Abstract
After more than 50 years of debates, the role of spatial and temporal gradients during cell chemotaxis is still a contentious matter. One major challenge is that when cells move in response to a heterogeneous chemical environment they are exposed to both spatial and temporal concentration changes. Even in the presence of perfectly stable chemical gradients, moving cells experience temporal changes of concentration simply by moving between locations with different chemical concentrations in a heterogeneous environment. Thus, the effects of the spatial and temporal stimuli cannot be dissociated and studied independently, hampering progress towards understanding the mechanisms of cell chemotaxis. Here we employ microfluidic and other engineering tools to build a system that accomplishes a function analogous to a treadmill at the cellular scale, holding a moving cell at a specified, unchanging location in a chemical gradient. Using this system, we decouple the spatial and temporal gradients around moving human neutrophils and find that temporal gradients are necessary for the directional persistence of human neutrophils during chemotaxis. Our results suggest that temporal chemoattractant changes are important during neutrophil migration and should be taken into account when deciphering the signalling pathways of cell chemotaxis.
Introduction
Neutrophils are the most effective barrier in preventing the invasion and spreading of microorganisms within the human body. Failure of neutrophils to promptly arrive at sites of infection or inflammation can result in uncontrollable infections1. At the same time, overzealous neutrophil infiltration can lead to chronic inflammation and unnecessarily damage normal tissues2 and impair organ function e.g. in severe forms of asthma3 and arthritis4, or ischemia-reperfusion injury5 While most of the neutrophils in the body reside in the blood and enter tissues only in response to local chemical gradients inside these tissues, better understanding of the response of neutrophils to these gradients could create new opportunities for effective therapies for infections and chronic inflammation that are more selective and more effective.
More than a hundred signalling molecules involved in different aspects of neutrophil motility have been identified in the past decades6. However, we still do not understand how these molecules work together to achieve neutrophil motility. Traditional methods that helped uncover the signalling molecules are not efficient in finding the systems that organize the same group of molecules. The bottom-up approach allowed by the traditional molecular biology methods can only help finding some relations between pairs of molecules but is limited when it comes to analyzing the larger systems of molecules7. A more appropriate strategy to address the systems-level problem is usually a top-down approach, where one starts by analyzing the response of neutrophils to precise perturbations of the gradients and begins to infer the organization of signalling networks8.
Two sensing mechanisms are commonly considered to optimize migratory responses of neutrophils in complex environments, but controversy exists on which of these is responsible for chemical gradient sensing9-11. A spatial sensing mechanism uses differences in chemokine concentration between the front and the back of the cell to direct their migration towards higher concentrations. A temporal mechanism relies on changes in concentration over time to direct cell migration towards areas with the largest change. Arguments for a spatial gradient come from observations of the pseudopod formation in a stationary neutrophil subsequent to gradient exposure11, or after the ability of a cell to move or extend protrusions has been inhibited by latrunculin12. On the other hand, temporal mechanisms are supported by observations of neutrophil responses to waves of chemoattractant13 and reversal of the direction of migration following concentration decrease of uniform chemotactic fields14 and are comparable to gradient sensing in bacteria15. Our earlier observations using fast switching of chemoattractant gradients suggest that neutrophils can employ at least some temporal sensing mechanisms to direct their responses to microenvironment changes16.
One limiting factor for advancing the knowledge on neutrophil migration in response to heterogeneous environments is the lack of adequate experimental systems17. In most experiments, the spatial and temporal components are coupled, because moving cells are exposed simultaneously to spatial and temporal gradients. Even in the presence of utmost stable chemical gradients18, cells moving in a spatial gradient of chemokine, by virtue of changing their location inside the gradient, are being exposed to temporal changes in concentration. Experiments that attempt to change the concentration of chemokines around cells, are limited by the physics of chemical diffusion which lead to the formation of simultaneous spatial gradients during any concentration change10. Moreover, experiments involving the immobilization of the neutrophils by disrupting the motility apparatus using chemical12 or physical restrictions (e.g. low temperature19) are difficult to interpret because they perturb the signalling pathways they attempt to resolve. Solving such problems requires new tools that can analyze cellular responses in fully functional neutrophils, with intact signalling networks, and in precise extra-cellular conditions.
Here we demonstrate a new tool that actively decouples the contribution of spatial and temporal gradients of a chemokine to neutrophil migration. For this, we continuously and precisely change the location of the cell within the gradient in synchronicity with the cell migration, to achieve a complete cancellation of the temporal stimulus that the moving neutrophil would otherwise experience. Such capability is not available in any of the current chemotaxis devices, in which the control of the gradient is passive, restricted by physical design of the device, and independent of cell migration. Observations enabled by the new technology provide unexpected insights into the mechanisms of neutrophils sensing the chemical gradients, and could eventually lead to the development of rational therapeutic approaches, capable of fine tuning the neutrophil responses appropriate to various diseases.
