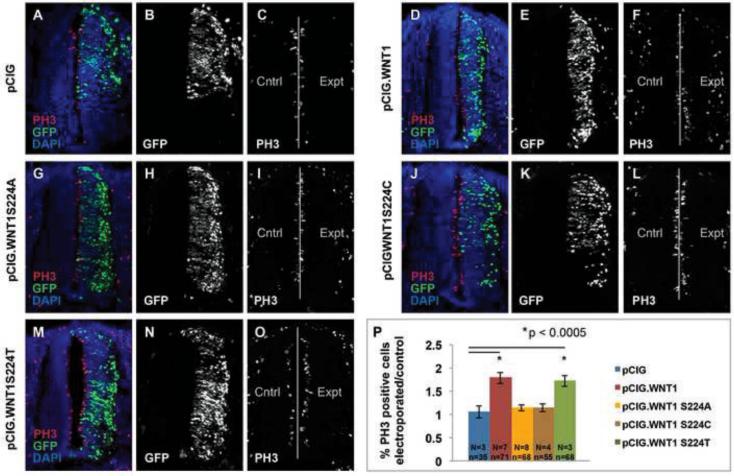
To test the role of WNT1 palmitoylation in vivo, we compared the biological activity of the WNT1 S224A, Wnt S224C, and WNT1 S224T variants to wild-type WNT1 in the developing spinal cord. As overexpression of WNT1 is well known to promote proliferation in the spinal cord [42, 53], we first tested the ability of wild-type and mutated WNT1 to promote proliferation. Consistent with previous reports, electroporation of chick embryos with pCIG.WNT1 (expressing WNT1 and GFP) stimulates proliferation in the spinal cord as compared to pCIG (GFP) alone (Fig 6). As predicted, neither pCIG.WNT1 S224A nor pCIG.WNT1 S224C have a significant effect on proliferation (Fig 6). WNT1 S224T, on the other hand, shows proliferative activity that is indistinguishable from wild-type WNT1 (Fig 6).
Figure 6. Overexpression of wild-type Wnt1 and Wnt1S224T significantly promote cell proliferation in the developing spinal cord.
HH stage 11-14 chick embryos were electroporated with pCIG, pCIG.WNT1, pCIG.WNT1 S224A, pCIG.WNT1 S224C, or pCIG.WNT1 SS24T and harvested at HH stage 20-22. Transverse sections from embryos were stained with anti-phosphohistone H3 to mark proliferative cells and DAPI to mark cell nuclei. After collecting images via confocal microscopy, each image was cut into left and right halves using NIH Image J. Cells were counted as described in materials and methods. A ratio was generated comparing cell proliferation on the experimental (right, electroporated) side cell to that on the control (left, non-electroporated) side. (A, D, G, J, M) Merged images of electroporated spinal cords visualized with GFP (green), anti-phospo histone H3 antibodies (red) and DAPI (blue) to mark electroporated cells, proliferative cells and cell nuclei, respectively. (B, E, H, K, N) Expression of GFP in spinal cords electroporated with pCIG, pCIG.WNT1, pCIG.WNT1 S224A, pCIG.WNT1 S224C, or pCIG.WNT1 S224T. (C, F, I, L, O) Expression of PH3 marker in spinal cords electroporated with pCIG, pCIG.WNT1, pCIG.WNT1 S224A, pCIG.WNT1 S224C, or pCIG.WNT1 S224T. (P) The total number of PH3+ proliferative cells was normalized back to the total number of cells to yield the fraction of proliferative cells on the experimental and control sides of the spinal cord. Values reported on the Y axis are the fraction of proliferative cells on the experimental side divided by that on the control side. N=number of embryos analyzed while and n=number of sections. Error bars represent +/− standard error. A Student’s t-test was used to assess statistical significance.
