Abstract
Serine proteases are critical for epidermal barrier homeostasis and their aberrant expression and/or activity is associated with chronic skin diseases. Elevated levels of the serine protease inhibitors SERPINB3 and SERPINB4 are seen in patients with atopic dermatitis and psoriasis. However their mechanistic role in the skin is unknown. To evaluate the contribution of Serpinb3a (mouse homolog of SERPINB3 and SERPINB4) in atopic dermatitis, we examined the effect of topical Aspergillus fumigatus extract exposure in wild-type and Serpinb3a null mice on transepidermal water loss (TEWL), sensitization and inflammation. Allergen exposure induced Serpinb3a expression in the skin, along with increased TEWL, epidermal thickness, and skin inflammation, all of which were attenuated in the absence of Serpinb3a. Attenuated TEWL correlated with decreased expression of the pro-inflammatory marker S100A8. Silencing of SERPINB3/B4 in human keratinocytes decreased S100A8 expression supporting a role for SERPINB3/B4 in initiation of the acute inflammatory response. RNA-Seq analysis following allergen exposure identified a network of pro-inflammatory genes induced in the wild type mice that was absent in the Serpinb3a null mice. In conclusion, Serpinb3a deficiency attenuates barrier dysfunction and the early inflammatory response following cutaneous allergen exposure, supporting a role for Serpinb3a (mice) and SERPINB3/B4 (humans) early in atopic dermatitis.
Keywords: Atopic dermatitis, SERPINB3/B4, Serpinb3a, TEWL, S100A8, inflammation
Introduction
The importance of protease function in maintaining skin architecture and regulating responses to pathogens and allergens is well established (Cork et al., 2009). Mass spectrometric analysis identified SERPINB4 in exudates from non-healing skin wounds but not in healing wounds (Eming et al., 2010). Subjects with elevated SERPINB3 serum levels show greater changes in TEWL in response to oleic acid application (Katagiri et al., 2010). SERPINB3/B4 expression is increased in the skin and serum of individuals with psoriasis (Suarez-Farinas et al., 2010; Takeda et al., 2002; Tian et al., 2012) and atopic dermatitis (AD) (Broccardo et al., 2009; Kawashima et al., 2000; Lu et al., 2009a; Lu et al., 2009b; Mitsuishi et al., 2005; Saaf et al., 2008). SERPINB3/B4 expression is detected in the horny layer of skin lesions from individuals with AD and the level of expression correlates with serum total IgE levels (Yamane et al., 2009). SERPINB3/B4 expression is higher in the involved skin of individuals with AD compared to uninvolved skin and levels correlate with disease severity (Kawashima et al., 2000; Mitsuishi et al., 2005) and levels return to baseline in AD patients who show improvement post-treatment (Kawashima et al., 2000). In vitro studies in keratinocytes and other cell types have demonstrated that overexpression of SERPINB3/B4 protected the cells from UV, gamma radiation, and TNF alpha induced cell death (Hashimoto et al., 2005; Katagiri et al., 2006; Murakami et al., 2001) further supporting the key role of SERPINB3/B4 in the skin. Thus, there is considerable evidence implicating SERPINB3/B4 in skin inflammation. However the role of these proteins in AD has yet to be defined.
We utilized Serpinb3a (mouse homolog of SERPINB3/B4) knockout mice that we have previously described (Sivaprasad et al., 2011) to directly examine the role of SERPINB3/B4 in mouse models of AD following mold exposure. We examined early and late effects of exposure to Aspergillus fumigatus extract. Serpinb3a KO mice had attenuated inflammatory phenotypes associated with less transepidermal water loss (TEWL) compared to their wild type counterparts. Furthermore, SERPINB3/B4 downregulation in human keratinocytes was associated with markedly attenuated expression of S100A8, a marker of early inflammation, implicating SERPINB3/B4 in initiating early inflammatory events that lead to chronic skin conditions like AD.