Materials and Methods
Preparation of human neutrophils and media
De-identified, fresh human blood samples from healthy, consenting adults were purchased from Research Blood Components (Alston, MA). The blood was collected into tubes containing sodium heparin (Vacutainer; Becton Dickinson). HetaSep separation buffer (STEMCELL Technologies, Vancouver, BC) was added to blood in 1:5 ratio and mixed gently. The resulting mixture was allowed to sediment for 45 minutes to separate nucleated cells from erythrocytes. The supernatant containing nucleated cells was pipetted into a separate tube and combined with 4 parts RoboSep buffer (STEMCELL Technologies, Vancouver, BC), then centrifuged at 1000 rpm for 5 minutes. Neutrophils were isolated using the EasySep Human Neutrophil Enrichment Kits (STEMCELL Technologies, Vancouver, BC) following the protocol provided by the manufacturer. After isolation, neutrophils were resuspended in 1 mL IMDM (ATCC, Manassas, VA) with 20% FBS (Sigma-Aldrich, St. Louis, MO). Neutrophils were used within 2 hours of isolation, and kept at 37°C and 5% CO2 until use.
Leukotriene B4 (LTB4) (Cayman Chemical, Ann Arbor, Michigan) was used as a chemoattractant. A final concentration of 100 nM LTB4 in Hank’s Balanced Salt Solution (HBSS) (Life Technologies, Grand Island, NY) and 0.2% human serum albumin (HAS) (Sigma-Aldrich, St. Louis, MO) was prepared. An additional 1 μM of fluorescein isothiocyanate (FITC) was added to this mixture to allow observation of the gradient. HBSS and 0.2% HSA was used as the ambient solution.
Design and fabrication of microfluidic device
Microfluidic devices were constructed of two layers of polydimethyl siloxane (PDMS, Dow Corning, Midland, MI) bonded together on top of a glass coverslip. First, PDMS was cast as a 1 mm thick layer onto a silicon wafer on which a 100 μm layer of photoresist (SU-8, Microchem, Newton, MA) was micro-patterned using standard photolithography techniques. Second, a layer of PDMS roughly 100 μm thick was deposited by spin-coating on a blank silicon wafer. Both PDMS layers were cured at 65°C for at least 24 hours. The thicker PDMS was carefully cut and peeled off of the micro-patterned wafer. A 0.75 mm hole-punch (Harris Uni-Core, Ted Pella, Redding, CA) was used to create fluid inlets and outlets for each device. Both this piece of PDMS and the thin layer on the blank wafer were exposed to an oxygen plasma for 35 seconds in a plasma asher (Nordson March, Concord, CA), then brought into contact to form a bond so that the channels on the thick layer were sealed against the PDMS of the thin layer, with the punched holes being the only inlets. The wafer with the bonded PDMS was set on a hot plate at 75°C for 10 minutes. The bonded pieces were then carefully cut and peeled off of the blank wafer and trimmed to make individual devices. At this point the ends of the channels were trimmed so that the inlets led to open channels that terminated roughly 100 μm from the bottom of the channel. The trimming was done at a steep angle to make the device as thin as possible at the open end. The oxygen plasma step was then repeated to bond devices on top of a glass cover slide to provide additional support during handling. The end of this glass cover slide was bonded far enough from the outlets of the channels to prevent the cover slide from interfering with channel positioning.
Generation of the chemical gradients
Pieces of flexible plastic tubing (Tygon, Greene Rubber Co., Woburn, MA) were cut to the lengths of 50 cm (×2) and 100 cm. One end of each piece of tubing was connected to a 30G blunt needle connected to a 5 mL plastic syringe (Fisher Scientific, Pittsburgh, PA). The other ends of the pieces of tubing were inserted into the punched inlet and outlet holes with the two 50 cm pieces being inserted into the inlet holes and the 100 cm piece of tubing being inserted into the outlet hole. The device was attached to a micromanipulator so that the outlet could be positioned near the centre of the microscope field of view and just above the surface of a 50 mm glass bottom dish (MatTek, Ashland, MA). The glass bottom dish was coated with 100 nM human-fibronectin (Sigma-Aldrich, St. Louis, MO) and filled with HBSS + 0.2% HSA. The two syringes for the inlet holes were raised above the device while the syringe attached to the outlet hole was lowered below the device to allow for gravity driven flow to control the delivery or withdrawal of fluid through the device channels. HBSS + 0.2% HSA containing 100 nM LTB4 and 1 μM FITC was perfused through the centre channel, and was taken back up by the surrounding two channels. By matching the total ejection rates of the centre and outermost channels, with the uptake in the drain channels, a spatial gradient of LTB4 is generated and precisely controlled. The role of the two outermost channels were used to perfuse additional HBSS + 0.2% HSA to keep the gradient spatially confined around the tip of the device and block the diffusion of LTB4 into the dish. This feature is critical to preventing artefacts from a temporal increase in the chemokine concentration for all the cells inside the dish, including the tracked ones.