Overexpression of WNT1 also causes an expansion of dorsal interneurons at the expense of ventral neuronal subtypes, including motor neurons (MNs) [54]. Therefore, we tested the effect of wild-type WNT1 and the two variants on the specification of DI3 interneurons and MNs. Consistent with previous work [54], electroporation of embryos with pCIG.WNT1 significantly enriched the DI3 population as compared to pCIG alone (Fig 7). By contrast, the percent of DI3 interneurons in embryos electroporated with WNT1 S224A was indistinguishable from control embryos (Fig 7). Despite the inability of PORCN to palmitoylate a cysteine residue, WNT1 S224C showed a dramatic inhibitory effect (Fig 7). Though the underlying mechanism for this reduction is not yet known, we suspect that the formation of higher order oligomers due to inappropriate disulfide bridges might be involved. Most importantly, however, electroporation of WNT1 S224T mimicked the effects of wild-type WNT1.
Figure 7. Wnt1S224T mimics wild-type Wnt1 neuron specification activity in the developing spinal cord.
HH stage 11-14 chick embryos were electroporated with pCIG, pCIG.WNT1, pCIG.WNT1 S224A, pCIG.WNT1 S224C, or pCIG.WNT1 SS24T and harvested at HH stage 20-22. Transverse sections from embryos electroporated with pCIG, pCIG.WNT1 wild-type, pCIG.WNT1 S224A, pCIG.WNT1 S224C, and pCIG.WNT1 SS24T were stained with Islet1 to mark dI3 interneuron (dorsal) and motor neurons (ventral) and DAPI to mark cell nuclei. After collecting images via confocal microscopy, each image was cut into left and right halves using NIH Image J. A cell count program was run on all populations (DAPI, Islet1) on each half. A ratio was generated comparing the number of Di3 interneurons or motor neurons on the experimental (electroporated) side cell to that on the control (non-electroporated) side. (A, D, G, J, M) Merged images of electroporated spinal cords visualized with GFP (green), anti-Islet1 immunostaining (red) and DAPI (blue) to mark electroporated cells, neuron subtypes (DI3 is dorsal, MNs are ventral), and cell nuclei, respectively. (B, E, H, K, N) Expression of GFP in spinal cords electroporated with pCIG, pCIG.WNT1, pCIG.WNT1 S224A, pCIG.WNT1 S224C or pCIG.WNT1 S224T. (C, F, I, L, O) Expression of Islet1 marker in spinal cords electroporated with pCIG, pCIG.WNT1, pCIG.WNT1 S224A, pCIG.WNT1 S224C, or pCIG.WNT1 S224T. (P) The total number of neuron subtypes was normalized back to the total number of cells and reported as fraction of Islet1+ cells (DI3s dorsally and MNs ventrally) on the experimental and control sides of the spinal cord. Values reported on the Y axis are the fraction of DI3 or MNs on the experimental side divided by that on the control side. N=number of embryos analyzed while and n=number of sections: pCIG alone (N=3, n=35); WNT1 (N=7, n=71); WNT1 S224A (N=8, n=68); WNT1 S224C (N=4 n=55); and WNT1 S224T (N=3, n=68). Error bars represent +/− standard error. A Student’s t-test was used to assess statistical significance. Abbreviations are as follows: ISL1=Islet1; DI3=dorsal interneuron 3; and MN=motor neurons.
Similar trends were observed for MNs. Whereas expression of WNT1 and GFP reduced the overall percentage of MNs as compared to GFP alone (Fig 6), neither WNT1 S224A nor WNT1S224C decreased the specification of MNs (Fig 6). Again, overexpression of WNT1 S224T recapitulated the effects of wild-type WNT1 (Fig 6). In sum, these data suggest that the S224T mutation is tolerated in vivo.
Determining the minimal length of the WNT1 substrate required for PORCN-dependent palmitoylation
To assess the conservation of residues surrounding the site of palmitoylation, we performed a Clustal Omega cross-species alignment of the sequences surrounding S224 from multiple Wnt family members (Fig 8A). This alignment shows only six absolutely conserved residues in the 31-residue peptide flanking the palmitoylated serine. These include four cysteines and two glycines. A shorter 21-residue peptide retains all of these conserved residues while an 11-residue peptide lacks the two outer cysteines. In XWNT8, the four cysteine residues form two disulfide bridges that result in the serine residue being fully exposed [30]. Based on the conservation of these cysteine residues, we predict that chick WNT1 has a similar secondary structure (Fig 8B).