Results
Serpinb3a is not required for the development of chronic atopic dermatitis
TEWL is a well-established measure of skin barrier function (Gupta et al., 2008; Sivaprasad et al., 2010) and elevated TEWL is a hallmark of atopic dermatitis (AD) in children. Following repeated cutaneous exposure to Aspergillus fumigatus extract using the more chronic conventional model (Figure 1A), wild type Balb/c (WT) and Serpinb3a null (KO) mice exhibited no differences in barrier function as assessed by TEWL at baseline. TEWL increased following exposure to Aspergillus fumigatus extract (ASP) after the second and third patches, there were no differences based on genotype (Figure 1B, C). Both WT and KO mice were effectively sensitized (Figure 1D, E) and lymph node cells cultured in the presence of ASP produced comparable levels of IL-4 (Figure 1F) and IL-17 (Figure 1G) independent of genotype. Together, the data indicate that the absence of Serpinb3a does not impact the development of an AD-like response in mice following allergen exposure over several weeks.
Figure 1. Serpinb3a is not required for the development of chronic AD.
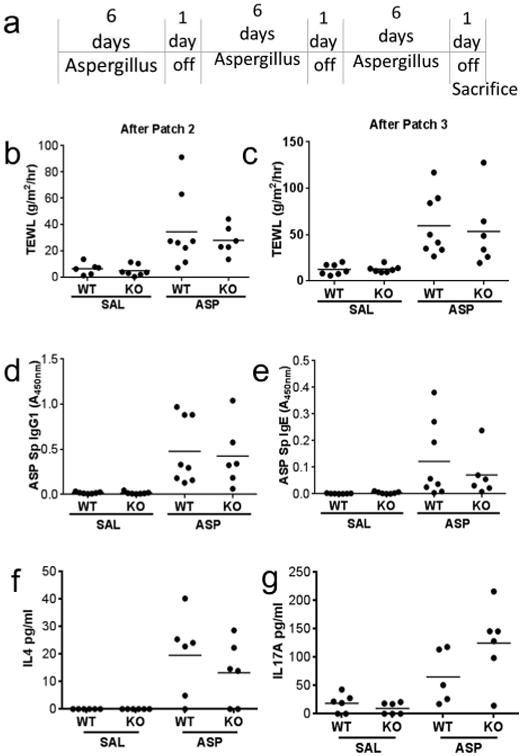
(a) Schematic describing our chronic AD model. TEWL measurements after the second (b) and third (c) patches. Sensitization to allergen measured by ASP-specific IgG1 (d) and ASP-specific IgE (e). Production of IL-4 (f) and IL-17 (g) in supernatants of lymph node cultures of ASP patched mice cultured with saline (SAL) or ASP for 5 days. Data shown are representative of 3 independent experiments (n=6-8 per group).
Serpinb3a contributes to barrier dysfunction early after allergen exposure
In asthma, SERPINB3/B4 expression is increased during an acute exacerbation (Guajardo et al., 2005). In AD, SERPINB3/B4 expression is associated with disease severity, and is also higher in involved skin compared to uninvolved skin (Kawashima et al., 2000; Mitsuishi et al., 2005). In wound exudates, SERPINB4 levels are higher in non-healing wounds compared to healing wounds (Eming et al., 2010). This led us to speculate that the role of SERPINB3/B4 (or their mouse homolog) is likely to be part of the acute response rather than a chronic effect. To address this, we used a shorter model of cutaneous allergen exposure (three patches, each applied for 3 days, with a 24 hour rest period after each; Figure 2A). In this model, TEWL was increased in WT mice exposed to ASP compared to saline (SAL) treated mice. This was significantly attenuated in the Serpinb3a KO mice treated with ASP after both the second and third patches (Figure 2B, C). The skin was visually evaluated and skin scores were assigned based on redness, skin thickening, and excoriations. Skin scores were increased following cutaneous ASP exposure in the WT mice, but were markedly attenuated in the Serpinb3a KO mice (Figure 2D). Consistent with the TEWL and the skin scores, epidermal thickness was increased following cutaneous ASP exposure in the WT mice, but was at only slightly elevated (close to saline treatment) in the Serpinb3a KO mice (Figure 2E). Collectively, the data indicate that Serpinb3a contributes to disruption of the skin barrier early in the response to allergen exposure.
Figure 2. Serpinb3a contributes to barrier dysfunction in early AD.