Feed-back control of neutrophil position during migration
Neutrophils were then loaded into the 50 mm glass bottom dish, allowed to settle for 5 minutes, and then exposed to the gradient. A heating stage (Carel, Manheim, PA) was used to maintain a temperature of 37°C within the dish. The neutrophils were imaged on an inverted microscope (Nikon TE2000) with DIC filters at 20× magnification using a digital camera (Retiga 2000R FAST, QImaging, Surrey, BC, Canada). A motorized stage (Proscan II, Prior Scientific, Rockland, MA) was used to control the movement of the stage. The position of the cell of interest in the microscope field was controlled using a modified version of the open-source Micro-Manager platform20. Our modification replaced the existing Tracker code with a system that provides multiple approaches to detecting cell displacement: a correlation-based method (as originally encoded), a fast gradient-based motion measurement algorithm with sub-pixel resolution21, 22, and a segmentation-based approach. Initial experiments revealed that the segmentation approach was the most accurate, so it was used for all results reported in this paper.
The segmentation approach works as follows. First, a region around the most recent cell position is selected. The mean and standard deviation of pixel brightness in this region are computed, and the region is thresholded at a user-specified multiple of standard deviations from the mean. The Erode operator of ImageJ is applied to the thresholded image twice in succession, followed by the Dilate operator twice. The Analyze Particles operator is used to identify individual regions remaining, and the largest of these is assumed to be the cell. Based on the expected direction of cell motion (user-defined), one edge of this region is defined as the leading edge. The position in the orthogonal direction is taken as the centre of the bounding box containing this region. Each operator in this process is a function built into ImageJ. This method was used to determine the position of a single cell of interest during each experiment. The collected images were later analyzed using the same method to measure the displacement of other cells relative to the target cell.
To adjust the stage position, two affine transforms were applied to the measured cell displacement. The first enabled adjustments of the scale and offset of the feedback, while the second corrected for scaling and rotation of the stage coordinates relative to camera coordinates. The resulting stage motion was given by
where the 3×3 matrix with lower-case variables defines the transformation from camera to stage coordinates, and the 2×3 matrix with upper-case variables defines the scaling and offset. To keep the cell in place, A=E=-1 and B=C=D=F=0. Other transformations, such as over- and under-compensation for cell motion, rotation, and constant cell velocity can be applied through appropriate values of these variables.
Neutrophil migration analysis
The stability of the gradient was determined by taking fluorescent images of the gradient over time, extracting a kymograph from the images in ImageJ, and determining the intensities in MATLAB (MathWorks, Natick, MA) to visualize the intensities of the gradient at specified points over the duration of the experiment.
To explain these results concisely we introduce several terms. Total Displacement refers to the integrated motion of the cell in any direction relative to its immediate surroundings. Effective Displacement refers to the component of Total Displacement that occurs in the direction of increasing chemokine concentration. Net Displacement refers to the integrated vector motion of the cell relative to its immediate surroundings. Gradient Displacement refers to the displacement of the cell in digital images relative to the source of chemokine gradient. For example, a cell which moves 4 μm in the direction of the gradient and 3 μm in an orthogonal direction, has a Total Displacement of 7 μm, Effective Displacement of 40 μm, Net Displacement of 5 μm . When the stage is then moved to bring the cell back to its starting location, the Gradient Displacement is 0 μm.
MATLAB was used to calculate the average lifetime velocity,
and directional persistence,
of each cell from the positions of the cell over time determined by the tracking algorithm.
For both feedback conditions and no feedback conditions, 10 cells were measured from 10 different experiments. Velocities were compared using the independent Student t-test. Differences were considered significant above a 95% confidence interval. The noise introduced by the system was determined by calculating the root mean squared of the error in the tracking algorithm.
Results and discussion
To decouple the influence of spatial and temporal gradients during neutrophils chemotaxis, we employed a system that combined a microfluidic gradient generator, a motorized stage, and a microscopy system in a computer-controlled feedback control loop (Fig.1A). The goal was to move the stage in response to neutrophil migration, such that the neutrophil remains in the same position relative to the spatial gradient and experience no change in concentration over time (Fig.1B). The time required for each computation and actuation loop was less than 2 seconds, faster than the shape changes of the neutrophils. The cell tracking algorithm worked well in phase contrast as well as differential interference contrast. This allowed us to avoid using fluorescent dyes for cell tracking, which could potentially interfere with cell migration when taking into account the large number of images required for implementing a robust feedback system (more than 30 images every minute for the duration of the experiment). This approach also has the advantage that it keeps all fluorescence channels of the microscope available for molecular studies during cell migration, or interface with physical measurements of cell migration e.g. traction force microscopy23.