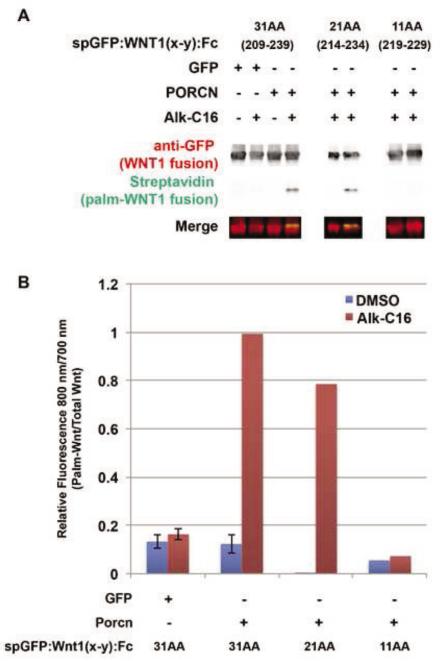
To determine the minimal length of the polypeptide chain surrounding the WNT1 S224 residue which that is sufficient for palmitoylation, we generated constructs encoding spGFP:WNT1 and spGFP:WNT1:Fc fusions with 31 (209-239), 21 (214-234), and 11 (219-229) amino acids and determined their level of palmitoylation by PORCN (Fig 9). HEK293T cells were co-transfected with these fusion constructs along with GFP or PORCN and then labeled with 3H-palmitate (not shown) or Alk-C16 (Fig 9). Alk-C16 labeled cell lysates were immunoprecipitated, subjected to click chemistry, and analyzed by western blot. The fusion proteins with the 31 and 21-residue WNT1 peptides incorporated Alk-C16 (Fig 9). However, there was no observed palmitoylation of the 11-residue peptide, which lacks the outer pair of cysteine residues (Fig 8B,C). These data suggest that the outer disulfide bridge plays a critical role in defining the PORCN recognition site.
Figure 9. The minimal length of the WNT1 substrate is 21 residues.
HEK293T cells were co-transfected spGFP:WNT1(x-y):Fc fusions proteins with either 31 (209-239), 21 (214-234), or 11 (219-229) amino acids flanking S224, along with GFP (control) or PORCN. Cells were metabolically labeled with DMSO carrier (− Alk-C16) or Alkyne-palmitate (+ Alk-C16) for 24hrs. After lysis, Fc containing proteins were precipitated with protein A/G beads. Proteins bound to the beads were subjected to bio-orthogonal click chemistry with biotin-azide, separated by SDS-PAGE and analyzed by Western Blot. Western Blots were probed with anti-GFP followed by Alexa680-conjugated secondary antibody (red) and IRDye800-conjugated streptavidin (green). The blot was scanned using a Licor Odyssey CLX scanner. (A) Only the 31 and 21 residue Wnt1 fragments are able to incorporate Alkyne palmitate. (B) Quantification of the blots (800 nm/700 nm) shows that the 21 residue fragment incorporates roughly 80% of the palmitate as does the 31 residue fragment. The 11 amino acid peptide does not incorporate any detectable palmitate. Data shown for spGFP:WNT1 (209-239) are the average of four independent replicates. Although data shown for spGFP:WNT1(214-234):Fc and spGFP:WNT1(219-229):Fc are from a single replicate, it should be noted that we also measured the incorporation of 3H-palmitate into spGFP:WNT1(209-239), (214-234), and (219-229) fusion proteins (data not shown). Similar results were obtained using both assays (incorporation of Alk-C16 or 3H-palmitate). Error bars represent +/− standard error. A Student’s t-test was used to assess statistical significance.
In sum, we have determined that the 21-residue WNT1 peptide spanning amino acids 214-234 is the shortest fragment that is sufficient to serve as a substrate for PORCN-dependent palmitoylation.