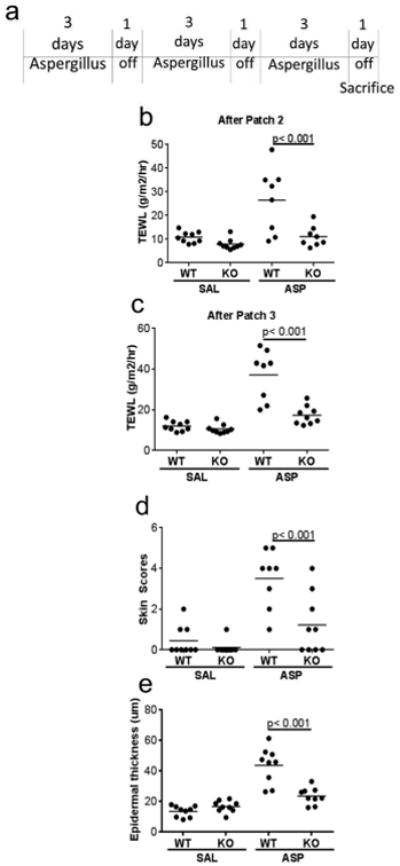
(a) Schematic describing our early AD model. TEWL measurements after the second (b) and third (c) patches. (d) Scoring of AD-like phenotypes (excoriations, redness and thickening). (e) Quantification of epidermal thickness after the third allergen patch. Data shown are representative of 3 independent experiments (n=6-8 per group).
Attenuated early inflammation in the absence of Serpinb3a
Histological examination of H&E stained skin sections revealed a significant increase in the influx of inflammatory cells into the epidermis following cutaneous ASP exposure in the WT mice (Figure 3A, C). This effect was attenuated in the Serpinb3a KO mice (Figure 3B, D). These findings were confirmed following staining for CD3 (T cells; Figure 3E, F) and Ly6G (neutrophils; data not shown), which revealed decreased influx of T cell and neutrophils into the skin of Serpinb3a KO mice.
Figure 3. Inflammation is attenuated in the absence of Serpinb3a.
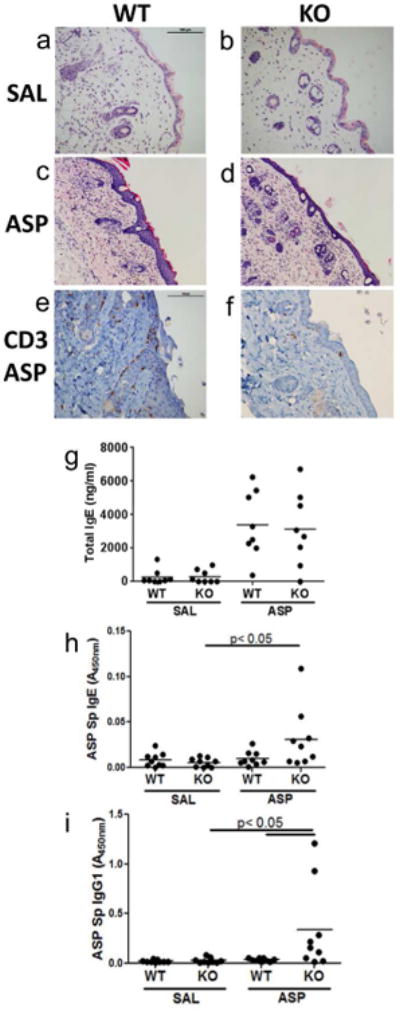
Representative H&E stained sections of patched skin (magnification 20×) from WT-SAL (a), KO-SAL (b), WT-ASP (c) and KO-ASP (d) mice. Representative CD3 stained sections of patched skin from WT-ASP (e) and KO-ASP (f) mice. Sensitization to allergen measured by total serum IgE levels (g), ASP-specific IgG1 (h) and ASP-specific IgE (i). Data shown are representative of 3 independent experiments. Scale bar=100μm.
Total IgE levels were increased in both WT and Serpin3a KO mice following cutaneous AF exposure and there were no significant differences between the two strains (Figure 3G). ASP-specific IgG1 and IgE levels were increased in the Serpinb3a KO mice compared to wild type mice suggesting that these mice may be more easily sensitized than WT Balb/c mice (Figure 3H, I). While statistically significant, in each replicate, only one or two mice had elevated allergen-specific antibody levels, making the biological relevance of this observation questionable.