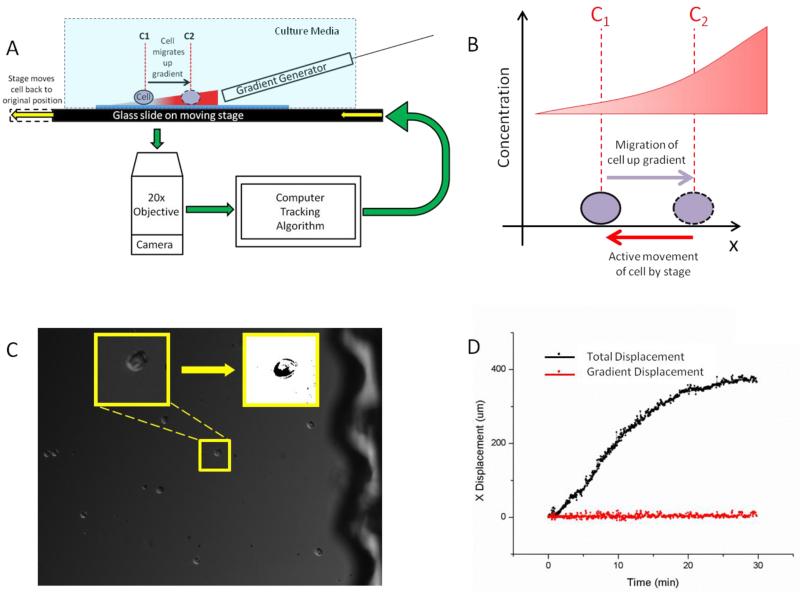
Fig 1.
Neutrophil treadmill platform (A) The platform consists of a gradient generator that expels a gradient onto a glass slide on a microscope stage without touching or being affected by stage movement. The cell on the glass slide is exposed to the gradient and begins to migrate. As the cell is migrating, the microscope is imaging the cell and a computer algorithm is determining the displacement of the cell for each time step. The cell movement information is fed into the microscope stage which is moved in an equal and opposite direction to the cell’s movement. This returns the cell to its original position relative to the gradient. Since the gradient generator is separate from the stage movement, the gradient remains in a constant position and whole system provides the feedback to maintain the cell’s position relative to the chemokine gradient. (B) As a cell migrates up a gradient it is exposed to a different concentration of chemokine. When feedback is applied, the cell is actively moved in an equal and opposite direction by the stage and, therefore, the local chemokine concentration around the cell is constant over time. (C) The camera captures a DIC image around the region of interest. The region of interest is processed through a series of image processing steps to allow for the algorithm’s determination of the cell’s position and subsequent feedback to the stage. (D) While a cell migrates in the direction of the chemokine gradient, the stage compensates equally and oppositely to the cell’s motion so that the cell remains in the same place relative to the gradient and the field of view just as a runner on a treadmill does not actually displace.
We verified the ability of the platform to track individual objects and move the microscope stage correspondingly, by applying the system first to a static object and then to a controlled dynamic object. When the system was applied to a stationary object we observed no stage drift over time. When a micromanipulator was used to precisely displace a bead the system was able to accurately track the movement and feed the information into the stage controllers so as the return the bead to its original position after each controlled displacement. We also verified the ability of the system to recognize human neutrophils undergoing shape changes during migration (Fig.1C), and track the motion of individual neutrophils and move the stage following the feedback algorithm (Fig.1D).
By varying the heights of the reservoirs and length of the tubing for the input and output solutions of the gradient generator (Fig.2A), flow rates of 10 μL/min, 20 μL/min, and 15 μL/min were achieved for the chemokine solution, drain, and distal buffer solution channels, respectively. These flow rates and the dimensions and distances between channels of the gradient generator caused a semi-circular gradient to be formed at the end of the gradient generator on the surface of the glass slide (Fig.2B). It is important to note that the gradient generated by this system is such that it completely avoids the build-up of chemokine on top of neutrophils at a distance from the gradient generator. Unlike classical micropipettes24, we took special care to balance the flows through the distal channels such that all the chemokine injected in the dish is recovered. In addition to matching the ejection and uptake flow rates, addressed previously by microfluidic designs like the push-pull probes25,26, multifunctional pipettes27, or quadropol gradient generators28, we implemented two more channels distal from the centre channels. Flow patterns between these and drain channels are such that they confine the chemokine diffusion at the tip of the device, by removing all chemokine diffusing away from the tip, before reaching the dish. This feature is key for preventing slow chemokine build-up in the dish over time during feed-back experiments. Moreover, we calculated the flow rates such that the shear stress for the cells in the gradient is within the limits previously demonstrated not to alter the migration of the neutrophils29.