DISCUSSION
Identification of a minimal WNT1 palmitoylation sequence
In this manuscript, we have identified a short WNT1 sequence that is sufficient for the palmitoylation of WNT1 by PORCN. First, we show that a serine or threonine residue is required as the fatty acyl acceptor. The minimal sequence is around 21 amino acids long and need not be glycosylated. In addition to the palmitoylated serine or threonine, there are 6 other completely conserved residues, 4 cysteines (C218, C220, C227, and C232) and 2 glycines (G222 and G225), which are likely required to maintain a structural epitope. Based on results obtained with XWNT8, we expect that C218 and C232 as well as C220 and C227 are linked by disulfide bridges [30] (Fig 8B). These two bridges create a fingerlike projection with the palmitoylated serine present at the tip of the finger. When this sequence is trimmed to 11 amino acids, the disulfide bond at the base of the finger (C218 - C232) is removed and the peptide is no longer an efficient substrate for PORCN palmitoylation. Thus, it seems likely that the structure provided by these two residues is key for recognition of WNT1 by PORCN. Our data agree well with those of two recent publications, which show that these two cysteines are required for the palmitoylation and biological activity of WNT3A [55].
As glycine residues are known to confer flexibility to a polypeptide chain, we further speculate that G222 and G225 permit necessary bending at the tip of the finger-like projection created by the two disulfide bridges. However, in the case of WNT3A, the glycine residue (G207) preceding the palmitoylation site (S209) is required for palmitoylation and biological activity whereas the glycine immediately after the palmitoylation site (G210) is not [56].
In sum, the identification of the WNT1 minimal sequence represents an important step forward for the design of short PORCN substrates, which can be used in enzymatic assays. Our studies also provide a model system for elucidating the substrate specificity of other MBOAT proteins.
Comparison of substrate specificity of HHAT, GOAT and PORCN
Members of the MBOAT family utilize a wide variety of acyl acceptors, including diacylglycerol, lysophospholipids, sterols, and proteins [9, 18, 21, 57-64]. To date, the only MBOATs known to acylate protein substrates are GOAT, HHAT/RASP, and PORCN [9, 18, 21, 58, 61, 64]. Though related in overall function, mouse PORCN shares only 22% identity with GOAT and HHAT. Thus, it is conceivable that the requirements for their peptide substrates are different. One of the striking differences is the minimal length of the peptide substrate. Whereas PORCN requires a peptide between 11 and 21 amino acids, HHAT requires 6 amino acids and GOAT requires only 4 [65]. One possible reason for this distinction is that the palmitoylated cysteine of SHH is located at the N-terminus while the octanoylated serine of Ghrelin is at the 3rd amino acid from the N-terminus [66]. By contrast, for Wnts, the palmitoylated serine is far from the N- and C-termini and is situated at the tip of a loop, which is stabilized by disulfide bonds, requiring additional residues for porcupine recognition.
Moreover, the type of linkage generated by GOAT, HHAT, and PORCN also varies. While GOAT and PORCN generate ester linkages to serine (GOAT, PORCN) and threonine (PORCN) residues [21, 38, 64], HHAT/RASP promote the formation of a thioester linkage on the N-terminal cysteine residue, which then rearranges to an amide linkage via an intramolecular S-to-N (or O-to-N) shift, producing an amide-linked palmitate [58, 65]. HHAT, but not RASP, can also utilize a serine in place of the cysteine [65].
The palmitoylation and biological activity of WNT1 are closely correlated
To test the correlation between WNT1 palmitoylation and biological activity, we assessed the biological activity of wild-type and mutant WNT1 variants in cell culture and in vivo. Consistent with previous work, WNT1S224A constructs were biologically inactive in cell culture and in vivo [19]. Along the same lines, the WNT1 S224C variant lacked biological activity in the 8xSuperTopFlash assay in cultured cells and in the proliferation and motor neuron specification assays in vivo. Surprisingly, this variant exhibited inhibitory activity in the DI3 specification assay. It is possible that the WNT1 S224C variant could form inappropriate disulfide linkages with wild-type Wnts, thus acting as a dominant negative inhibitor [55]. However, it is challenging to conjure up mechanisms that would explain its inhibitory activity in just one of three in vivo assays. On the other hand, we observed that WNT1 S224T mutant was palmitoylated in a PORCN-dependent manner and retained biological activity in the 8xSuperTopFlash assay and in vivo. These data support the idea that palmitoylation of Moon Jelly Wnt which contains a threonine residue instead of serine could generate biologically active Wnt.