Correlation between TEWL and the pro-inflammatory S100A8 gene
The onset of acute AD lesions in humans is associated with increased expression of the S100 family of proteins (specifically S100A7, A8, and A9) (Gittler et al., 2012). Studies have shown that S100A9 expression is regulated in an S100A8 dependent manner (Riva et al., 2013). Thus, we determined the impact of Serpinb3a deficiency on S100A8 mRNA expression. Consistent with our previous published data (Brandt et al., 2013), S100A8 (Figure 4A) and Serpinb3a (Figure 4B) mRNA levels were significantly induced in WT mice following ASP exposure. In our shorter model, TEWL correlated with S100A8 mRNA expression levels (r=0.571, p=0.0185; Figure 4C). Induction of S100A8 was markedly attenuated in the Serpinb3a KO mice (Figure 4A). S100A8 expression correlated with Serpinb3a levels (r=0.7516, p=0.0008; Figure 4D) in wild type mice indicating that Serpinb3a may impact S100A8 expression.
Figure 4. Serpinb3a expression correlates with S100A8 expression in mouse skin.
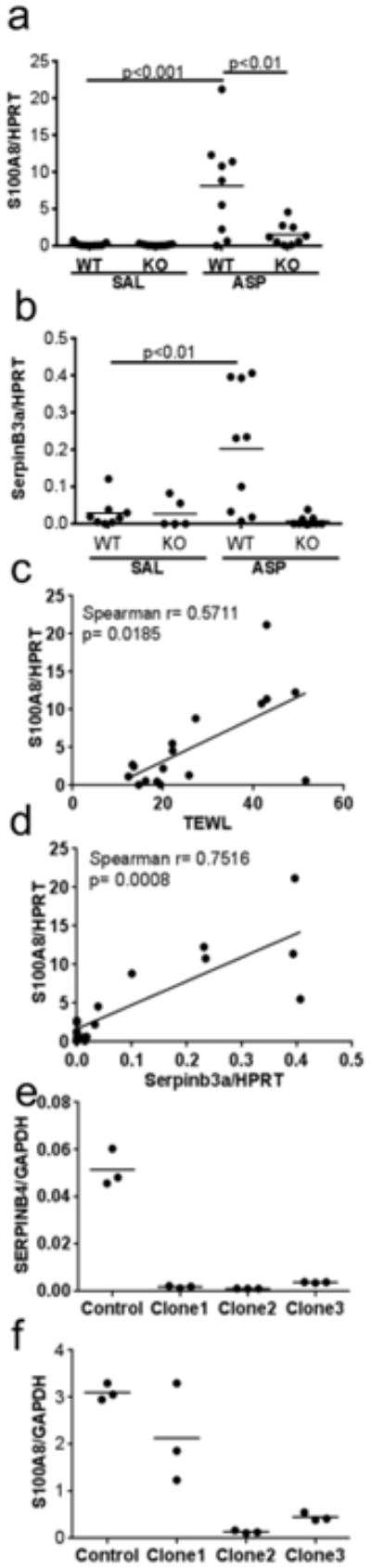
Quantitative real-time PCR for S100A8 (a) and Serpinb3a (b) normalized to HPRT for the indicated groups. Correlation plots between TEWL and S100A8/HPRT (c) and between Serpinb3a/HPRT and S100A8/HPRT (d). Expression of SERPINB4 (e) and S100A8 (f) in three independent clones (Clone1-3) stably transfected with shRNA targeting SERPINB3/B4 compared to a clone generated with the empty vector (control), normalized to GAPDH.
SERPINB3/B4 down-regulation results in decreased S100A8 expression in human keratinocytes
To determine if Serpinb3a directly impacted expression of S100A8 expression in the epithelium versus an indirect effect on expression due to fewer inflammatory cells in the skin, we generated stable clones expressing shRNAs targeting SERPINB3/B4 in HaCaT cells, a human keratinocyte cell line and examined the effect on S100A8 expression. The shRNA clones showed decreased SERPINB4 mRNA by over 90% (Figure 4E). Depletion of SERPINB3/B4 alone was sufficient to decrease S100A8 mRNA levels in three independent shRNA transduced stable cell lines (Figure 4F) supporting a role for SERPINB3/B4 in the induction of expression of S100A8.
