Abstract
Members of the Casitas B-Lineage Lymphoma (Cbl) family (Cbl, Cbl-b and Cbl-c) of ubiquitin ligases serve as negative regulators of receptor tyrosine kinases (RTKs). An essential role of Cbl-family protein-dependent ubiquitination for efficient ligand-induced lysosomal targeting and degradation is now well-accepted. However, a more proximal role of Cbl and Cbl-b as adapters for CIN85-endophilin recruitment to mediate ligand-induced initial internalization of RTKs is supported by some studies but refuted by others. Overexpression and/or incomplete depletion of Cbl proteins in these studies is likely to have contributed to this dichotomy. To address the role of endogenous Cbl and Cbl-b in the internalization step of RTK endocytic traffic, we established Cbl/Cbl-b double-knockout (DKO) mouse embryonic fibroblasts (MEFs) and demonstrated that these cells lack the expression of both Cbl-family members as well as endophilin A, while they express CIN85. We show that ligand-induced ubiquitination of EGFR, as a prototype RTK, was abolished in DKO MEFs, and EGFR degradation was delayed. These traits were reversed by ectopic human Cbl expression. EGFR endocytosis, assessed using the internalization of 125I-labeled or fluorescent EGF, or of EGFR itself, was largely retained in Cbl/Cbl-b DKO compared to wild type MEFs. EGFR internalization was also largely intact in Cbl/Cbl-b depleted MCF-10A human mammary epithelial cell line. Inducible shRNA-mediated knockdown of CIN85 in wild type or Cbl/Cbl-b DKO MEFs had no impact on EGFR internalization. Our findings, establish that, at physiological expression levels, Cbl, Cbl-b and CIN85 are largely dispensable for EGFR internalization. Our results support the model that Cbl-CIN85-endophilin complex is not required for efficient internalization of EGFR, a prototype RTK.
INTRODUCTION
The Casitas B-lineage Lymphoma (Cbl) family members, Cbl, Cbl-b and Cbl-c, are RING finger type ubiquitin ligases (or E3s) that function, selectively, in the negative regulation of protein tyrosine kinase (PTK) signaling (Mohapatra et al. 2013). The PTK selectivity of Cbl proteins is imparted by the mechanism of their recruitment to PTKs, which requires binding of the conserved N-terminal Tyrosine Kinase-Binding (TKB) domain of Cbl proteins to specific phospho-tyrosine peptide motifs on PTKs generated upon their activation (Lill et al. 2000; Meng et al. 1999). Interaction with PTKs leads to phosphorylation of a highly conserved tyrosine in the linker helix region of Cbl proteins and this event is required for their activation as E3s (Dou et al. 2012; Kales, Ryan, Lipkowitz 2012; Lipkowitz and Weissman 2011; Miyake et al. 1998). These attributes have established Cbl-family proteins as activation-induced negative feedback regulators of PTKs (Mohapatra et al. 2013).
Both receptor tyrosine kinases (RTKs) and non-receptor PTKs have been demonstrated as targets of Cbl proteins (Mohapatra et al. 2013). Mechanistically, however, regulation of RTKs follows a distinct pathway. Early studies established that Cbl promotes the ubiquitination of PDGF receptor (Joazeiro et al. 1999; Miyake et al. 1998; Miyake et al. 1999) and EGFR (Levkowitz et al. 1998; Lill et al. 2000). Based on yeast genetic identification of the vacuolar protein sorting pathway (Katzmann, Odorizzi, Emr 2002), it was soon established that ubiquitination of RTKs targets them for recognition by the mammalian counterparts of the Endosomal Sorting Complex Required for Transport (ESCRT) complexes (Henne, Buchkovich, Emr 2011; MacGurn, Hsu, Emr 2012; Rusten, Vaccari, Stenmark 2011; Saksena et al. 2007; Wegner, Rodahl, Stenmark 2011). The ESCRT machinery is essential for sorting of receptors with cytoplasmic domain ubiquitin tags into inner vesicles of the multi-vesicular body (MVB; vacuole in yeast) and to subject them to lysosomal degradation (Henne, Buchkovich, Emr 2011; Rusten, Vaccari, Stenmark 2011; Saksena et al. 2007; Wegner, Rodahl, Stenmark 2011). This mechanism predicted that Cbl proteins will be required for lysosomal degradation of endocytosed RTKs. Indeed, expression of E3-deficient mutant Cbl protein expression, which impairs the RTK ubiquitination, was shown to reduce the ligand-induced degradation of EGFR (Levkowitz et al. 1998; Lill et al. 2000) and c-MET (Peschard et al. 2001). Studies using mouse embryonic fibroblast (MEF) lines isolated from two independently-derived Cbl-null mouse models demonstrated that Cbl was required for efficient ligand-induced degradation of EGFR (Duan et al. 2003), PDGFR (Reddi et al. 2007) and FGFR (Mason et al. 2004). Other studies used RNAi knockdown to further confirm the requirement of Cbl in ligand-induced RTK degradation (Pennock and Wang 2008; Reddi et al. 2007). Furthermore, combined knockdown of Cbl and Cbl-b in immortal human mammary epithelial cells was shown to be more efficient than Cbl knockdown alone at reducing the ligand-induced degradation of EGFR (Duan et al. 2011). These studies have established an important role of Cbl and Cbl-b in ligand-induced degradation of EGFR and other RTKs. However, these studies used cell models where the expression of all Cbl proteins was not completely absent. For example, RNAi knockdown studies suffer from the residual low level protein expression that might be sufficient for certain phenotypes. Expression of Cbl-b in Cbl-null MEFs used in earlier studies (Duan et al. 2003) raises a similar issue. Similarly, in studies using combined Cbl and Cbl-b knockdown (Duan et al. 2011), expression of endogenous Cbl-c could provide partial compensation since Cbl-c is capable of promoting EGFR ubiquitination (Ryan et al. 2010; Yarden and Sliwkowski 2001).
Previous studies demonstrated that mono-ubiquitin modification of cytoplasmic tails of yeast surface receptors provided a motif for their internalization from the cell surface (Hicke and Riezman 1996). Use of CHO cells bearing a temperature-sensitive ubiquitin-activating enzyme (E1), showed a requirement of the ubiquitin pathway for ligand-induced internalization of human growth hormone receptor even though the receptor itself was not ubiquitinated (Govers et al. 1999). These findings have raised the possibility that Cbl proteins may also be important in the initial internalization step (endocytosis) of RTK endocytic traffic. Consistent with such a possibility, Cbl-dependent ubiquitination of EGFR was shown to begin at the cell surface and to continue until the receptor entered the late endosome/lysosomal compartment (Longva et al. 2002). Furthermore, EGFR fused at the C-terminus to a chain-elongation-deficient ubiquitin mutant was shown to constitutively traffic to lysosomes (Bertelsen et al. 2011; Mosesson et al. 2003). Other studies have revealed a mechanism by which Cbl or Cbl-b can function as adapters to mediate ligand-stimulated RTK internalization. In this model, shown for EGFR and c-MET (Petrelli et al. 2002; Soubeyran et al. 2002), Cbl proteins recruit a CIN85-endophilin A complex, bound to proline-rich region on Cbl, to activated receptors to allow endophilin to facilitate endocytic bud formation (Dikic 2002). The conclusions were based primarily on heterologous overexpression of Cbl, CIN85 and endophilin, and use of mutant forms of Cbl and endophilin (Petrelli et al. 2002; Soubeyran et al. 2002).
Contrary to above conclusions, relatively more physiological approaches have revealed that Cbl proteins are dispensable for ligand-induced RTK internalization. Ligand-induced internalization of EGFR was unaltered in Cbl-null MEFs (Duan et al. 2003), or upon combined knockdown of Cbl and Cbl-b in 293T cells, BT20 breast cancer cell line (Pennock and Wang 2008) or immortal mammary epithelial cells (Duan et al. 2011). However, Cbl-family E3-dependent ubiquitination was found to be crucial for lysosomal sorting and degradation of RTKs. Complementing these studies, it has been demonstrated that mutation of 15 lysine residues within the cytoplasmic region of EGFR eliminated its ligand-induced ubiquitination but did not impair the ligand-induced internalization (Huang, Goh, Sorkin 2007). In fact, inhibition of the ubiquitin machinery in CHO cells with temperature-sensitive E1 left EGFR endocytosis intact (Duan et al. 2003). Despite predominant reliance on depletion rather than overexpression approaches in the above studies, it remains possible that deletion of a single family member in knockout approaches or partial depletion in combined Cbl/Cbl-b knockdown studies may account for the lack of an effect on RTK internalization. Studies using epithelial cell models could also be complicated by the expression of Cbl-c (Pennock and Wang 2008). Thus, analyses in cell systems with a complete absence of the expression of all Cbl family members will help provide a definitive test of the contradictory notions on the role of Cbl proteins in ligand-induced initial internalization (endocytosis) of RTK.
Aside from contradictory findings on the role of Cbl proteins, recent studies have also cast doubt on the role of Cbl-CIN85 complex in promoting RTK internalization. Knockdown of CIN85 has been shown to leave ligand-induced internalization of EGFR unperturbed while impairing its ubiquitination and degradation (Schroeder et al. 2010). In one study (Ronning et al. 2011), expression of dominant-negative CIN85 was shown to impede EGFR internalization, similar to previous studies (Soubeyran et al. 2002), but notably CIN85 knockdown only impaired EGFR degradation without altering its internalization. Additional studies have shown that CIN85 interacts with Cbl in post-endocytic endosomal compartments (Schroeder et al. 2010; Zhang et al. 2009). It was also reported that ARAP1 interacts with CIN85 in endosomes, in competition with Cbl, and impairs EGFR degradation (Schmidt et al. 2004). Another study showed that Src-mediated CIN85 phosphorylation reduces its interaction with Cbl and impairs EGFR degradation but not the endocytosis step (Schroeder et al. 2012). Consistent with these recent studies, even one of the original studies that proposed a Cbl-CIN85-endophilin-mediated RTK internalization model found the endogenous Cbl-CIN85 interaction to require c-MET activation and to peak at substantially late time points (Petrelli et al. 2002).