Our results support the notion that WNT1 palmitoylation and biological activity are closely correlated and are consistent with recent studies involving WNT3A [56]. In addition to fatty acylating human, mouse and Xenopus WNTs, PORCN mediates palmitoylation of chick WNT1 via the formation of an ester linkage at a serine residue. Finally, our data establish PORCN as an O-acyl transferase for chick WNT1 and lay the foundation for developing PORCN enzymatic assays utilizing short substrate mimics.
HIGHLIGHTS.
PORCN palmitoylates chick WNT1 on Ser224.
WNT1 S224T is palmitoylated and biologically active; WNT1 S224C is not.
Glycosylation of WNT1 promotes palmitoylation, but is not required.
WNT1 residues 209-239 are sufficient for PORCN-dependent palmitoylation.
ACKNOWLEDGEMENTS
We thank Dr. Annette Chan of the Cell and Molecular Imaging Center for her assistance with confocal microscopy (SFSU). Matilde Miranda was supported by an NIH MARC fellowship while Michael Enriquez was supported by an NSF REU fellowship. Linda Szabo was supported by NSF RUI IOS-0950892. This research was made possible by NSF RUI MCB-1244602, NSF RUI IOS-0950892, NIH 2 SO6 GM52588, NIH 1R15HD070206-01A1, and CSUPERB grants to Dr. Laura Burrus and a NIH-RIMI (P20MD000262) grant to San Francisco State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Axelrod JD. Progress and challenges in understanding planar cell polarity signaling. Semin Cell Dev Biol. 2009;20:964–71. doi: 10.1016/j.semcdb.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Cadigan KM, Peifer M. Wnt signaling from development to disease: insights from model systems. Cold Spring Harb Perspect Biol. 2009;1:a002881. doi: 10.1101/cshperspect.a002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chien AJ, Conrad WH, Moon RT. A Wnt survival guide: from flies to human disease. J Invest Dermatol. 2009;129:1614–27. doi: 10.1038/jid.2008.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coombs GS, Covey TM, Virshup DM. Wnt signaling in development. disease and translational medicine, Curr Drug Targets. 2008;9:513–31. doi: 10.2174/138945008784911796. [DOI] [PubMed] [Google Scholar]
- 5.van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–14. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 6.Brown AM, Papkoff J, Fung YK, Shackleford GM, Varmus HE. Identification of protein products encoded by the proto-oncogene int-1. Mol Cell Biol. 1987;7:3971–7. doi: 10.1128/mcb.7.11.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papkoff J, Brown AM, Varmus HE. The int-1 proto-oncogene products are glycoproteins that appear to enter the secretory pathway. Mol Cell Biol. 1987;7:3978–84. doi: 10.1128/mcb.7.11.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cha SW, Tadjuidje E, White J, Wells J, Mayhew C, Wylie C, Heasman J. Wnt11/5a complex formation caused by tyrosine sulfation increases canonical signaling activity. Curr Biol. 2009;19:1573–80. doi: 10.1016/j.cub.2009.07.062. [DOI] [PubMed] [Google Scholar]
- 9.Gao X, Hannoush RN. Single-cell imaging of Wnt palmitoylation by the acyltransferase porcupine. Nat Chem Biol. 2014;10:61–8. doi: 10.1038/nchembio.1392. [DOI] [PubMed] [Google Scholar]
- 10.Komekado H, Yamamoto H, Chiba T, Kikuchi A. Glycosylation and palmitoylation of Wnt-3a are coupled to produce an active form of Wnt-3a. Genes Cells. 2007;12:521–34. doi: 10.1111/j.1365-2443.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- 11.Kurayoshi M, Yamamoto H, Izumi S, Kikuchi A. Post-translational palmitoylation and glycosylation of Wnt-5a are necessary for its signalling. Biochem J. 2007;402:515–23. doi: 10.1042/BJ20061476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason JO, Kitajewski J, Varmus HE. Mutational analysis of mouse Wnt-1 identifies two temperature-sensitive alleles and attributes of Wnt-1 protein essential for transformation of a mammary cell line. Mol Biol Cell. 1992;3:521–33. doi: 10.1091/mbc.3.5.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papkoff J. Inducible overexpression and secretion of int-1 protein. Mol Cell Biol. 1989;9:3377–84. doi: 10.1128/mcb.9.