SERPINB3/B4 expression correlates with S100A8 expression in AD skin
To determine whether our observations demonstrating an effect of SERPINB3/B4 on S100A8 expression in a mouse model of skin inflammation were also evident in humans, we examined published microarray data comparing normal skin to lesional skin from subjects with AD (Guttman-Yassky et al., 2009). When looking at probes specific for SERPINB3 and/or SERPINB4, SERPINB3 and SERPINB4 expression were significantly increased in AD skin compared to normal skin (Figure 5A-C). In addition, S100A8 expression was also significantly upregulated in AD skin (Figure 5D). In all cases, there was significant correlation between SERPINB3/B4 expression and S100A8 levels in AD skin (Figure 5E-G). Together these data support a role for Serpinb3a in mice and SERPINB3/B4 in humans in regulating expression of S100A8.
Figure 5. Serpinb3a expression correlates with S100A8 expression in human subjects with AD.
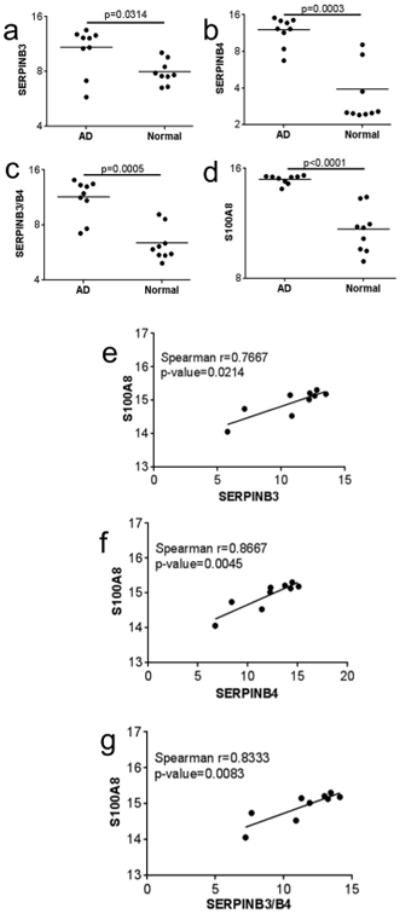
Expression of (a) SERPINB3 (probe 209719_x_at), (b) SERPINB4 (probe 211906_s_at), or both (c) SERPINB3/B4 (probe 210413_x_at) and (d) S100A8 (probe 202917_s_at) in the skin of subjects with AD (AD) and normal control skin (Normal) derived from GSE16161 expression array data (GUTTMAN YASSKY REF). Correlation plots between SERPINB3 and S100A8 (e), SERPINB4 and S100A8 (f) and SERPINB3/B4 and S100A8 (g).
RNAseq analysis shows differential early expression of pro-inflammatory genes between WT and Serpinb3a null mice
Based on our observations, it is clear that Serpinb3a acts early in response to cutaneous allergen exposure. To identify very early changes caused by Serpinb3a, we used RNAseq followed by Ingenuity Pathways Analysis (IPA) to study global changes in gene expression in WT and Serpinb3a KO mice following a single application of allergen. For both genotypes, we identified genes whose expression was significantly induced or repressed in the ASP treated group compared to SAL controls, filtered the lists to those with a 2.5 fold or greater change (increase or decrease), and performed IPA analysis independently for the WT and KO mice. In the WT mice, the primary network of genes regulated by ASP identified by IPA was related to hematological system development and function, tissue morphology, and cellular movement. This included upregulation of the pro-inflammatory cytokines (IL-4, EBI3) and chemokines (CXCL13, CCL24, CCL25), downregulation of an inhibitor of T cell apoptosis (NR4A1) , and differential expression of regulators of development and differentiation (CTLA2α, INHBB, CSDC2, ALPNR), and metabolism (KHK, NR1H4) (Figure 6A). IPA analysis also identified inflammation and immune responses as the key processes affected in the WT mice following allergen exposure. When we examined expression of the genes in this same network in the Serpinb3a KO mice, only a small fraction were differentially expressed (Table I).
Figure 6. Primary network of ASP affected genes generated by Ingenuity Pathways Analysis (IPA).