To rigorously test the requirement of Cbl proteins in ligand-induced RTK internalization, the present study used a genetic approach in which knockout MEFs with engineered deletions of both Cbl and Cbl-b genes were utilized and demonstrated to lack of the expression of Cbl and Cbl-b. Cbl-c, which is known to be primarily expressed in epithelial tissues (Keane et al. 1999; Mohapatra et al. 2013), and lacks the CIN85 interacting proline-rich region (Keane et al. 1999; Ryan et al. 2010; Soubeyran et al. 2002) is not expressed in these cells. Hence, the cell system used here completely lacks the expression of all Cbl-family proteins and hence Cbl-family protein-dependent CIN85-endophilin recruitment to promote RTK internalization can be excluded. We show that EGFR does not undergo any detectable EGF-induced ubiquitination in Cbl/Cbl-b double knockout (DKO) MEFs, as anticipated, but EGFR internalization in these cells is comparable to that in wild-type MEFs. These studies show that Cbl proteins and consequent ubiquitination are not required for EGFR internalization. We also demonstrate a lack of expression of endophilin-1A in both WT and DKO MEFs, and show that CIN85 depletion in WT or DKO MEFs has no impact on EGFR internalization. Collectively, our studies resolve the contradictory conclusions in the field and demonstrate clearly that the Cbl-CIN85-Endophilin complex is dispensable for ligand-induced internalization of a prototype RTK, EGFR.
EXPERIMENTAL PROCEDURES
Reagents
The sources for reagents were as follows: Chemicals: Enzyme immunoassay assay-grade BSA, paraformaldehyde (PFA), saponin, purified mouse EGF (used for endogenous mouse EGFR studies in MEFs) and human EGF (used for hEGFR overexpressing MEFs and MCF-10A cells) were from Sigma-Aldrich (St. Louis, MO). Alexa Fluor 488-conjugated EGF was from Molecular Probes (Eugene, OR). OptiMEM (reduced serum media) and Lipofectamine as well as MEF culture media were from Life Technologies (Grand Island, NY).
Antibodies
The following antibodies were obtained from commercial sources: rabbit polyclonal (pAb) anti-EGFR (1005), pAb anti-Cbl (C-15), mouse monoclonal antibody (mAb) anti-HSC70 (B-6), and mAb anti-spleen tyrosine kinase (Syk) (4D10) from Santa Cruz Biotechnology Inc. (Santa Cruz, CA); mAb anti-β-actin (A5441) from Sigma (St. Louis, MO); pAb anti-Endophilin-1A from Synaptic Systems (159002); mAb anti-Ubiquitin (P4D1) from Covance Research Products (Denver, PA); Rabbit monoclonal anti-pEGFR (D7A5), anti-CIN85 (D1A4), anti-Cbl-b (D3C12) and anti-EEA1 (C45B10) from Cell Signaling Inc. (Danvers, MA); mAb anti-EGFR (clone 528; ATCC) was Protein G-purified in-house from serum-free hybridoma supernatant. HRP conjugated species-specific secondary antibodies used for immunoblotting were from Zymed Laboratories (South San Francisco, CA), and those for immuno-staining were from Invitrogen-Molecular Probes (Eugene, OR).
DNA Constructs
MSCV-puro human EGFR (Duan et al. 2003), and pGCDNsamIRESGFP Flag-tagged human CBL (Sanada et al. 2009) retroviral expression vectors have been previously described. A doxycycline-inducible lentiviral mouse CIN85 knockdown construct was generated by moving CIN85 shRNA sequences (Open Biosystems clone ID: V3LHS_320424) from GIPZ into TRIPZ backbone according to the Open Biosystems protocol, and empty TRIPZ was used as a control.
Cell Culture and viral transduction
MEF lines were derived using previously described methods (Duan et al. 2003) from day 13.5 embryos of Cblfl/fl; Cbl-b−/− or WT genotype (C57BL/6 background), followed by the 3T3 protocol from passage 3 to 25 (Todaro and Green 1966). Cbl−/−(GU) MEFs have been described previously (Duan et al. 2003); these represent germline Cre-mediated deletion of floxed Cbl. Cblflox/flox; Cbl-b-null conditional DKO MEFs were derived from Cblflox/flox; Cbl-b−/− embryos (Naramura et al. 1998). These cells were grown in minimal essential-α medium with supplements as described previously (Duan et al. 2003). Conditional DKO MEFs were infected with adenoviruses encoding the enhanced green fluorescent protein (EGFP) (control) or EGFP-Cre (University of Iowa Gene Transfer Vector Core) at an MOI of 50-100, cells with highest fluorescence isolated by sorting on a fluorescence-activated cell sorter (FACS; FACSAria), and used as polyclonal lines.
Retroviral supernatants for hEGFR and hCbl expression were generated by transient transfection into ecotropic packaging cell line Phoenix (Provided by Dr. Garry P. Nolan, Stanford University USA) using the calcium phosphate co-precipitation method. Supernatants of Phoenix cells transfected with MSCV-puro-hEGFR, pGCDNsamIRESGFP-hCbl or vector alone (control) were applied on target MEFs every 6 hours for 24h (4 rounds). The TRIPZ CIN85-shRNA vector or control vector was transfected into 293FT packaging cells (Provided by Dr. Garry P. Nolan, Stanford University USA) to generate lentiviral supernatants. Lentiviral infection was carried out in the presence of 8 μg/ml Polybrene (Sigma) and transductants selected in 2 μg/ml puromycin. CIN85 shRNA was induced with doxycycline (2 μg/ml) for 4 days prior to use in experiments.
PCR-based genotyping
DNA was isolated from MEFs after proteinase K (Roche) digestion. Genotyping was performed using a duplex PCR using the primers for Cbl and Cbl-b shown in Supplementary Table S1.
RNA Extraction, Semi-Quantitative and Real-time qPCR
2 μg of total RNA isolated from MEFs using the RNeasy Mini kit (Qiagen) was subjected to reverse transcription using iScript™ reverse transcriptase (BIO RAD). PCR amplification was performed by using the primers listed in Supplementary Table S1. Semi-quantitative analysis of PCR amplicons on agarose gels were performed by using Taq polymerase from Thermo scientific (Pittsburgh PA) for 30 cycles. Expression level of GAPDH served as loading control. Real-time PCR was performed on cDNA generated as above in triplicates using SYBR Green PCR master mix (Applied Biosystems). Expression levels were normalized against GAPDH signals.
EGF stimulation, preparation of cell lysates, Immunoprecipitation, Immunoblotting and Confocal Immunofluorescence Microscopy
These procedures have been described previously (Duan et al. 2003) and brief explanations are included in figure legends.
Flow cytometry
Control MEFs or those transduced with adenovirus, retrovirus or lentivirus constructs were seeded at 300,000 cells per 100-mm dish for 48h, released with trypsin/EDTA (Invitrogen) followed by excess trypsin inhibitor. Cells were washed and used as such or following immuno-staining with specific antibodies in cold FACS buffer (2% fetal bovine serum in PBS) and two washes. The samples were analyzed on a BD FACScalibur flow cytometer. Data were analyzed using the CellQuest® software.
125I-EGF Internalization Assay
Purified mouse and human EGF was labeled with 125I (using 125I-sodium iodide; specific activity 1mCi 125I /10μl) to a specific activity of approximately 120 μCi/μg [2 × 108 counts per minute (cpm)] using the chloramine-T method, as described (Rizzino, Ruff, Kazakoff 1988). 125I-EGF internalization assay was performed as previously described (Huang, Goh, Sorkin 2007; Kassel et al. 2007). Cells were seeded at 1×105 per well in 6-well dishes with 3 wells per sample. Confluent growth factor-deprived (0.5% FBS-containing medium for 48h) cells were rinsed once with ice-cold binding medium (DMEM with 0.1% BSA) and then incubated at 4°C in 1 ml of binding medium containing 1 ng/ml 125-EGF or 125I-EGF and 100× molar excess of unlabeled EGF (to quantify non-specific binding) to allow EGF binding to cell surface EGFR. The cells were washed thrice with 1 ml binding medium to remove unbound EGF. The cells were then switched to 37°C and incubated for various time points to allow internalization of 125I-EGF-EGFR complexes. Non-internalized 125I-EGF was removed by a 2-minute rinse at 4 °C in sodium acetate buffer (0.2 M acetic acid/0.5 M NaCl, pH 2.8), followed by a rinse washed with sodium acetate buffer; both rinses were saved for γ-counting as surface-bound 125I-EGF. The cells were then lysed in 1N NaOH to determine the amount of internalized 125I-EGF. Specific 125I-EGF binding represents total minus non-specific binding, and % internalized 125I-EGF was calculated as internalized counts divided by the sum of surface-bound and internalized counts multiplied by 100, as described previously (Sorkin and Duex 2010) .
Statistical Analysis
Statistical significance was determined using one-way ANOVA with the Newman-Keuls, Tukey’s, or Dunnett’s post-tests with the help of GraphPad Prism (GraphPad Software Inc., San Diego, CA). Data are presented as mean ± SEM from at least three separate experiments unless otherwise indicated.