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Port F, Basler K. Wnt trafficking: new insights into Wnt maturation. secretion and spreading, Traffic. 2010;11:1265–71. doi: 10.1111/j.1600-0854.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 15.Smolich BD, McMahon JA, McMahon AP, Papkoff J. Wnt family proteins are secreted and associated with the cell surface. Mol Biol Cell. 1993;4:1267–75. doi: 10.1091/mbc.4.12.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang X, Wu Y, Belenkaya TY, Huang Q, Ray L, Qu J, Lin X. Roles of N-glycosylation and lipidation in Wg secretion and signaling. Dev Biol. 2012;364:32–41. doi: 10.1016/j.ydbio.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto H, Awada C, Hanaki H, Sakane H, Tsujimoto I, Takahashi Y, Takao T, Kikuchi A. The apical and basolateral secretion of Wnt11 and Wnt3a in polarized epithelial cells is regulated by different mechanisms. J Cell Sci. 2013;126:2931–43. doi: 10.1242/jcs.126052. [DOI] [PubMed] [Google Scholar]
- 18.Galli LM, Barnes TL, Secrest SS, Kadowaki T, Burrus LW. Porcupine-mediated lipid-modification regulates the activity and distribution of Wnt proteins in the chick neural tube. Development. 2007;134:3339–48. doi: 10.1242/dev.02881. [DOI] [PubMed] [Google Scholar]
- 19.Galli LM, Burrus LW. Differential palmit(e)oylation of Wnt1 on C93 and S224 residues has overlapping and distinct consequences. PLoS One. 2011;6:e26636. doi: 10.1371/journal.pone.0026636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rios-Esteves J, Resh MD. Stearoyl CoA Desaturase Is Required to Produce Active, Lipid-Modified Wnt Proteins. Cell Rep. 2013;4:1072–81. doi: 10.1016/j.celrep.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H, Takao T, Takada S. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev Cell. 2006;11:791–801. doi: 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, 3rd, Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–52. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 23.Zhai L, Chaturvedi D, Cumberledge S. Drosophila Wnt-1 undergoes a hydrophobic modification and is targeted to lipid rafts; A process that requires porcupine. The Journal of biological chemistry. 2004;279:33220–33227. doi: 10.1074/jbc.M403407200. [DOI] [PubMed] [Google Scholar]
- 24.van den Heuvel M, Harryman-Samos C, Klingensmith J, Perrimon N, Nusse R. Mutations in the segment polarity genes wingless and porcupine impair secretion of the wingless protein. Embo J. 1993;12:5293–302. doi: 10.1002/j.1460-2075.1993.tb06225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bänziger C, Soldini D, Schutt C, Zipperlen P, Hausmann G, Basler K. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006;125:509–22. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 26.Bartscherer K, Boutros M. Regulation of Wnt protein secretion and its role in gradient formation. EMBO Rep. 2008;9:977–82. doi: 10.1038/embor.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coombs GS, Yu J, Canning CA, Veltri CA, Covey TM, Cheong JK, Utomo V, Banerjee N, Zhang ZH, Jadulco RC, Concepcion GP, Bugni TS, Harper MK, Mihalek I, Jones CM, Ireland CM, Virshup DM. WLS-dependent secretion of WNT3A requires Ser209 acylation and vacuolar acidification. J Cell Sci. 2010;123:3357–67. doi: 10.1242/jcs.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodman RM, Thombre S, Firtina Z, Gray D, Betts D, Roebuck J, Spana EP, Selva EM. Sprinter: a novel transmembrane protein required for Wg secretion and signaling. Development. 2006;133:4901–11. doi: 10.1242/dev.02674. [DOI] [PubMed] [Google Scholar]
- 29.Herr P, Basler K. Porcupine-mediated lipidation is required for Wnt recognition by Wls. Dev Biol. 2012;361:392–402. doi: 10.1016/j.ydbio.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrott JJ, Cash GM, Smith AP, Barrow JR, Murtaugh LC. Deletion of mouse Porcn blocks Wnt ligand secretion and reveals an ectodermal etiology of human focal dermal hypoplasia/Goltz syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12752–7. doi: 10.1073/pnas.1006437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofmann K. A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem Sci. 2000;25:111–2. doi: 10.1016/s0968-0004(99)01539-x. [DOI] [PubMed] [Google Scholar]