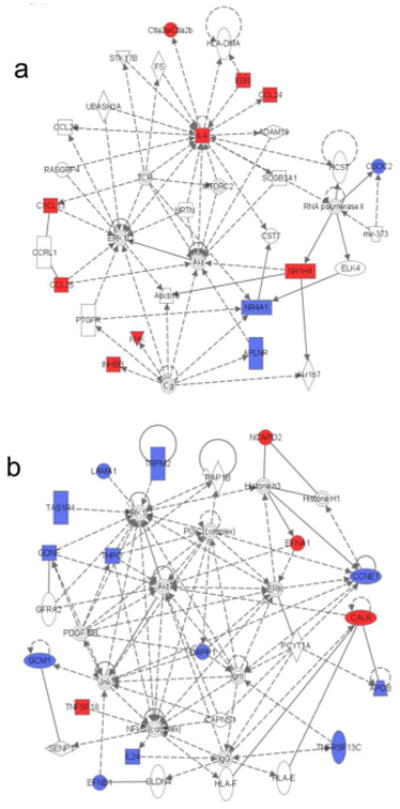
A list of differentially expressed genes (≥2.5 fold up- or down-regulated) when comparing ASP patched to SAL patched mice was generated for WT mice (a) and Serpinb3a KO mice (b) separately. Each list of genes was submitted independently subjected to IPA to identify the top networks affected in the WT and Serpinb3aKO mice. Genes marked in in red were upregulated while genes in blue were down-regulated in the ASP-patched mice when compared to SAL-patched mice.
Table I. IPA Analysis of Differentially Expressed Genes.
| Name | P-value | # in WT* | # in KO* |
|---|---|---|---|
| Diseases and Disorders | |||
| Inflammatory Response | 6e-5-0.05 | 14 | 2 |
| Gastrointestinal Disease | 0.001-0.05 | 7 | 1 |
| Hematological Disease | 0.001-0.05 | 5 | 1 |
| Immunological Disease | 0.001-0.05 | 10 | 1 |
| Inflammatory Disease | 0.001-0.05 | 9 | 1 |
| Physiological System Development and Function | |||
| Hematological System Development and Function | 6e-5-0.05 | 11 | 2 |
| Humoral Immune Response | 6e-5-0.04 | 3 | 2 |
| Immune Cell Trafficking | 6e-5-0.04 | 6 | 2 |
| Cell Mediated Immune Response | 6e-5-0.05 | 7 | 2 |
| Organ Morphology | 0.001-0.05 | 10 | 1 |
# in WT and # in KO refer to the number of genes in each category that were differentially expressed in either the WT or KO mice following ASP challenge. P-values are for the significance of association with the indicated disease/disorder in the WT mice.
In contrast, the primary network identified by IPA in the Serpinb3a KO mice included genes regulating cell morphology, cellular assembly and organization, and cellular development. They contained genes that regulate or are regulated by MAPK, PI3K, NFκB, or AKT signaling pathways (Figure 6B) that have all been implicated in mediating allergic responses (Choi et al., 2013; Sharon et al., 2011). These include genes that regulate T cell survival and signaling (TNFSF18, DAPP1), B cell survival (TNFSF13C), neutrophil infiltration and activation (TRPM2, IL24), along with a variety of genes involved in development (LAMA1, GDNF, GCM1, EFNB1, THPO, TAS1R1, EFNA1, CALR) and cell cycle (CCNE1, NCAPD2). It is important to highlight that expression of most of the pro-inflammatory genes is downregulated in the absence of Serpinb3a. Since the differences in expression of signaling pathway proteins was observed after a single patch of allergen, this observation, combined with the increased expression of pro-inflammatory genes in the WT mice supports a role for Serpinb3a in early inflammatory responses in the skin following allergen exposure.
Discussion
In this study, using an approach that integrates mouse models of atopic dermatitis and in vitro knockdown studies in human keratinocytes with analysis of human disease expression data, we have identified a role for the serine protease inhibitor Serpinb3a following allergen exposure. Serpinb3a regulates expression of the pro-inflammatory gene S100A8, with a significant impact on barrier function as assessed by TEWL. Collectively, our data reveal that human SERPINB3/B4 (Serpinb3a in mice) contributes to the cutaneous inflammatory response during the pathogenesis of more chronic skin diseases.