RESULTS
Generation and characterization of Cbl/Cbl-b double knockout Mouse Embryonic Fibroblasts
To unambiguously assess the requirement of endogenous Cbl proteins for ligand-induced RTK internalization, we generated a cell model expected to completely lack the expression of all three Cbl-family proteins. Since Cbl-c has been shown to be expressed primarily in epithelia (Keane et al. 1999), we generated MEFs in which both Cbl alleles carry loxP sites flanking exon 4 and both Cbl-b alleles have been deleted (Cblflox/flox; Cbl-b−/−), together with wild type MEFs as a control. Ex vivo deletion of loxP-flanked sequences was achieved using Adeno-Cre-GFP followed by sorting for GFP+ population.
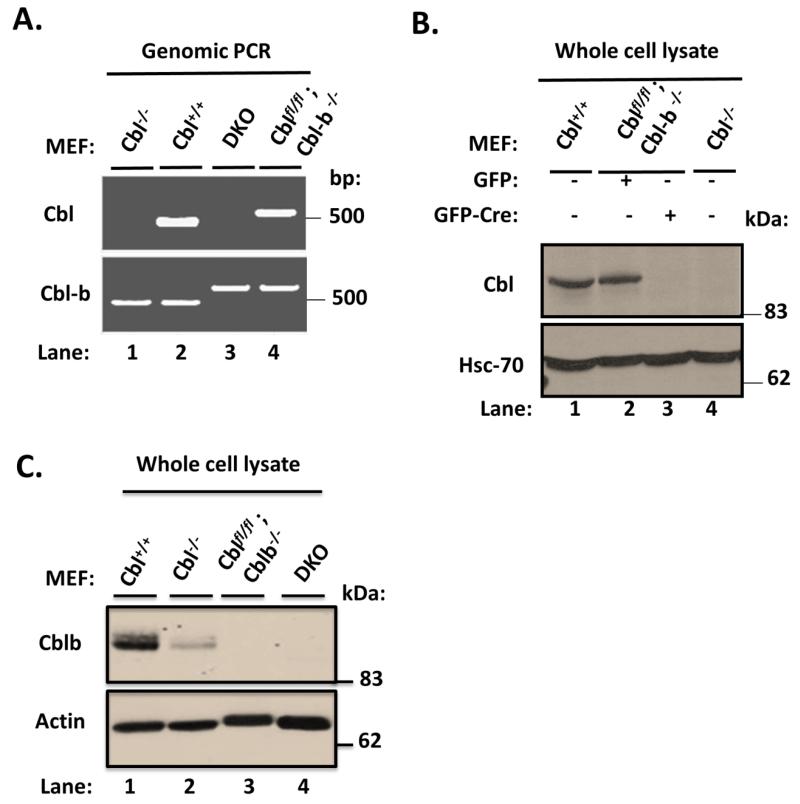
PCR analysis of genomic DNA with specific primers for Cbl and Cbl-b confirmed the deletion of floxed Cbl region in Cblflox/flox; Cbl-b−/− MEFs infected with Adeno-Cre-GFP but not with control Adeno-GFP (Fig. 1A); WT and Cbl-null MEFs provided negative and positive controls. Immunoblotting of whole cell lysates of the same panel of cell lines with an anti-Cbl antibody demonstrated that Adeno-Cre-GFP infection but not the control Adeno-GFP infection led to complete loss of Cbl protein expression, with WT and Cbl-null MEFs serving as negative and positive controls, respectively (Fig. 1B). By using a Cbl-b specific antibody, we confirmed the expected loss of Cbl-b protein expression in both Adeno-Cre-GFP and Adeno-GFP infected Cblflox/flox; Cbl-b−/− MEFs but Cbl-b was detectable in both WT and Cbl-null MEFs, as expected. The substantially lower levels of Cbl-b protein expression in Cbl-null MEFs (Fig. 1C) are expected based on their previous characterization (Duan et al. 2003).
FIGURE 1. Characterization of Cbl/Cblb DKO MEFs.
A) Cblflox/flox; Cbl-b−/− MEFs were infected with Adeno-GFP-Cre to derive DKO MEFs. Genomic DNA isolated from parental Cblflox/flox; Cbl-b−/− MEFs, their Adeno-GFP-Cre infected DKO derivative, as well as control Cbl−/− and wildtype (WT) MEFs was subjected to PCR using specific primer pairs indicated in Supplementary Table S1. Absence of the floxed Cbl band in DKO MEFs confirmed the deletion. B) 100 μg aliquots of Triton X-100 lysate protein from WT, Cblflox/flox; Cbl-b−/−, adeno-Cre infected Cblflox/flox; Cbl-b−/− (DKO) and Cbl−/− MEFs were subjected to Western blotting with anti-Cbl and anti-HSC-70 (loading control) antibodies. Molecular weight markers in kilodaltons (kDa) are indicated. C) 100 μg aliquots of Triton X-100 lysate protein from WT, Cblflox/flox; Cbl-b−/−, adeno-Cre infected Cblflox/flox; Cbl-b−/− (DKO) and Cbl−/− MEFs were subjected to Western blotting with anti-Cbl-b and anti-β-actin (loading control) antibodies. Molecular weight markers in kilodaltons (kDa) are indicated.
Thus, the DKO MEFs generated here represent a cell model with complete lack the expression of all Cbl-family proteins, making them eminently suitable to address the requirement of Cbl-family proteins in EGFR internalization.
Impaired ligand-induced downregulation of EGFR in Cbl/Cbl-b DKO MEFs
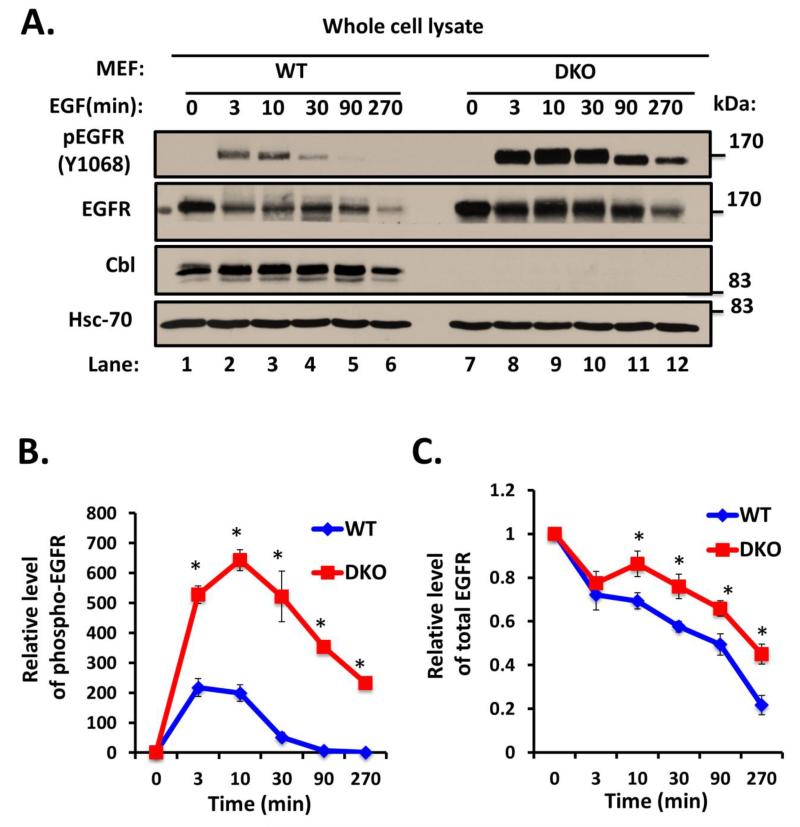
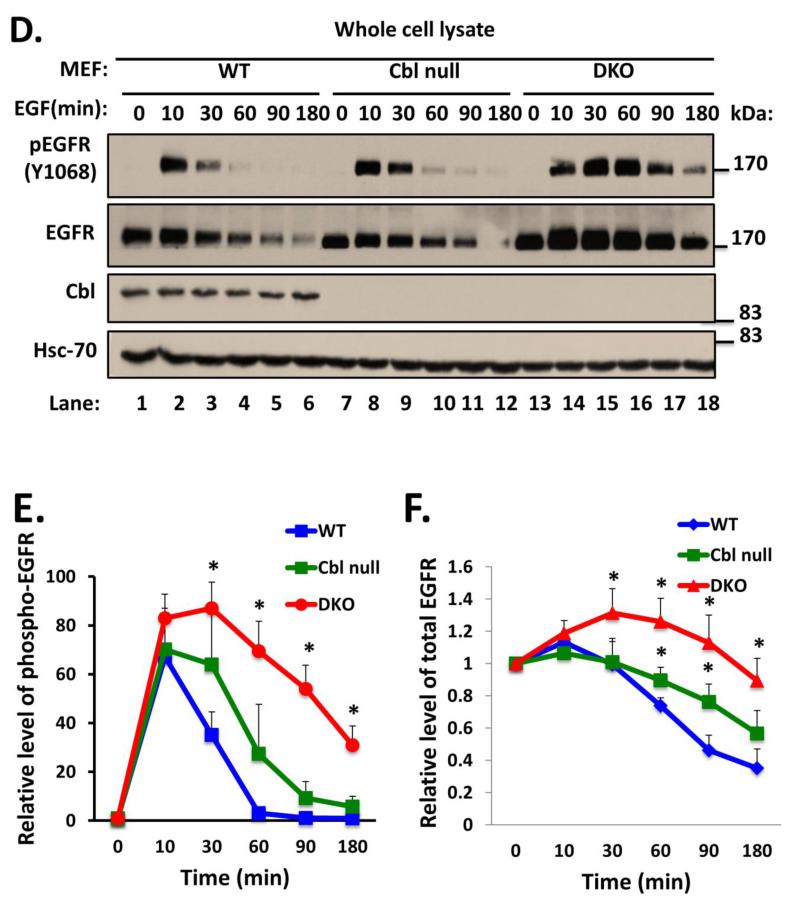
To examine the requirement of endogenous Cbl proteins in RTK endocytic traffic, we used EGFR as a prototype RTK as it is most widely studied in the context of Cbl proteins. To confirm the expected retardation of ligand-induced degradation of EGFR, WT and Cbl/Cbl-b DKO MEFs were serum-starved and stimulated with 100 ng/ml EGF for various time points. Anti-phospho-EGFR (Y1068) blotting of these lysates revealed substantial and significant delay in the loss of phospho-EGFR signals in DKO MEFs compared to WT MEFs (Fig. 2A and B). Immunoblotting of whole cell lysates with an anti-EGFR antibody showed a slower EGF-induced loss of endogenous EGFR in DKO MEFs as compared to that in WT MEFs (Fig. 2A and C). We have previously shown the deletion of Cbl alone to impair ligand-induced EGFR downregulation (Duan et al. 2003). To assess if additional loss of Cbl-b has a larger impact on EGFR downregulation, as we anticipated, we compared Cbl/Cbl-b DKO MEFs to previously-established Cbl-null MEFs, with WT MEFs as control. Ligand-induced EGFR downregulation as well as the downregulation of phospho-EGFR signals was substantially more impaired in Cbl/Cbl-b DKO MEFs (Fig. 2D, E and F). To rule out any differences in EGFR levels at the level of mRNA expression, we performed RT-PCR on mRNA isolated from unstimulated and 270-minute EGF-stimulated WT and DKO MEFs, using mouse EGFR-specific primers. The RT-PCR showed comparable signals at both time points between WT and DKO MEFs (Fig. 3S).