- 33.Barrow JR, Howell WD, Rule M, Hayashi S, Thomas KR, Capecchi MR, McMahon AP. Wnt3 signaling in the epiblast is required for proper orientation of the anteroposterior axis. Dev Biol. 2007;312:312–20. doi: 10.1016/j.ydbio.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 34.Biechele S, Cockburn K, Lanner F, Cox BJ, Rossant J. Porcn-dependent Wnt signaling is not required prior to mouse gastrulation. Development. 2013;140:2961–71. doi: 10.1242/dev.094458. [DOI] [PubMed] [Google Scholar]
- 35.Biechele S, Cox BJ, Rossant J. Porcupine homolog is required for canonical Wnt signaling and gastrulation in mouse embryos. Dev Biol. 2011 doi: 10.1016/j.ydbio.2011.04.029. [DOI] [PubMed] [Google Scholar]
- 36.Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat Genet. 1999;22:361–5. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 37.Tortelote GG, Hernandez-Hernandez JM, Quaresma AJ, Nickerson JA, Imbalzano AN, Rivera-Perez JA. Wnt3 function in the epiblast is required for the maintenance but not the initiation of gastrulation in mice. Dev Biol. 2013;374:164–73. doi: 10.1016/j.ydbio.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grzeschik KH, Bornholdt D, Oeffner F, Konig A, del Carmen Boente M, Enders H, Fritz B, Hertl M, Grasshoff U, Hofling K, Oji V, Paradisi M, Schuchardt C, Szalai Z, Tadini G, Traupe H, Happle R. Deficiency of PORCN, a regulator of Wnt signaling, is associated with focal dermal hypoplasia. Nat Genet. 2007;39:833–5. doi: 10.1038/ng2052. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Reid Sutton V, Omar Peraza-Llanes J, Yu Z, Rosetta R, Kou YC, Eble TN, Patel A, Thaller C, Fang P, Van den Veyver IB. Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nat Genet. 2007;39:836–8. doi: 10.1038/ng2057. [DOI] [PubMed] [Google Scholar]
- 40.Bornholdt D, Oeffner F, Konig A, Happle R, Alanay Y, Ascherman J, Benke PJ, Boente Mdel C, van der Burgt I, Chassaing N, Ellis I, Francisco CR, Della Giovanna P, Hamel B, Has C, Heinelt K, Janecke A, Kastrup W, Loeys B, Lohrisch I, Marcelis C, Mehraein Y, Nicolas ME, Pagliarini D, Paradisi M, Patrizi A, Piccione M, Piza-Katzer H, Prager B, Prescott K, Strien J, Utine GE, Zeller MS, Grzeschik KH. PORCN mutations in focal dermal hypoplasia: coping with lethality. Hum Mutat. 2009;30:E618–28. doi: 10.1002/humu.20992. [DOI] [PubMed] [Google Scholar]
- 41.Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell. 2003;5:367–77. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 42.Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–98. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka K, Okabayashi K, Asashima M, Perrimon N, Kadowaki T. The evolutionarily conserved porcupine gene family is involved in the processing of the Wnt family. Eur J Biochem. 2000;267:4300–11. doi: 10.1046/j.1432-1033.2000.01478.x. [DOI] [PubMed] [Google Scholar]
- 44.Fokina VM, Frolova EI. Expression patterns of Wnt genes during development of an anterior part of the chicken eye. Dev Dyn. 2006;235:496–505. doi: 10.1002/dvdy.20621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–9. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 46.Hannoush RN, Arenas-Ramirez N. Imaging the lipidome: omega-alkynyl fatty acids for detection and cellular visualization of lipid-modified proteins. ACS chemical biology. 2009;4:581–7. doi: 10.1021/cb900085z. [DOI] [PubMed] [Google Scholar]
- 47.Gao X, Arenas-Ramirez N, Scales SJ, Hannoush RN. Membrane targeting of palmitoylated Wnt and Hedgehog revealed by chemical probes. FEBS Lett. 2011 doi: 10.1016/j.febslet.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 48.Gao X, Hannoush RN. Method for cellular imaging of palmitoylated proteins with clickable probes and proximity ligation applied to Hedgehog, tubulin, and Ras. Journal of the American Chemical Society. 2014;136:4544–50. doi: 10.1021/ja410068g. [DOI] [PubMed] [Google Scholar]