In a mouse model of asthma, we have previously shown that Serpinb3a KO mice have attenuated AHR and goblet cell hyperplasia. In this model, there was very little impact of Serpinb3a on inflammation after 3 weeks of exposure to dust mite. However Serpinb3a deficient mice treated with IL-13 alone for one week showed attenuated inflammation in addition to attenuated AHR and mucus production (Sivaprasad et al., 2011). Similar to our current findings in the skin, these data suggest that Serpinb3a promotes inflammation early in the pathogenesis of allergic disease. With continued allergen exposure, AD developed even in the absence of Serpinb3a indicating that other compensatory pathways are activated to restore the full inflammatory response. This is further supported by our RNAseq data, which revealed that expression of genes in several pro-inflammatory signaling pathways induced early during allergic responses is attenuated in the absence of Serpinb3a, consistent with attenuated skin inflammation phenotype. SERPINB3/B4 has been implicated in these pro-inflammatory responses previously. Transfection of human epidermal keratinocytes with SERPINB4 was sufficient to induce expression of IL-1alpha (Titapiwatanakun et al., 2005), which is known to induce expression of S100A8/A9 in a p38 MAPK dependent manner (Bando et al., 2013). Our microarray data did not detect increased expression of S100A8 in the WT mice. This is likely because the time point chosen for the RNAseq (with a view to identifying very early changes that drive the decreased expression of S100A8) was too short to see any induction.
The role of IL-4 in regulating TEWL and barrier function has been well established (Brandt and Sivaprasad, 2011; Sehra et al., 2010). Our RNAseq data in the WT mice also suggest that the more robust increase in TEWL in the WT mice is driven by an early increase in pro-TH2 responses.
Numerous studies have shown that SERPINB3/B4 protect cells from apoptosis in response to various triggers (de Koning et al., 2011; Katagiri et al., 2006; Murakami et al., 2001; Song et al., 2012; Suminami et al., 2000). None of our studies, either in vitro or in vivo, have identified an obvious impact on apoptosis in the absence of Serpinb3a/SERPINB3/B4. One explanation is that ASP extract may not induce similar responses as DNA damaging triggers (UV, gamma radiation), or TNFα. SERPINB3 has been shown to have a protective effect in lysosomal injury-induced necrosis and an enhancing effect on ER stress-induced apoptosis (Ullman et al., 2011). How exactly these SERPINB3 regulated processes contribute to the development and/or persistence of AD remains to be evaluated.
Examination of the RNAseq data for mechanistic explanation provides few answers. IL-24 is known to increase skin inflammation induced by TNF in the absence of NFκB activity (Kumari et al., 2013). TRPM2 is a Ca2+ permeable cation channel that is required for T cell proliferation and pro-inflammatory cytokine secretion as well as other phenotypes in an experimental model of autoimmune encephalomyelitis (EAE). It would be interesting to speculate that downregulation of one or both of these molecules could explain the attenuated inflammation in the absence of Serpinb3a. However the relevance of these findings remains to be established.
Serine proteases are important regulators of the balance between basal cell proliferation and desquamation to form the stratum corneum of the skin (Cork et al., 2009). One possible role for Serpinb3a in the skin is to modulate the function of one of the proteases involved in this process. Thus, it was surprising to us that the deletion of a serine protease inhibitor (implying increased serine protease activity in the skin) did not result in an exacerbation of the disease phenotype. In our studies, histological comparison of skin sections of WT and Serpinb3a null mice revealed no significant differences in gross skin architecture, however, subtle differences in epidermal structure cannot be excluded.
Recent genetic studies have identified single nucleotide polymorphisms (SNPs) in SERPINB3/B4. A single nucleotide polymorphism in SERPINB3 that increased the protease inhibitory function of the protein was associated with liver cirrhosis (Turato et al., 2009). Another study identified a functional SNP in the SERPINB3 promoter associated with skin prick test positivity in asthmatic children that impacted binding of the transcription factors GATA2 and GATA3 after IL-4 and IL-13 treatment (Karaaslan et al., 2013). The impact of increased expression or activity of a protease inhibitor resulting in worse disease would suggest that the serine protease targeted by SERPINB3 would serve an anti-inflammatory role in the skin. This hypothesis is consistent with our finding of attenuated inflammation in the skin following depletion of Serpinb3a. Studies in our laboratory are currently ongoing to determine the association of SNPs in the SERPINB3/B4 locus with atopic dermatitis.