FIGURE 2. Delayed ligand-induced downregulation of EGFR in Cbl/Cbl-b DKO MEFs.
A) Wildtype (WT) and Cbl/Cbl-b DKO (DKO) MEFs were serum-starved for 48h, and then left unstimulated or were stimulated with 100 ng/ml EGF for the indicated time points in minutes (min). 100 μg aliquots of Triton X-100 lysate protein were subjected to Western blotting with anti-phospho-EGFR (Y1068) antibody to visualize phospho-EGFR (top panel), anti-EGFR antibody to visualize total EGFR (middle panel), anti-Cbl antibody to confirm lack of Cbl expression in DKO MEFs, and with anti-HSC-70 antibody as a loading control. The bands corresponding to phospho-EGFR (B) and total EGFR (C) were quantified using Image J software, and normalized relative to band intensities for the corresponding HSC-70 loading controls. The bars represent mean ± SEM of data from three independent experiments. D). WT, Cbl-null (Cbl-/-) and Cbl/Cbl-b DKO (DKO) MEFs were serum-starved for 48h, and then left unstimulated or were stimulated with 100 ng/ml EGF for the indicated time points. 100 μg aliquots of Triton X-100 lysate protein were subjected to Western blotting with anti-phospho EGFR (Y1068) antibody to visualize phospho-EGFR (top panel), anti-EGFR antibody to visualize total EGFR (middle panel), anti-Cbl antibody to confirm lack of Cbl expression in Cbl-null (Cbl-/-) and DKO MEFs and with anti-HSC-70 antibody as a loading control. The bands corresponding to phospho-EGFR (E) and total EGFR (F) were quantified using Image J software, and normalized relative to band intensities for the corresponding HSC-70 loading controls. The bars represent mean ± SEM of data from three independent experiments.
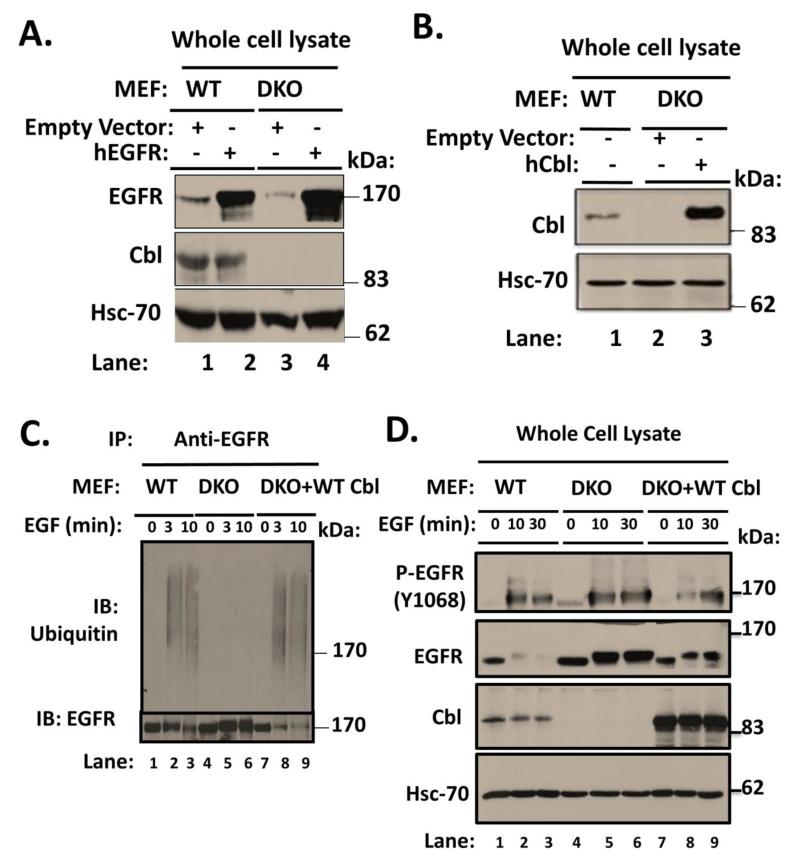
FIGURE 3. Restoration of defective ligand-induced EGFR turnover in Cbl/Cbl-b DKO MEFs by ectopically-expressed human Cbl.
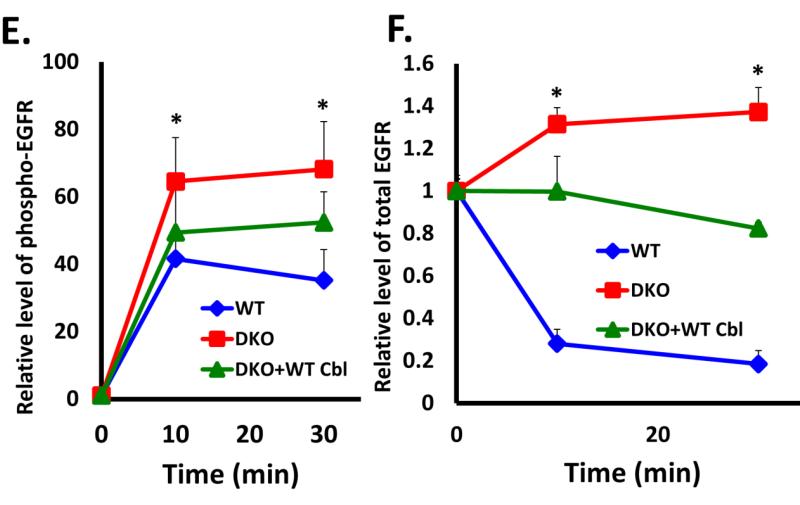
A) Wildtype (WT) or Cbl/Cbl-b DKO (DKO) MEF lines retrovirally transduced with human EGFR or the corresponding vector control were derived. 100 μg aliquots of Triton X-100 lysate protein from these lines were subjected to Western blotting with an anti-EGFR antibody to assess the increased EGFR levels in hEGFR-expressing lines (upper panel), anti-Cbl antibody to confirm lack of Cbl expression in DKO MEFs and anti-HSC-70 antibody (loading control). B) The human EGFR-expressing Cbl/Cbl-b DKO MEF lines in (A) were further transduced with a retrovirus encoding human Cbl and GFP (separated by IRES), and successfully transduced cells were FACS sorted to obtain stably transduced lines. 100 μg aliquots of Triton X-100 lysate protein from these lines and WT were subjected to Western blotting with antibodies against Cbl (top panel) and HSC-70 (loading control). C) WT, Cbl/Cbl-b DKO (DKO) and hCbl-reconstituted DKO MEFs were serum-starved for 48 hours and then stimulated with 100 ng/ml EGF for the indicated time points. Anti-EGFR immunoprecipitations from 1 mg aliquots of RIPA lysate protein were subjected to Western blotting with anti-ubiquitin (upper panel) and anti-EGFR antibodies. D) WT, Cbl/Cbl-b DKO (DKO) and human Cbl-reconstituted DKO MEFs were serum-starved and stimulated as in (C) for the indicated time points. 100 μg aliquots of Triton X-100 lysate protein from these lines were subjected to Western blotting with antibodies against phospho EGFR (Y1068) to visualize phospho-EGFR (top panel), anti-EGFR to visualize total EGFR (second panel), Cbl (third panel) and HSC-70 (bottom panel; loading control). The bands corresponding to phospho-EGFR (E) and total EGFR (F) were quantified using Image J software, and normalized relative to band intensities for the corresponding HSC-70 loading controls. The bars represent mean ± SEM of data from three independent experiments.