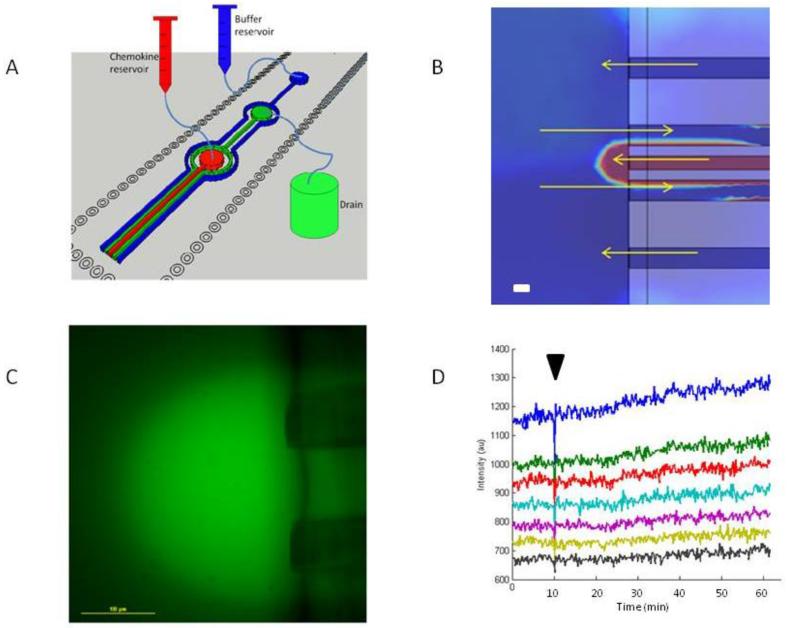
Fig 2.
Design and characterization of the floating microfluidic gradient generator (A) A 3D image of the gradient generator channels. The centre red channel (100 × 100 × 100 μm3) has an inlet at its top allowing for chemokine to be flowed into the channel and out its end onto a glass slide. The inner two green centre channels (100 × 100 × 100 μm3) come together at an outlet from which solution is removed from the slide. The outermost two blue channels (100 × 100 × 100μm3) share an inlet into which buffer can be flowed into the channel and out the ends onto the glass slide to contain the chemokine from the centre channel. The flow within the device is gravity driven with reservoirs of chemokine and buffer positioned above the gradient generator and a reservoir for the drain being positioned below. (B) A COMSOL simulation of the concentration of chemokine in the gradient generated from the device. A semi-circular gradient can be seen at the end of the centre channel. The flow patterns from the two outermost channels to the drain channels create a functional boundary to help confine the chemokine around the centre channel. In the absence of the outermost channels, chemokine could diffuse continuously into the dish and result in a temporal increase in the chemokine concentration for all the cells, including the tracked ones.(C) A FITC fluorescent dye was loaded into the centre channel to characterize the semi-circular gradient formed by the chemical in the centre channel of the gradient generator. Scale bar 100 μm. (D) A kymograph showing the linear intensity of the gradient at various distances from the centre channel to the edge of the gradient against time demonstrates the stability of the gradient over time. A perturbation in the environment was introduced 10 minutes into the experiment (marked by a black arrow) by moving the stage over 100 μm to demonstrate that the gradient returns to a stable state after external disturbances.
To validate the gradient, we added 1 μM of fluorescein isothiocyanate (FITC) to the chemokine solution (Fig.2C). The chemical gradient was stable over 60 minutes, verified by the analysis of a kymograph demonstrating the stability over time of the fluorescent signal at different points in the gradient (Fig.2D). When we applied a short perturbation to the stage by moving the stage 100 μm the disturbance in the gradient was corrected in less than 30 seconds. The displacement used in this test was at least 100 times larger than the 1μm distance for the fastest neutrophil moving in the 2 seconds between imaging during the experiment, which represents the maximum displacement of the stage at one time point. The stability of the gradient to perturbation is important in order to assure that accurate corrections of stage position do not interfere with the position of the gradient.
We calculated that during the experiments, the majority of tracked neutrophils experience concentrations between 1 and 50 nM of LTB4. Studies of binding kinetics in human neutrophils have reported dissociation constant for LTB4 in the range of 10-15 nM30, suggesting that the receptors are not saturated. In addition, rapid receptor recycling31 assures that the neutrophils remain sensitive to spatial gradients of LTB432. Moreover, spatial conditions around a moving cell do not change during cell migration, the main purpose of the feedback system.
With the feedback system on, the spatial conditions at the new position of the cell are identical with those at the start location. One limitation of this approach is that even though multiple cells may be present in the field of view, only one cell per experiment is selected for the feedback algorithm. Because not all cells move at the same speed, or with the same persistence, all other cells are not used in the analysis. For these cells, a temporal gradient is present and uncorrelated to their motility because of the mismatch between migration and the movement of the stage, limiting the overall throughput of the experiments.