- 49.Dodge ME, Moon J, Tuladhar R, Lu J, Jacob LS, Zhang LS, Shi H, Wang X, Moro E, Mongera A, Argenton F, Karner CM, Carroll TJ, Chen C, Amatruda JF, Lum L. Diverse chemical scaffolds support direct inhibition of the membrane bound O-acyltransferase Porcupine. The Journal of biological chemistry. doi: 10.1074/jbc.M112.372029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic acids research. 2010;38:W695–9. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Resh MD. Palmitoylation of ligands. receptors, and intracellular signaling molecules, Sci STKE. 2006;2006:re14. doi: 10.1126/stke.3592006re14. [DOI] [PubMed] [Google Scholar]
- 53.Dickinson ME, Krumlauf R, McMahon AP. Evidence for a mitogenic effect of Wnt-1 in the developing mammalian central nervous system. Development. 1994;120:1453–71. doi: 10.1242/dev.120.6.1453. [DOI] [PubMed] [Google Scholar]
- 54.Alvarez-Medina R, Cayuso J, Okubo T, Takada S, Marti E. Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development. 2008;135:237–47. doi: 10.1242/dev.012054. [DOI] [PubMed] [Google Scholar]
- 55.MacDonald BT, Hien A, Zhang X, Iranloye O, Virshup DM, Waterman ML, He X. Disulfide bond requirements for active wnt ligands. The Journal of biological chemistry. 2014;289:18122–36. doi: 10.1074/jbc.M114.575027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rios-Esteves J, Haugen B, Resh MD. Identification of Key Residues and Regions Important for Porcupine-mediated Wnt Acylation. The Journal of biological chemistry. 2014;289:17009–19. doi: 10.1074/jbc.M114.561209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bosson R, Jaquenoud M, Conzelmann A. GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol Biol Cell. 2006;17:2636–45. doi: 10.1091/mbc.E06-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buglino JA, Resh MD. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. The Journal of biological chemistry. 2008;283:22076–88. doi: 10.1074/jbc.M803901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cases S, Smith SJ, Zheng YW, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ, Erickson SK, Farese RV., Jr. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase. a key enzyme in triacylglycerol synthesis, Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13018–23. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang CC, Huh HY, Cadigan KM, Chang TY. Molecular cloning and functional expression of human acyl-coenzyme A:cholesterol acyltransferase cDNA in mutant Chinese hamster ovary cells. The Journal of biological chemistry. 1993;268:20747–55. [PubMed] [Google Scholar]
- 61.Gutierrez JA, Solenberg PJ, Perkins DR, Willency JA, Knierman MD, Jin Z, Witcher DR, Luo S, Onyia JE, Hale JE. Ghrelin octanoylation mediated by an orphan lipid transferase. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6320–5. doi: 10.1073/pnas.0800708105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hishikawa D, Shindou H, Kobayashi S, Nakanishi H, Taguchi R, Shimizu T. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2830–5. doi: 10.1073/pnas.0712245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shindou H, Shimizu T. Acyl-CoA:lysophospholipid acyltransferases. The Journal of biological chemistry. 2009;284:1–5. doi: 10.1074/jbc.R800046200. [DOI] [PubMed] [Google Scholar]
- 64.Yang J, Brown MS, Liang G, Grishin NV, Goldstein JL. Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell. 2008;132:387–96. doi: 10.1016/j.cell.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 65.Hardy RY, Resh MD. Identification of N-terminal residues of Sonic Hedgehog important for palmitoylation by Hedgehog acyltransferase. The Journal of biological chemistry. 2012;287:42881–9. doi: 10.1074/jbc.M112.426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402:656–60. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]