Restoration of ligand-induced EGFR downregulation in Cbl/Cbl-b DKO MEFs by ectopic human Cbl
To demonstrate that our findings of impaired ligand-induced EGFR turnover in Cbl/Cbl-b DKO MEFs are not an artifact of MEF immortalization process but rather a consequence of the lack of expression of Cbl-family proteins, we assessed the ability of ectopically expressed human Cbl to complement the defective EGFR ubiquitination and turnover in Cbl/Cbl-b DKO MEFs. The available anti-mouse EGFR antibodies however proved inadequate for this purpose. Therefore, similar to previous studies (Thien, Walker, Langdon 2001), we first introduced human EGFR (hEGFR) into WT and DKO MEFs, selected the cell lines in puromycin and confirmed the expression of hEGFR by FACS analysis with anti-human EGFR mAb 528; the levels of retrovirally transduced hEGFR were higher in DKO MEFs compared to those on WT MEFs (data not shown). Immunoblotting confirmed the increased EGFR levels in hEGFR-transduced lines, with higher levels in the hEGFR-expressing DKO MEFs (Fig. 3A). Next, we retrovirally introduced human Cbl into hEGFR-expressing DKO MEFs, and FACS sorted the hCbl-expressing cells by virtue of their GFP-positivity as the vector contains an EGFP coding region separated from Cbl sequences by an internal ribosome entry sequence (IRES) (Sanada et al. 2009). Immunoblotting for Cbl revealed that DKO MEFs reconstituted with hCbl express a substantially higher level of Cbl compared to WT MEF controls (Fig. 3B).
To assess if ectopic hCbl indeed reverses the impact of loss of Cbl proteins, we serum-starved the hEGFR-expressing WT, DKO and hCbl-reconstituted DKO MEFs, and stimulated these with 100 ng/ml EGF for various time points. Cell lysates were subjected to immunoprecipitation with anti- human EGFR mAb followed by immunoblotting with an anti-Ub antibody. As expected, robust ligand-induced ubiquitination (as a smear) was readily detected in WT MEFs but was undetectable in DKO MEFs (Fig. 3C). Notably, hCbl-reconstituted DKO MEFs exhibited robust ligand-induced EGFR ubiquitination. Anti-EGFR blotting of IPs indicated that EGF stimulation robustly reduced the EGFR levels in hCbl-expressing DKO MEFs (Fig. 3C). Immunoblotting of whole cell lysates further demonstrated that the impaired ligand-induced loss of phospho-EGFR (Fig. 3D [first panel] and 3E) and total EGFR was restored by hCbl expression (Fig. 3D [second panel] and 3F). These results establish that the MEF system characterized here exhibits a specific loss of function of Cbl-family proteins, and therefore provides an ideal tool to examine the requirement of Cbl proteins in RTK internalization.
EGFR internalization is largely unimpaired in Cbl/Cbl-b DKO MEFs
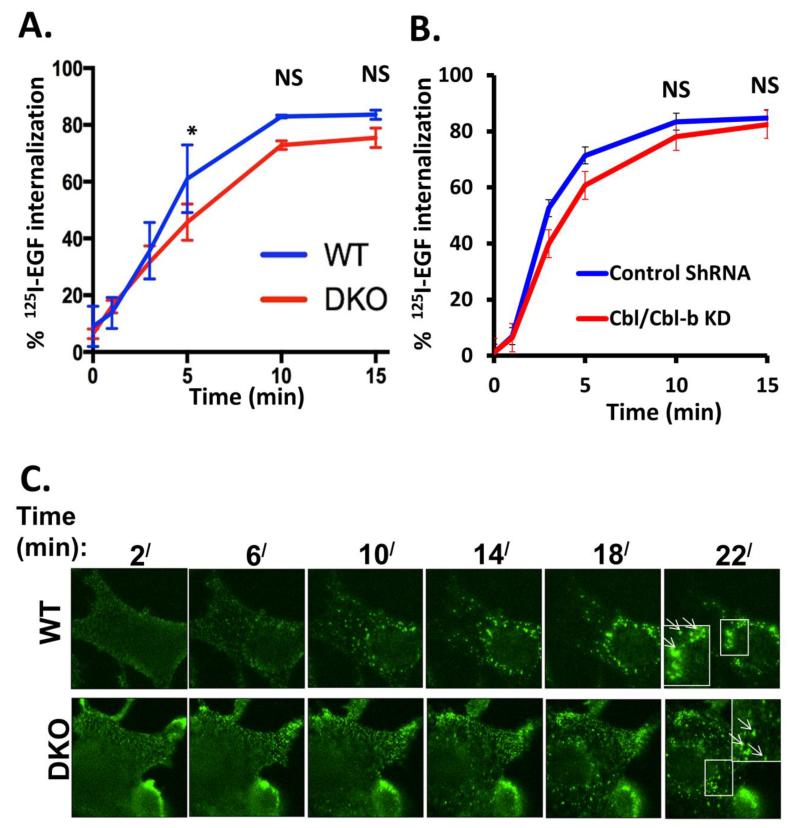
To assess the impact of complete absence of Cbl proteins on ligand-induced EGFR endocytosis, we first used the well-established 125I-EGF internalization assay (Kassel et al. 2007) to quantify the rate and extent of EGFR internalization in WT vs. Cbl/Cbl-b DKO MEFs expressing only the endogenous mouse EGFR; the hEGFR-expressing MEFs were avoided for these studies given different levels of hEGFR expression in WT vs. DKO MEFs. As shown in Figure 4A, internalization of EGF proceeds rapidly in MEFs, with about 80% of cell surface pre-bound 125I-EGF internalized in WT MEFs in 15 minutes. Notably, the rate and extent of 125I-EGF internalization in Cbl/Cbl-b DKO MEFs was essentially identical at early time points and was only marginally lower at later time points.
FIGURE 4. Complete lack of Cbl-family protein expression does not impair ligand-induced EGFR internalization.
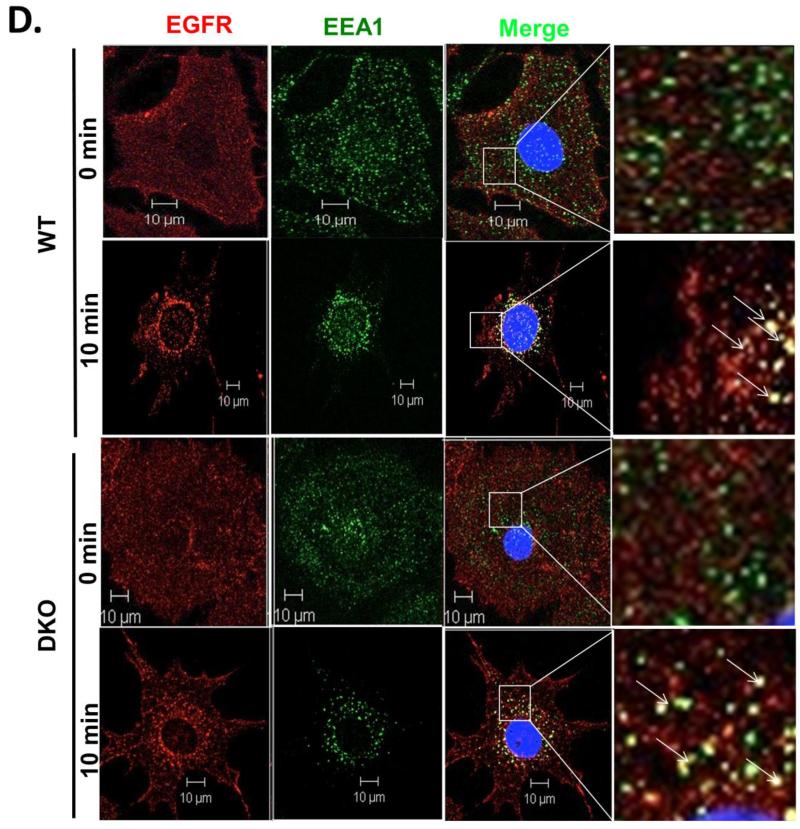
A) Triplicate wells of 6-well plates seeded with wildtype (WT) and Cbl/Cbl-b DKO (DKO) MEFs, and serum-starved for 24h, were incubated with 1 ng/ml 125I-EGF (about 200,000 cpm) at 4 °C for 120 minutes to allow EGF binding. Following washes, the cells were incubated at 37°C for the indicated times and the % of internalized relative to total 125I-EGF was plotted against time. The data points are mean ± SEM of data from three independent experiments. B) Control and Cbl/Cbl-b double knockdown MCF-10 human mammary epithelial cells described previously (Duan et al. 2011) and confirmed via anti-Cbl immunoblotting (Supplementary Fig. S4) were used for 125I-EGF internalization as in (A). The data points are mean ± SEM of three independent experiments. C) WT (upper panel) and Cbl/Cbl-b DKO (DKO; lower panel) MEFs expressing hEGFR (Fig. 3A) were serum-starved for 24h and Alexa Fluor® 488-conjugated EGF was allowed to bind to the surface of live cells at 4°C for 1h. The cells were then switched to 37°C and confocal images captured at 2 minutes intervals to assess the transfer of surface fluorescence into intracellular endocytic vesicles. D) WT (upper two panels) and Cbl/Cbl-b DKO (DKO; lower two panels) MEFs expressing human EGFR (Fig. 3A) were serum-starved for 24h, and either left unstimulated or stimulated with EGF (10 ng/ml) for 10 minutes. Cells were fixed and stained for human EGFR and EEA1. Co-localization was assessed in merged images using ZEN 2010 software. The panels on right are magnified images of the insets in third column, as shown. The arrows indicate the EGFR co-localization with the early endosome marker EEA1 (Scale bars: 10 μm).
