Abstract
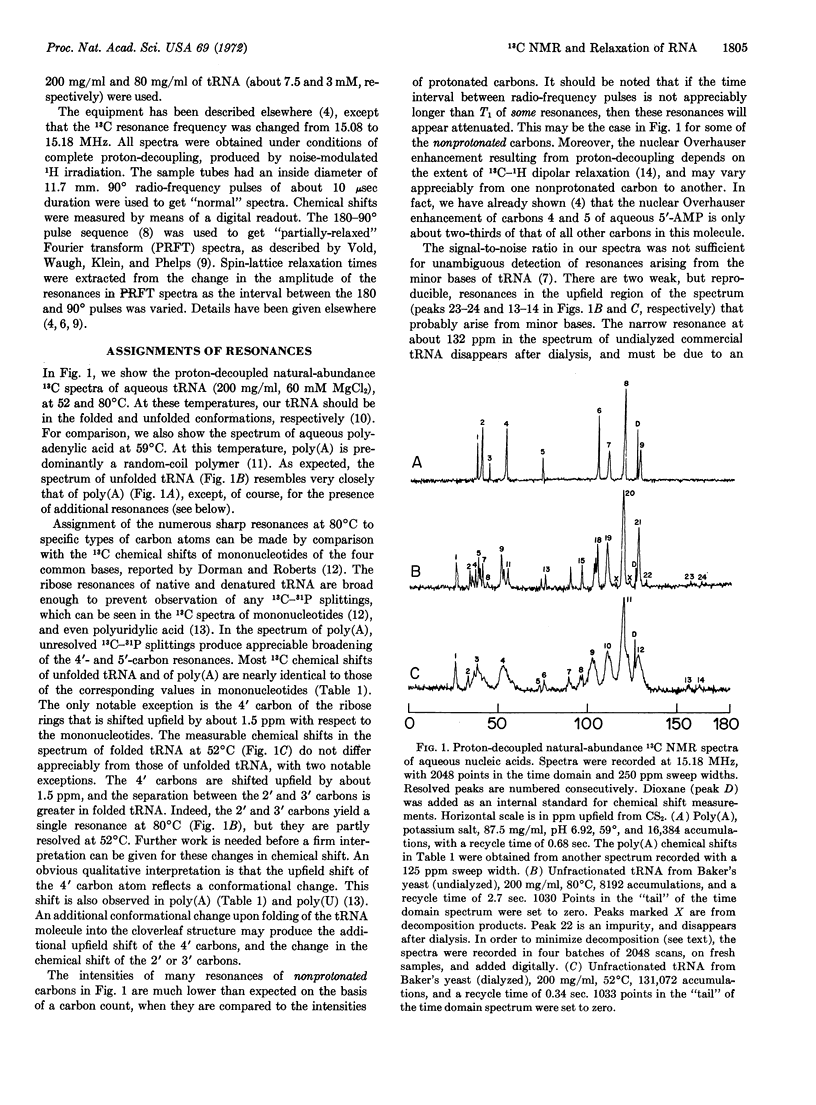
High-resolution Fourier-transform nuclear magnetic resonance at 15.18 MHz was used to observe the proton-decoupled natural-abundance 13C spectra of aqueous unfractionated tRNA from baker's yeast in the presence of Mg+2 (8 ions per tRNA molecule), as a function of temperature in the range of 27-82°C. The spectrum of thermally denatured tRNA at 82°C showed numerous sharp resonances, which were assigned to specific types of carbon atoms by comparison with the 13C spectra of mononucleotides. Only the resonance of carbon 4′ of the ribose rings was appreciably shifted (by about 1.5 ppm upfield) with its average position in the mononucleotides. This effect was also seen in the spectrum of poly(A). In the spectrum of folded tRNA (52°C), carbon 4′ was further shifted upfield by about 1.5 ppm, and carbons 2′ and 3′, which yielded a single resonance at 82°C, now showed two partly resolved peaks.