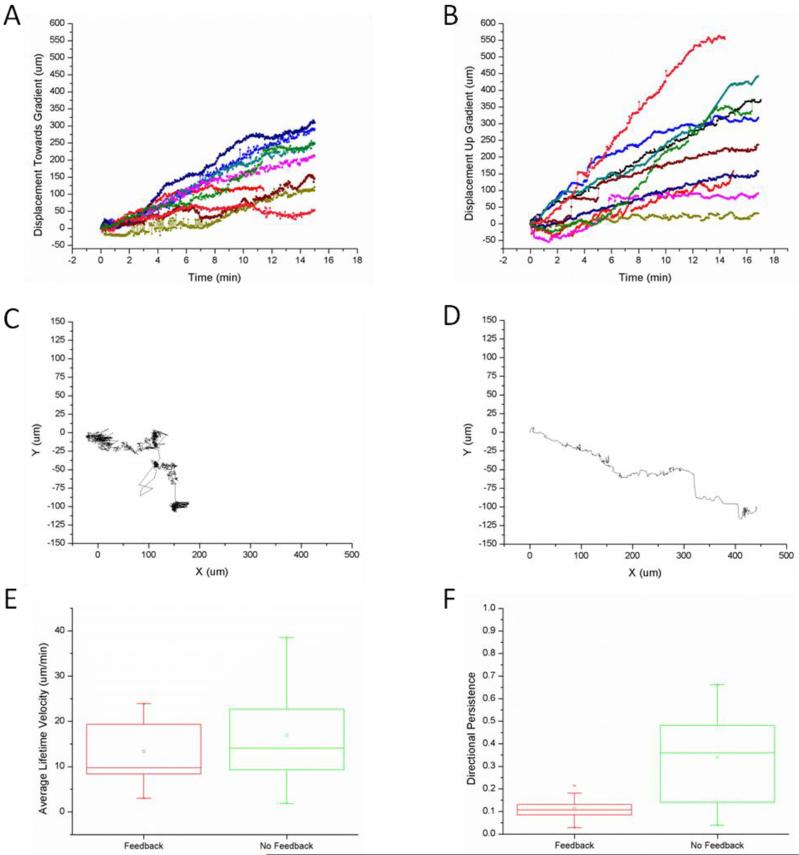
We employed the treadmill platform to compare the effective displacement with and without feedback (Fig.3A,B). Typical tracks of neutrophils with feedback are shorter and often include overlapping tracks (Fig.3C), compared with those of neutrophils without feedback, which display typical migration trajectories and serve as controls (Fig.3D). We compared the average lifetime velocities of neutrophils with and without feedback (N=10 for each condition) and found that these were comparable (Fig.3E, p > 0.05). However, when we calculated the directional persistence of the cells, we found significantly lower directional persistence in the presence compared to the absence of the feedback (0.04 vs. 0.66, Fig.3F, p < 0.05). These results show that both the spatial and temporal gradients are necessary for persistent migration during neutrophil chemotaxis. Moreover, cells moving in the absence of feedback maintain the same average speed and persistence at different locations in the gradient, as they reach higher concentrations, consistent with previous observations of human neutrophil migration on flat surfaces in the presence of linear chemoattractant gradients33. It is thus unlikely that concentration alone, in the range of concentrations experienced in our assay, could explain the differences in persistence. While the spatial gradients are important for the initiation and directionality of the migration, the temporal concentration changes are important for sustaining the persistent migration towards a target.
Fig 3.
Neutrophil migration in the presence and absence of feed-back. (A) The Effective Displacement of the cell was tracked over the time in 10 experiments where feedback was applied and the stage was moved in equal and opposite distances of the cell’s movement. (B) The Effective Displacement of the cell was tracked over the time in 10 experiments where feedback was not applied and no stage movement occurred. (C) A trajectory of a cell that had low directional persistence (DP = 0.03). (D) A trajectory of a cell that had high directional persistence (DP = 0.66). (E) The average lifetime velocities of the tracked cells were calculated. No significant difference was found between the velocities of the cells that were kept in the same place relative to the gradient versus the cells that did not experience any stage feedback and moved up the gradient (p > 0.05). (F) The directional persistence of the cells was calculated by dividing the displacement of the cell in the direction of the gradient by the total displacement of the cell. The cells that experienced stage feedback and were kept in the same place relative to the gradient had significantly lower directional persistence than the cells that did not experience stage feedback and migrated up the gradient (p < 0.05).
Our finding of distinct and complementary roles for the spatial and temporal gradients, enabled by new tools, provides an unexpected resolution for the long standing controversy regarding the role of spatial and temporal stimuli of cell migration11. Unlike spatial gradients of chemokines, which are generally accepted as key for establishing the front-back cell polarity towards the higher concentration, the role of temporal concentration changes has been repeatedly challenged10, 11 For example, recent studies comparing the differences in neutrophil-like HL60 cells prowess to migrate in linear and exponential concluded that chemical gradients are interpreted purely spatially34. In contrast to these, however, motile bacteria rely exclusively on temporal sensing to accumulate towards the highest concentrations of a chemoattractant15. Moreover, in real word spatial and temporal gradient exist simultaneously35. Suggesting a possible solution to this dilemma, our study provides support to the role of temporal stimuli in neutrophils for enforcing the persistent migration in one direction after the initiation of migration. Unlike bacteria, which start moving in random directions upon stimulation, neutrophils can also take advantage of a spatial sensing mechanism and orient their migration in the direction of higher concentration.