To extend these results to a non-MEF cell type, we carried out a similar 125I-EGF internalization assay using Cbl/Cbl-b shRNA-expressing MCF10A immortal mammary epithelial cell line vs. its control parental line (Fig. 4B, Supplementary Fig. S4). We have previously used confocal imaging to conclude that EGFR internalization in these cells was not impaired by Cbl/Cbl-b knockdown but these analyses were not quantitative (Duan et al. 2011). Similar to the MEF cell system, the rate and extent of 125I-EGF internalization in Cbl/Cbl-b knockdown MCF10A cells was intact and only marginally less compared to parental MCF10A cells (Fig. 4B).
As a complementary approach to quantitative 125I-EGF internalization assays, we also assessed the ability of fluorescent EGF to reach the endosomal compartments, using live-cell confocal imaging (Fig 4C). Serum-starved WT or Cbl/Cbl-b DKO MEFs overexpressing hEGFR were incubated with Alexa Fluor® 488-conjugated EGF at 4°C for 1 hour and switched to 37°C followed by capture of live-cell images every 2 minutes for 22 minutes. In WT MEFs, labeled EGF, initially concentrated at the cell surface, was seen to rapidly enter intracellular vesicles; towards the end of the assay, most fluorescent EGF was intracellular, including larger vesicles in the perinuclear area (Fig. 4C, upper panel). Comparable results were seen with Cbl/Cbl-b DKO MEFs (Fig. 4C, lower panel).
Direct imaging of endogenous mouse EGFR was not successful using available antibodies. We therefore tracked the ligand-induced entry of EGFR into endosomal compartments using hEGFR-expressing WT and Cbl/Cbl-b DKO MEFs for these studies. Serum-starved MEFs, left unstimulated or stimulated with EGF (10 ng/ml) for various time points, were fixed and stained for EGFR (mAb 528) and early endocytic compartment marker EEA1. In both WT and Cbl/Cbl-b DKO MEFs, EGFR was localized exclusively at the cell surface in unstimulated cells, with little co-localization with EEA1. Upon EGF stimulation, surface EGFR staining was reduced over time while intracellular vesicular staining co-localizing with EEA1 was seen in both WT and Cbl/Cbl-b DKO MEFs (Fig 4D). Overall, our results using a MEF-based system with complete lack of expression of all Cbl-family proteins unequivocally establish that Cbl-family proteins are largely dispensable for the endocytosis step of a prototype RTK, EGFR.
CIN85 is dispensable for Ligand-induced Internalization of EGFR
Given the dispensability of Cbl proteins for EGFR internalization demonstrated above, we wished to assess whether this might reflect a Cbl-independent role of CIN85-endophilin complex in our MEF system. We first used western blotting to assess the expression of endophilin-1A isoform (Giachino et al., 1997; Milosevic et al., 2011; Ringstad, Nemoto, & De Camilli, 1997) in MEF lines, with brain and testis tissue lysates as positive controls. These analyses revealed that endophilin-A was essentially undetectable in MEFs (Fig. S1). These results suggest that MEFs are deficient in EGFR endocytosis-relevant isoform of endophilin-A.
Expression of substantial levels of CIN85 proteins was detected by western blotting in MEFs (Fig. S2). The lack of expression of RTK endocytosis-relevant isoform of endophilin-A in our MEF system suggested that a CIN85-endophilin complex was unlikely to mediate the ligand-induced EGFR internalization. However, as MEFs did express CIN85, we assessed whether CIN85 might still be required for EGFR internalization.
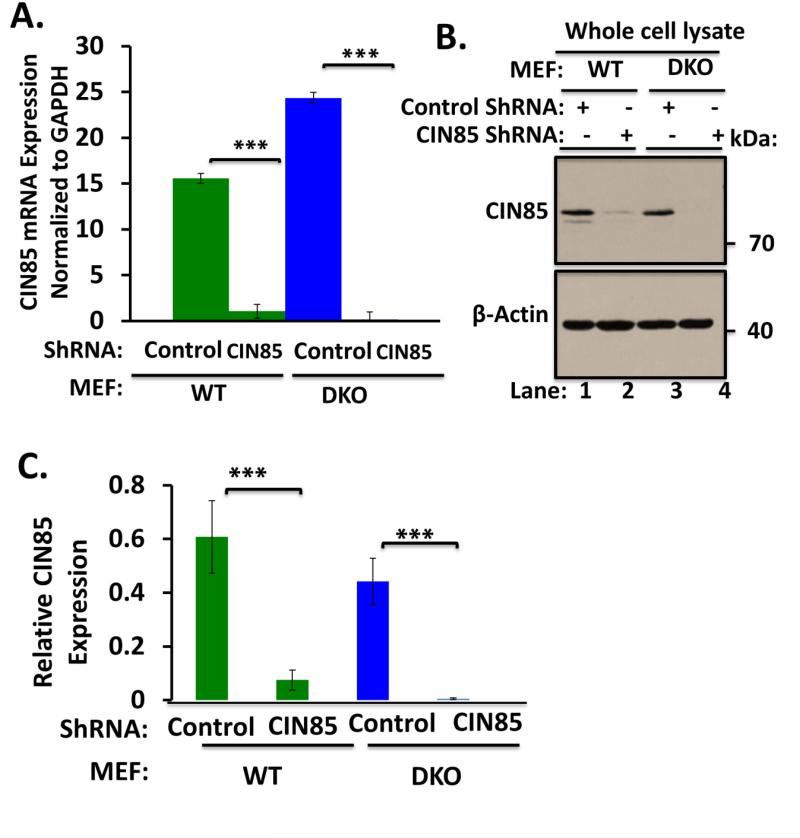
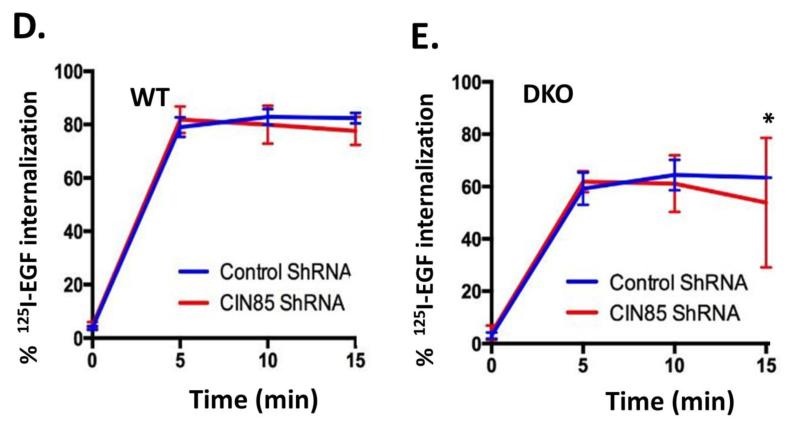
To investigate the role of CIN85 in ligand-induced internalization of EGFR in MEFs, we used a knockdown approach to deplete endogenous CIN85. As siRNA transfection of MEFs was quite inefficient, we used a doxycycline-inducible TRIPZ lentiviral system to deplete CIN85 in WT and Cbl/Cbl-b DKO MEFs. Both quantitative real-time PCR and Western blotting demonstrated efficient knockdown of CIN85 in doxycycline-treated MEFs transduced with CIN85 shRNA, compared to those expressing the vector alone (Fig. 5A, B and C). Analyses using the 125I-EGF internalization assay in these cells demonstrated that there was no impact of CIN85 knockdown on the rate and extent of internalization in either WT MEFs (Fig. 5D) or in Cbl/Cbl-b DKO MEFs (Fig. 5E). These results indicate that endogenous CIN85 is not required for ligand-induced internalization of EGFR.
FIGURE 5. CIN85 knockdown does not impair EGFR internalization.
(A) Wildtype (WT) and Cbl/Cbl-b DKO (DKO) MEFs stably expressing the doxycycline-inducible control or CIN85 shRNA were induced with doxycycline for 4 days and real-time qPCR analysis was performed using the primers indicated in Supplementary Table S1 to confirm the CIN85 knockdown. GAPDH was used as a normalization control. B) WT and Cbl/Cbl-b DKO MEFs were induced with doxycycline as in (A) and 100 μg aliquots of Triton X-100 lysate protein from these cell lines were subjected to Western blotting with antibodies against CIN85 and β-actin (loading control). C) The bands corresponding to CIN85 in (B) were quantified using Image J software, and normalized relative to band intensities for the corresponding β-actin loading controls. The bars represent mean ± SEM of data from three independent experiments. D and E) WT (D) and Cbl/Cbl-b DKO (E) MEFs were induced with doxycycline as in (A), and used for 125I-EGF internalization as in Fig. 4A. The data points are mean ± SEM of three independent experiments.
DISCUSSION
While we and other investigators have clearly established a requirement of endogenous Cbl and Cbl-b proteins in promoting lysosomal degradation of RTKs (Mohapatra et al. 2013), the role of Cbl proteins as physiologically-required elements in the first step of endocytic traffic, that of internalization from the cell surface into endosomes (endocytosis), has remained unclear, with evidence for and against a role of Cbl proteins. Evidence for a role of Cbl proteins in RTK internalization has come particularly from studies demonstrating a Cbl-CIN85-endophilin complex that is recruited to the activated EGFR through Cbl (Soubeyran et al. 2002). Given the ability of endophilins to modulate membrane curvature, this complex has been proposed as a mechanism of RTK internalization involving a ubiquitination-independent adapter role of Cbl proteins. The results presented here using a novel MEF system with complete lack of expression of all Cbl proteins, demonstrate conclusively that endogenous Cbl proteins are dispensable for ligand-induced internalization of a prototype RTK, EGFR. By further demonstrating intact ligand-induced EGFR internalization upon CIN85 knockdown in this cell system, which also lacks the expression of relevant Endophilin partner of CIN85, our results support the idea that Cbl-CIN85-endophilin-A complex is dispensable for EGFR internalization.