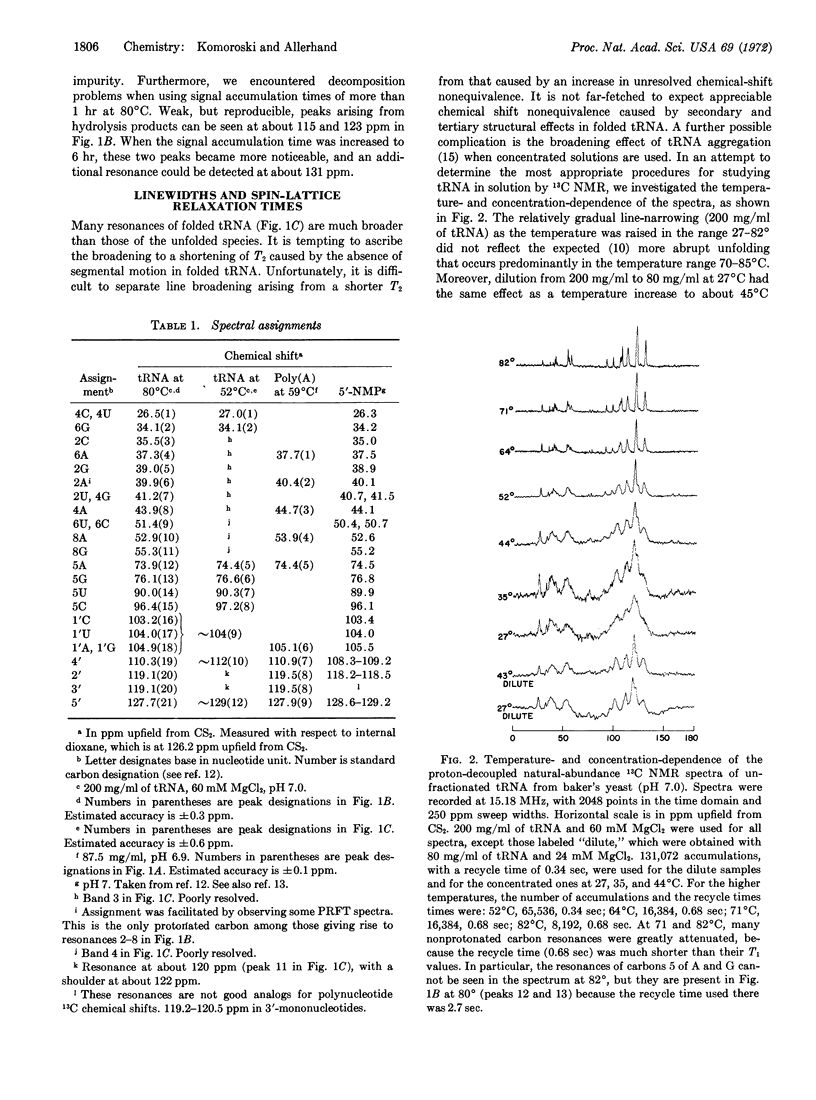
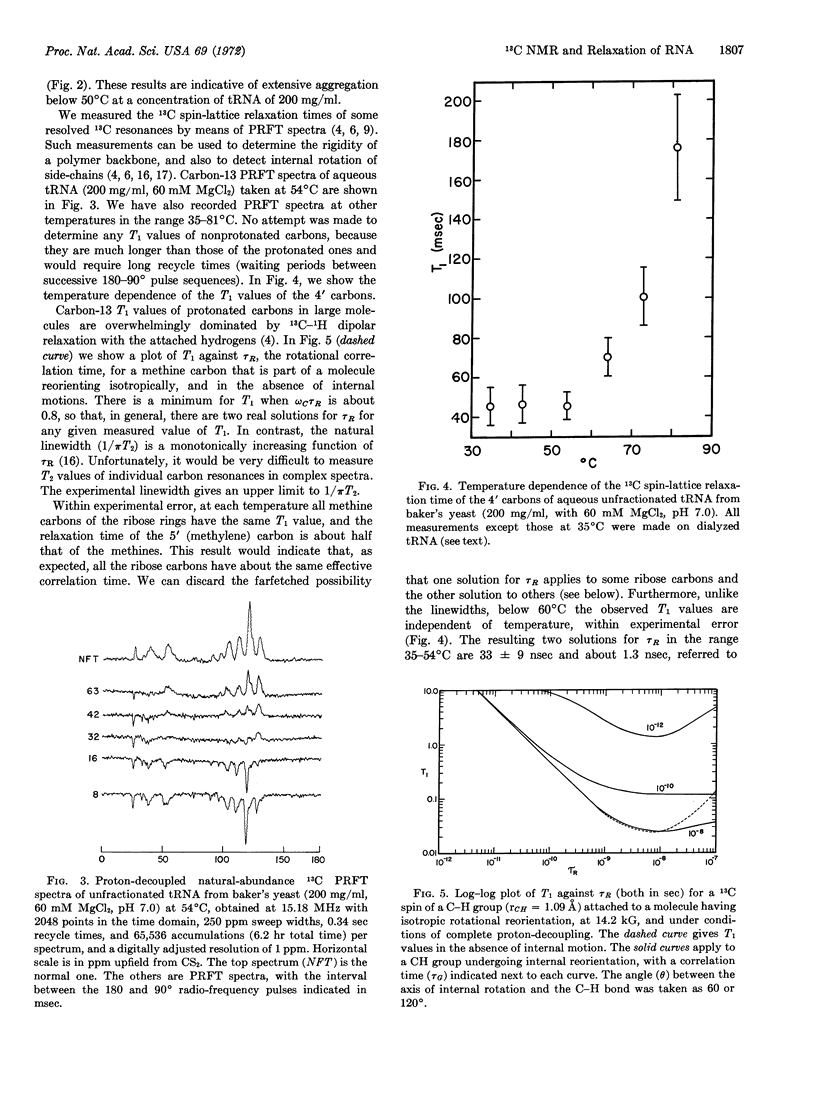
The variation of linewidths with temperature (200 mg/ml of tRNA) was gradual in the range 27-82°C, and did not reflect the expected unfolding behavior of tRNA. Moreover, dilution to 80 mg/ml at 27°C had the same effect as an increase in temperature to about 45°C. The line-width changes below 60° were ascribed to tRNA aggregation. In contrast to the behavior of the linewidths, the 13C spin-lattice relaxation times (T1) of individual ribose carbon atoms, measured by means of partially relaxed Fourier-transform spectra were practically independent of temperature up to about 60°C, and increased rapidly at higher temperatures. The T1 values indicated that the backbone of thermally denatured tRNA is undergoing rapid segmental motion, with an effective correlation time of (2.6 ± 0.5) × 10-10 sec. The T1 values of folded tRNA yielded no evidence of segmental motion. The correlation time for overall rotational reorientation is about (3 ± 1) × 10-8 sec in the range 35-54°C.
Within experimental error, the T1 values of methine carbons of the bases were equal to those of the methine carbons on the backbone at all temperatures. Only an upper limit to the rate of internal rotation of the bases could be established.
Keywords: mg++, temperature, concentration, linewidths, aggregation
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allerhand A., Cochran D. W., Doddrell D. Carbon-13 Fourier transform nuclear magnetic resonance. II. Ribonuclease. Proc Natl Acad Sci U S A. 1970 Nov;67(3):1093–1096. doi: 10.1073/pnas.67.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allerhand A., Doddrell D., Glushko V., Cochran D. W., Wenkert E., Lawson P. J., Gurd F. R. Conformation and segmental motion of native and denatured ribonuclease A in solution. Application of natural-abundance carbon-13 partially relaxed Fourier transform nuclear magnetic resonance. J Am Chem Soc. 1971 Jan 27;93(2):544–546. doi: 10.1021/ja00731a053. [DOI] [PubMed] [Google Scholar]