The interplay between spatial and temporal stimuli during neutrophil chemotaxis suggests that the molecular mechanisms of cell migration combine the ability to compare different levels of receptor stimulation in different regions of the cells, with a dynamic memory for the levels of stimulation at different time16. Such models consider not only the requirements for symmetry breaking in the presence of a gradient36-38 and incorporate stochastic and transient features that benefit from temporal concentration changes around the cells16. Additional cues for the molecular substrate of temporal sensing may come from observations of neutrophils that display persistent migration inside small channels in the presence of constant, polar stimulation39, 40 or even in the presence of decreasing concentrations during retrotaxis41. During the migration inside channels, the mechanical confinement may compensate for the absence of temporal stimuli, possibly by elements of the cytoskeleton e.g. microtubules which are maintained in stable configuration inside the small channels16, 39.
In the future, the treadmill technology for cell migration will facilitate the study of cell motility and open new opportunities for understanding the effects of temporal changes on motile cells. Historically, feed-back based systems have enabled critical insights into important biological processes, including the propagation of electrical potential42, while the decoupling of tonic and length effects on the muscle activity, led to better understanding of the mechanisms of actin-myosin contraction43, 44. Similarly, clamping spatial and temporal chemotactic gradients while exposing cells to other manipulations can provide new insights into the molecular mechanisms that drive neutrophil chemotaxis. Ultimately, identifying these motility mechanisms could create new opportunities for effective treatments targeting the trafficking of white blood cells through the body in acute and chronic inflammation.
Conclusions
We designed a system to decouple the spatial and temporal gradients during cell chemotaxis. We found that temporal gradients are responsible for the persistence of human neutrophils migration in the response to chemical gradients. Our findings could help understand the mechanisms that guide the neutrophils to sites of infection or inflammation.
Acknowledgements
All fabrication procedures were performed at the BioMEMS Resource Center (National Institutes of Health / National Institute of Biomedical Imaging and Bioengineering grant EB002503). Funding was provided in part from National Institutes of Health / National Institute for General Medicine (grant GM092804) and National Institute of Allergy and Infectious Diseases (grant AI076760).
Notes and references
- 1.Manz MG, Boettcher S. Nature reviews. Immunology. 2014;14:302–314. doi: 10.1038/nri3660. [DOI] [PubMed] [Google Scholar]
- 2.Nathan C. Nature reviews. Immunology. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 3.Turato G, Baraldo S, Zuin R, Saetta M. Thorax. 2007;62:465–466. doi: 10.1136/thx.2006.070656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominical VM, Bertolo MB, Almeida CB, Garrido VT, Miguel LI, Costa FF, Conran N. Scandinavian journal of immunology. 2011;73:309–318. doi: 10.1111/j.1365-3083.2011.02503.x. [DOI] [PubMed] [Google Scholar]
- 5.Chaturvedi S, Yuen DA, Bajwa A, Huang YW, Sokollik C, Huang L, Lam GY, Tole S, Liu GY, Pan J, Chan L, Sokolskyy Y, Puthia M, Godaly G, John R, Wang C, Lee WL, Brumell JH, Okusa MD, Robinson LA. Journal of the American Society of Nephrology: JASN. 2013;24:1274–1287. doi: 10.1681/ASN.2012090890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gambardella L, Vermeren S. Journal of leukocyte biology. 2013;94:603–612. doi: 10.1189/jlb.1112564. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Ding SJ, Wang W, Jacobs JM, Qian WJ, Moore RJ, Yang F, Camp DG, 2nd, Smith RD, Klemke RL. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8328–8333. doi: 10.1073/pnas.0701103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ku CJ, Wang Y, Weiner OD, Altschuler SJ, Wu LF. Cell. 2012;149:1073–1083. doi: 10.1016/j.cell.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourne HR, Weiner O. Nature. 2002;419:21. doi: 10.1038/419021a. [DOI] [PubMed] [Google Scholar]
- 10.Lauffenburger D, Farrell B, Tranquillo R, Kistler A, Zigmond S. Journal of cell science. 1987;88(Pt 4):415–416. doi: 10.1242/jcs.88.4.415. [DOI] [PubMed] [Google Scholar]
- 11.Zigmond SH. Nature. 1974;249:450–452. doi: 10.1038/249450a0. [DOI] [PubMed] [Google Scholar]
- 12.Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8951–8956. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geiger J, Wessels D, Soll DR. Cell motility and the cytoskeleton. 2003;56:27–44. doi: 10.1002/cm.10133. [DOI] [PubMed] [Google Scholar]