Studies in a number of laboratories have established the Cbl-family proteins as primary mediators of ligand-induced ubiquitination of a number of RTKs, with EGFR serving as a prototype in a majority of such studies (Mohapatra et al. 2013). Consistent with the essential role of ubiquitination of endocytosed cell surface receptors in targeting them for ESCRT-mediated sorting at the MVB for final degradation in the lysosome (MacGurn, Hsu, Emr 2012; Wegner, Rodahl, Stenmark 2011), previous studies using Cbl-null MEFs (Duan et al. 2003) as well knockdown of Cbl and/or Cbl-b (Duan et al. 2011; Pennock and Wang 2008) have demonstrated that loss of Cbl-family protein function impairs the ligand-induced EGFR degradation. Our findings in the MEF cell system, which is completely devoid of expression of all Cbl-family proteins, provide strong genetic confirmation of the previous studies of Cbl and Cbl-b depletion and completely eliminate any concerns of residual expression or compensation by family members. Importantly, the cell system established here provides a facile tool to examine the importance of Cbl proteins in the context of other RTKs as well as non-RTK targets without resort to overexpression strategies.
In contrast to a general consensus on the role of Cbl proteins in RTK targeting to lysosomes, their involvement in the first step of ligand-induced RTK internalization from the cell surface has remained unresolved. Studies using RTK mutants that did not interact with Cbl proteins (Lill et al. 2000; Reddi et al. 2007; Waterman et al. 2002; Yarden and Sliwkowski 2001) carried concerns of indirect recruitment of Cbl proteins, and partial retention of ubiquitination on such EGFR mutants was consistent such an interpretation. Similarly, more physiological approaches using Cbl-null cells (Duan et al. 2003), Cbl/Cbl-b knockdown (Duan et al. 2011; Pennock and Wang 2008) or temperature-regulated inhibition of ubiquitination in a CHO cell system (Duan et al. 2003) carried the caveat that low levels of ubiquitination due to remaining Cbl-family members or incomplete knockdown could be sufficient for ubiquitin-dependent internalization. Our results that ligand-induced internalization of EGFR can proceed essentially intact while ubiquitination was abolished strongly support the idea that ubiquitin signals are not a requirement for ligand-induced EGFR internalization. Consistent with this idea, mutation of all lysine residues that were demonstrated to be ubiquitinated in EGFR did not abrogate its ligand-induced internalization (Huang, Goh, Sorkin 2007). The ability of C-terminally fused ubiquitin to promote kinase activity-independent EGFR internalization does however suggest that ubiquitin moiety “can” function as an RTK internalization motif in mammalian cells, similar to the role of ubiquitin in growth hormone receptor internalization (Govers et al. 1999) and the well-established role of ubiquitin as an endocytic motif for non-RTK membrane receptors defined through studies in yeast (Hicke and Riezman 1996). It is conceivable that certain cell types/tissues in mammals might use this mechanism physiologically, but this remains to be demonstrated.
The role of Cbl-family proteins as ubiquitin ligase activity-independent adapters to recruit a CIN85/Endophilin complex for ligand-induced internalization of EGFR (Soubeyran et al. 2002) and c-MET (Petrelli et al. 2002) was demonstrated many years ago and has remained a widely-discussed model. In this regard, it is notable that the major pieces of evidence for the role of Cbl/CIN85/endophilin complex as an important determinant of RTK internalization relied on overexpression (Petrelli et al. 2002; Soubeyran et al. 2002). Even in these studies, it was noted that maximal endogenous Cbl-CIN85 association was not induced until a substantially late time point after c-MET stimulation, which would be inconsistent with the conclusion that CIN85 had a primary role in receptor internalization. Furthermore, the interaction of Cbl with CIN85 requires the C-terminal proline-rich region of Cbl (Dikic 2002; Dikic 2003), and a truncated Cbl protein lacking the CIN85 interaction sites would be expected to be unable to promote RTK internalization. Earlier studies (Levkowitz et al. 1999; Lill et al. 2000), demonstrating that such a truncated protein promotes EGFR ubiquitination and lysosomal degradation, were inconsistent with the CIN85-based RTK model. Our studies in the MEF model demonstrate unambiguously that the Cbl/CIN85-endophilin complex cannot be the primary mediator of EGFR internalization. This is supported by the facts that our cell line system lacks the expression of all Cbl-family proteins, the relevant endophilin isoform is not expressed, and the depletion of endogenous CIN85 in either WT MEFs or Cbl/Cbl-b DKO MEFs has no impact on EGFR internalization.
Our conclusion is consistent with recent results from multiple laboratories that have used knockdown approaches to assess the importance of endogenous CIN85 and concluded that CIN85 is dispensable for RTK internalization and instead support its role in the regulation of sorting decisions after RTK internalization (Ronning et al. 2011; Schroeder et al. 2010; Schroeder et al. 2012). Some of these sorting decisions have been linked to the ability of CIN85 to bind to Cbl, and certain CIN85-interacting partners that compete with Cbl alter RTK sorting in endosomes (Schmidt et al. 2004; Schroeder et al. 2010; Zhang et al. 2009). However this remains an active area that requires further investigation. The cell system characterized here, where Cbl proteins are present or absent, and CIN85 can be inducibly depleted, should allow future studies to more clearly characterize the roles of CIN85 in RTK endocytic traffic.
Importantly, while our conclusions on the dispensability of CIN85 and Cbl proteins, and hence of the Cbl-CIN85 complex, in RTK internalization are based on a MEF cell system which allows the flexibility of gene deletion with ease, studies of other cell types support the generality of these conclusions. Recent studies using Hela, Hep2, B lymphoma cell line BJAB and porcine aortic endothelial cell lines have arrived at a similar conclusion using a CIN85 knockdown approach (Niiro et al. 2012; Ronning et al. 2011; Schroeder et al. 2010; Schroeder et al. 2012).
CONCLUSION
In conclusion, our studies establish conclusively that ligand-induced internalization of EGFR, as a prototype RTK, does not require Cbl-family proteins and a Cbl-CIN85 complex.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Brian Druker (Oregon Health Science University) for a gift of 4G10 antibody used in initial studies. We also thank members of the Band Lab for numerous discussions and feedback. This manuscript is dedicated in fond memory of Patricia Lorenzo, Ph.D., a former member of the Molecular Oncogenesis NIH study section and the University of Hawaii Cancer Center, who brightened deliberations with her compassion, affability, and dedication. Her remarkable courage in the face of terminal cancer has been inspirational.