- Dorman D. E., Roberts J. D. Nuclear magnetic resonance spectroscopy: 13C spectra of some common nucleotides. Proc Natl Acad Sci U S A. 1970 Jan;65(1):19–26. doi: 10.1073/pnas.65.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J. T., Vallee B. L. Differences between the conformation of arsanilazotyrosine 248 of carboxypeptidase A in the crystalline state and in solution. Proc Natl Acad Sci U S A. 1971 Oct;68(10):2532–2535. doi: 10.1073/pnas.68.10.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng M., Felsenfeld G. A study of polyadenylic acid at neutral pH. J Mol Biol. 1966 Feb;15(2):455–466. doi: 10.1016/s0022-2836(66)80121-3. [DOI] [PubMed] [Google Scholar]
- Mantsch H. H., Smith I. C. Fourier-transformed 13 C NMR spectra of polyuridylic acid, uridine, and related nucleotides--the use of 31 POC 13 C couplings for conformational analysis. Biochem Biophys Res Commun. 1972 Jan 31;46(2):808–815. doi: 10.1016/s0006-291x(72)80213-4. [DOI] [PubMed] [Google Scholar]
- Roberts G. C., Jardetzky O. Nuclear magnetic resonance spectroscopy of amino acids, peptides, and proteins. Adv Protein Chem. 1970;24:447–545. doi: 10.1016/s0065-3233(08)60246-6. [DOI] [PubMed] [Google Scholar]
- Seno T., Kobayashi M., Nishimura S. Characteristic behavior of 4-thiouridine region of individual amino acid-specific Escherichia coli tRNA's upon heat denaturation. Biochim Biophys Acta. 1969 Jan 21;174(1):71–85. doi: 10.1016/0005-2787(69)90230-5. [DOI] [PubMed] [Google Scholar]
- Smith I. C., Yamane T., Shulman R. G. Proton magnetic resonance of transfer RNA. Science. 1968 Mar 22;159(3821):1360–1361. doi: 10.1126/science.159.3821.1360. [DOI] [PubMed] [Google Scholar]
- Zachau H. G. Transfer ribonucleic acids. Angew Chem Int Ed Engl. 1969 Oct;8(10):711–727. doi: 10.1002/anie.196907111. [DOI] [PubMed] [Google Scholar]