- 14.Albrecht E, Petty HR. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5039–5044. doi: 10.1073/pnas.95.9.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Block SM, Segall JE, Berg HC. Cell. 1982;31:215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- 16.Irimia D, Balazsi G, Agrawal N, Toner M. Biophysical journal. 2009;96:3897–3916. doi: 10.1016/j.bpj.2008.12.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irimia D. Annual review of biomedical engineering. 2010;12:259–284. doi: 10.1146/annurev-bioeng-070909-105241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Jeon N, Baskaran H, Dertinger SK, Whitesides GM, Van de Water L, Toner M. Nature biotechnology. 2002;20:826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 19.Vicker MG, Lackie JM, Schill W. Journal of cell science. 1986;84:263–280. doi: 10.1242/jcs.84.1.263. [DOI] [PubMed] [Google Scholar]
- 20.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. In: Current protocols in molecular biology. Ausubel Frederick M., et al., editors. 2010. Chapter 14, Unit14 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horn BP, Weldon EJ., Jr. Int J Comput Vision. 1988;2:51–76. [Google Scholar]
- 22.Davis QC, Freeman DM. Optical Engineering. 1998;37:1290–1298. [Google Scholar]
- 23.Smith LA, Aranda-Espinoza H, Haun JB, Dembo M, Hammer DA. Biophysical journal. 2007;92:L58–60. doi: 10.1529/biophysj.106.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerisch G, Keller HU. Journal of cell science. 1981;52:1–10. doi: 10.1242/jcs.52.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Momotenko D, Cortes-Salazar F, Lesch A, Wittstock G, Girault HH. Analytical chemistry. 2011;83:5275–5282. doi: 10.1021/ac2006729. [DOI] [PubMed] [Google Scholar]
- 26.Autebert J, Kashyap A, Lovchik RD, Delamarche E, Kaigala GV. Langmuir: the ACS journal of surfaces and colloids. 2014;30:3640–3645. doi: 10.1021/la500875m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ainla A, Jeffries GD, Brune R, Orwar O, Jesorka A. Lab on a chip. 2012;12:1255–1261. doi: 10.1039/c2lc20906c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qasaimeh MA, Gervais T, Juncker D. Nature communications. 2011;2:464. doi: 10.1038/ncomms1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker GM, Sai J, Richmond A, Stremler M, Chung CY, Wikswo JP. Lab on a chip. 2005;5:611–618. doi: 10.1039/b417245k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldman DW, Goetzl EJ. J Immunol. 1982;129:1600–1604. [PubMed] [Google Scholar]
- 31.Boneschansker L, Yan J, Wong E, Briscoe DM, Irimia D. Nature communications. 2014;5:4787. doi: 10.1038/ncomms5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brom J, Konig W. Immunology. 1989;68:479–485. [PMC free article] [PubMed] [Google Scholar]
- 33.Irimia D, Liu SY, Tharp WG, Samadani A, Toner M, Poznansky MC. Lab on a chip. 2006;6:191–198. doi: 10.1039/b511877h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herzmark P, Campbell K, Wang F, Wong K, El-Samad H, Groisman A, Bourne HR. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13349–13354. doi: 10.1073/pnas.0705889104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vicker MG. Journal of cell science. 1989;92(Pt 1):1–4. doi: 10.1242/jcs.92.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Turing AM. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 1952;237:37–72. [Google Scholar]
- 37.Levchenko A, Iglesias PA. Biophysical journal. 2002;82:50–63. doi: 10.1016/S0006-3495(02)75373-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori Y, Jilkine A, Edelstein-Keshet L. Biophysical journal. 2008;94:3684–3697. doi: 10.1529/biophysj.107.120824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ambravaneswaran V, Wong IY, Aranyosi AJ, Toner M, Irimia D. Integrative biology: quantitative biosciences from nano to macro. 2010;2:639–647. doi: 10.1039/c0ib00011f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irimia D, Charras G, Agrawal N, Mitchison T, Toner M. Lab on a chip. 2007;7:1783–1790. doi: 10.1039/b710524j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamza B, Wong E, Patel S, Cho H, Martel J, Irimia D. Integrative biology: quantitative biosciences from nano to macro. 2014;6:175–183. doi: 10.1039/c3ib40175h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hodgkin AL, Huxley AF, Katz B. The Journal of physiology. 1952;116:424–448. doi: 10.1113/jphysiol.1952.sp004716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huxley AF, Niedergerke R. Nature. 1954;173:971–973. doi: 10.1038/173971a0. [DOI] [PubMed] [Google Scholar]
- 44.Huxley H, Hanson J. Nature. 1954;173:973–976. doi: 10.1038/173973a0. [DOI] [PubMed] [Google Scholar]