The abbreviations used are
- EGFR
epidermal growth factor receptor
- CIN85
Cbl interacting protein of 85 kDa
- Cbl
Casitas B-lineage lymphoma
- EEA1
early endosome antigen 1
- FACS
fluorescence-activated cell sorting
- PFA
paraformaldehyde
- mAb
monoclonal antibody
- pAb
polyclonal antibody
- MEF
mouse embryonic fibroblast
- HSC70
heat shock protein cognate 70
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bertelsen V, Sak MM, Breen K, Rodland MS, Johannessen LE, Traub LM, Stang E, Madshus IH. A chimeric pre-ubiquitinated EGF receptor is constitutively endocytosed in a clathrin-dependent, but kinase-independent manner. Traffic. 2011;12(4):507–20. doi: 10.1111/j.1600-0854.2011.01162.x. [DOI] [PubMed] [Google Scholar]
- Dikic I. Mechanisms controlling EGF receptor endocytosis and degradation. Biochem Soc Trans. 2003;31(Pt 6):1178–81. doi: 10.1042/bst0311178. [DOI] [PubMed] [Google Scholar]
- Dikic I. CIN85/CMS family of adaptor molecules. FEBS Lett. 2002;529(1):110–5. doi: 10.1016/s0014-5793(02)03188-5. [DOI] [PubMed] [Google Scholar]
- Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition and phosphorylation-dependent activation of c-cbl. Nat Struct Mol Biol. 2012;19(2):184–92. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Raja SM, Chen G, Virmani S, Williams SH, Clubb RJ, Mukhopadhyay C, Rainey MA, Ying G, Dimri M, et al. Negative regulation of EGFR-Vav2 signaling axis by cbl ubiquitin ligase controls EGF receptor-mediated epithelial cell adherens junction dynamics and cell migration. J Biol Chem. 2011;286(1):620–33. doi: 10.1074/jbc.M110.188086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Miura Y, Dimri M, Majumder B, Dodge IL, Reddi AL, Ghosh A, Fernandes N, Zhou P, Mullane-Robinson K, et al. Cbl-mediated ubiquitinylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J Biol Chem. 2003;278(31):28950–60. doi: 10.1074/jbc.M304474200. [DOI] [PubMed] [Google Scholar]
- Giachino C, Lantelme E, Lanzetti L, Saccone S, Bella Valle G, Migone N. A novel SH3-containing human gene family preferentially expressed in the central nervous system. Genomics. 1997;41(3):427–34. doi: 10.1006/geno.1997.4645. [DOI] [PubMed] [Google Scholar]
- Govers R, ten Broeke T, van Kerkhof P, Schwartz AL, Strous GJ. Identification of a novel ubiquitin conjugation motif, required for ligand-induced internalization of the growth hormone receptor. EMBO J. 1999;18(1):28–36. doi: 10.1093/emboj/18.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21(1):77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84(2):277–87. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Huang F, Goh LK, Sorkin A. EGF receptor ubiquitination is not necessary for its internalization. Proc Natl Acad Sci U S A. 2007;104(43):16904–9. doi: 10.1073/pnas.0707416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c-cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286(5438):309–12. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- Kales SC, Ryan PE, Lipkowitz S. Cbl exposes its RING finger. Nat Struct Mol Biol. 2012;19(2):131–3. doi: 10.1038/nsmb.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassel KM, Schulte NA, Parker SM, Lanik AD, Toews ML. Lysophosphatidic acid decreases epidermal growth factor receptor binding in airway epithelial cells. J Pharmacol Exp Ther. 2007;323(1):109–18. doi: 10.1124/jpet.107.120584. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3(12):893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Keane MM, Ettenberg SA, Nau MM, Banerjee P, Cuello M, Penninger J, Lipkowitz S. Cbl-3: A new mammalian cbl family protein. Oncogene. 1999;18(22):3365–75. doi: 10.1038/sj.onc.1202753. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y. C-cbl/sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12(23):3663–74. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-cbl/sli-1. Mol Cell. 1999;4(6):1029–40. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Lill NL, Douillard P, Awwad RA, Ota S, Lupher ML, Jr, Miyake S, Meissner-Lula N, Hsu VW, Band H. The evolutionarily conserved N-terminal region of cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J Biol Chem. 2000;275(1):367–77. doi: 10.1074/jbc.275.1.367. [DOI] [PubMed] [Google Scholar]
- Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11(9):629–43. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J Cell Biol. 2002;156(5):843–54. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Emr SD. Ubiquitin and membrane protein turnover: From cradle to grave. Annu Rev Biochem. 2012;81:231–59. doi: 10.1146/annurev-biochem-060210-093619. [DOI] [PubMed] [Google Scholar]
- Mason JM, Morrison DJ, Bassit B, Dimri M, Band H, Licht JD, Gross I. Tyrosine phosphorylation of sprouty proteins regulates their ability to inhibit growth factor signaling: A dual feedback loop. Mol Biol Cell. 2004;15(5):2176–88. doi: 10.1091/mbc.E03-07-0503. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Meng W, Sawasdikosol S, Burakoff SJ, Eck MJ. Structure of the amino-terminal domain of cbl complexed to its binding site on ZAP-70 kinase. Nature. 1999;398(6722):84–90. doi: 10.1038/18050. [DOI] [PubMed] [Google Scholar]
- Milosevic I, Giovedi S, Lou X, Raimondi A, Collesi C, Shen H, Paradise S, O’Toole E, Ferguson S, Cremona O, et al. Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron. 2011;72(4):587–601. doi: 10.1016/j.neuron.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S, Lupher ML, Jr, Druker B, Band H. The tyrosine kinase regulator cbl enhances the ubiquitination and degradation of the platelet-derived growth factor receptor alpha. Proc Natl Acad Sci U S A. 1998;95(14):7927–32. doi: 10.1073/pnas.95.14.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S, Mullane-Robinson KP, Lill NL, Douillard P, Band H. Cbl-mediated negative regulation of platelet-derived growth factor receptor-dependent cell proliferation. A critical role for cbl tyrosine kinase-binding domain. J Biol Chem. 1999;274(23):16619–28. doi: 10.1074/jbc.274.23.16619. [DOI] [PubMed] [Google Scholar]
- Mohapatra B, Ahmad G, Nadeau S, Zutshi N, An W, Scheffe S, Dong L, Feng D, Goetz B, Arya P, et al. Protein tyrosine kinase regulation by ubiquitination: Critical roles of cbl-family ubiquitin ligases. Biochim Biophys Acta. 2013;1833(1):122–39. doi: 10.1016/j.bbamcr.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson Y, Shtiegman K, Katz M, Zwang Y, Vereb G, Szollosi J, Yarden Y. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J Biol Chem. 2003;278(24):21323–6. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- Naramura M, Kole HK, Hu RJ, Gu H. Altered thymic positive selection and intracellular signals in cbl-deficient mice. Proc Natl Acad Sci U S A. 1998;95(26):15547–52. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niiro H, Jabbarzadeh-Tabrizi S, Kikushige Y, Shima T, Noda K, Ota S, Tsuzuki H, Inoue Y, Arinobu Y, Iwasaki H, et al. CIN85 is required for cbl-mediated regulation of antigen receptor signaling in human B cells. Blood. 2012;119(10):2263–73. doi: 10.1182/blood-2011-04-351965. [DOI] [PubMed] [Google Scholar]
- Pennock S, Wang Z. A tale of two cbls: Interplay of c-cbl and cbl-b in epidermal growth factor receptor downregulation. Mol Cell Biol. 2008;28(9):3020–37. doi: 10.1128/MCB.01809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschard P, Fournier TM, Lamorte L, Naujokas MA, Band H, Langdon WY, Park M. Mutation of the c-cbl TKB domain binding site on the met receptor tyrosine kinase converts it into a transforming protein. Mol Cell. 2001;8(5):995–1004. doi: 10.1016/s1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. The endophilin-CIN85-cbl complex mediates ligand-dependent downregulation of c-met. Nature. 2002;416(6877):187–90. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- Reddi AL, Ying G, Duan L, Chen G, Dimri M, Douillard P, Druker BJ, Naramura M, Band V, Band H. Binding of cbl to a phospholipase Cgamma1-docking site on platelet-derived growth factor receptor beta provides a dual mechanism of negative regulation. J Biol Chem. 2007;282(40):29336–47. doi: 10.1074/jbc.M701797200. [DOI] [PubMed] [Google Scholar]
- Ringstad N, Nemoto Y, De Camilli P. The SH3p4/Sh3p8/SH3p13 protein family: Binding partners for synaptojanin and dynamin via a Grb2-like src homology 3 domain. Proc Natl Acad Sci U S A. 1997;94(16):8569–74. doi: 10.1073/pnas.94.16.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzino A, Ruff E, Kazakoff P. Isolation and characterization of A-431 cells that retain high epidermal growth factor binding capacity and respond to epidermal growth factor by growth stimulation. Cancer Res. 1988;48(9):2377–81. [PubMed] [Google Scholar]
- Ronning SB, Pedersen NM, Madshus IH, Stang E. CIN85 regulates ubiquitination and degradative endosomal sorting of the EGF receptor. Exp Cell Res. 2011;317(13):1804–16. doi: 10.1016/j.yexcr.2011.05.016. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Vaccari T, Stenmark H. Shaping development with ESCRTs. Nat Cell Biol. 2011;14(1):38–45. doi: 10.1038/ncb2381. [DOI] [PubMed] [Google Scholar]
- Ryan PE, Sivadasan-Nair N, Nau MM, Nicholas S, Lipkowitz S. The N terminus of cbl-c regulates ubiquitin ligase activity by modulating affinity for the ubiquitin-conjugating enzyme. J Biol Chem. 2010;285(31):23687–98. doi: 10.1074/jbc.M109.091157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32(12):561–73. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, Tamura A, Honda H, Sakata-Yanagimoto M, Kumano K, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460(7257):904. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Hoeller D, Yu J, Furnari FB, Cavenee WK, Dikic I, Bogler O. Alix/AIP1 antagonizes epidermal growth factor receptor downregulation by the cbl-SETA/CIN85 complex. Mol Cell Biol. 2004;24(20):8981–93. doi: 10.1128/MCB.24.20.8981-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B, Srivatsan S, Shaw A, Billadeau D, McNiven MA. CIN85 phosphorylation is essential for EGFR ubiquitination and sorting into multivesicular bodies. Mol Biol Cell. 2012;23(18):3602–11. doi: 10.1091/mbc.E11-08-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B, Weller SG, Chen J, Billadeau D, McNiven MA. A Dyn2-CIN85 complex mediates degradative traffic of the EGFR by regulation of late endosomal budding. EMBO J. 2010;29(18):3039–53. doi: 10.1038/emboj.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, Duex JE. Quantitative analysis of endocytosis and turnover of epidermal growth factor (EGF) and EGF receptor. Curr Protoc Cell Biol Chapter. 2010;15 doi: 10.1002/0471143030.cb1514s46. Unit 15.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature. 2002;416(6877):183–7. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- Thien CB, Walker F, Langdon WY. RING finger mutations that abolish c-cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol Cell. 2001;7(2):355–65. doi: 10.1016/s1097-2765(01)00183-6. [DOI] [PubMed] [Google Scholar]
- Todaro GJ, Green H. High frequency of SV40 transformation of mouse cell line 3T3. Virology. 1966;28(4):756–9. doi: 10.1016/0042-6822(66)90261-3. [DOI] [PubMed] [Google Scholar]
- Waterman H, Katz M, Rubin C, Shtiegman K, Lavi S, Elson A, Jovin T, Yarden Y. A mutant EGF-receptor defective in ubiquitylation and endocytosis unveils a role for Grb2 in negative signaling. EMBO J. 2002;21(3):303–13. doi: 10.1093/emboj/21.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner CS, Rodahl LM, Stenmark H. ESCRT proteins and cell signalling. Traffic. 2011;12(10):1291–7. doi: 10.1111/j.1600-0854.2011.01210.x. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–37. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zheng X, Yang X, Liao K. CIN85 associates with endosomal membrane and binds phosphatidic acid. Cell Res. 2009;19(6):733–46. doi: 10.1038/cr.2009.51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.