Current treatments for atopic dermatitis are focused mostly on skin hydration and topical corticosteroids. Better understanding of the molecular basis for skin inflammation will facilitate new drug development (Schakel et al., 2014). Our studies have identified one such pathway, driven by SERPINB3/B4. It is tempting to speculate that early intervention with treatment that attenuates SERPINB3/B4 expression or activity may be beneficial for primary prevention, attenuation, or delayed onset of chronic skin diseases like AD. This pathway might also serve as an important predictor of the natural history of AD. Alternatively, studies could examine the contribution of this pathway to sensitivity to the number and/or type of AD triggers.
Materials and Methods
Mice
Serpinb3a null mice (Sivaprasad et al., 2011) and BALB/c mice (Harlan Laboratories, Indianapolis, IN) were kept in a pathogen-free environment. All procedures were performed in accordance with the ethical guidelines in the Guide for the Care and Use of Laboratory Animals of the Institutional Animal Care and Use Committee approved by the Veterinary Services Department of the Cincinnati Children's Hospital Medical Center (CCHMC).
Epicutaneous (EC) antigen sensitization and challenge
The conventional cutaneous inflammation protocol has been previously described (Sivaprasad et al., 2010). Briefly, for both models (summarized in Figure 1A and 2A respectively), the mice were shaved and 24 hours later epicutaneously treated with 200 μg of Aspergillus fumigatus (ASP) extract (Greer Laboratories, Lenior, NC), in 200 μl of sterile saline or saline alone as control. The allergen was applied to a 2 cm2 gauze patch and taped to the back of the mice with tagaderm, a Band Aid, and water resistant tape. For the long model, the patch was removed on day 6 and 24 hours later a new patch re-applied, and repeated one more time, for a total of three patches. For the short protocol the patch was removed on day 3 and the patch re-applied 24 hours later, and repeated one more time, for a total of three patches. TEWL was measured 24 hours after the second and third patches. 24 hours after the last patch the mice were euthanized following standard procedures.
Measurement of TEWL
TEWL was measured by using the Dermalab instrument DermaLab USB module (Cortex Technology, Hadsund, Denmark) as previous described (Sivaprasad et al., 2010). An average of the two readings was used for each mouse and all measurements were made by the same investigator.
Skin scoring system
Mice were visually assessed for excoriations, erythema and skin thickening in the 2 cm2 area covered by the patch as previously described (Sivaprasad et al., 2010).
Tissue processing and measurement of epidermal thickness
Skin tissues were fixed in 10% formalin, cut into 5 μm sections and stained with hematoxylin and eosin (H&E) per the manufacturer's protocol. Epidermal thickness was measured by first capturing 8 images per section at 100× magnification with a Nikon (Melville, NY) 80i microscope, and then taking 8 measurements per image using the Nikon Elements software program. Data are plotted as the average of the 64 measurements per section for each mouse. CD3 staining was performed by the Pathology Core at CCHMC.
ELISAs
Plasma was diluted 1:10 for ASP Specific IgE and IgG1, 1:500 for total IgE, and 1:2 for IL-4 and IL-17. ELISAs for were performed using the appropriate kit from BD Biosciences (IgG1 and IgE) or Biolegend (IL-4 and IL-17A). For ASP-specific IgE and IgG1, wells were coated with 0.01% ASP extract and the rest of the protocol was carried out as recommended in the kits.
Generation of stable shRNA clones
SERPINB4 shRNA Lentiviral clones TRCN52408 and TRCN52409 of the MISSION shRNA library (Sigma Aldrich, St. Louis, MO) were purchased from the Lenti-shRNA Library Core at Cincinnati Children's Hospital Medical Center. HaCaT cells (kindly provided by Dr. T. Bowden, University of Arizona, Tuscon, AZ) were transduced using standard methods (Woods et al., 2009). 72 hours post-transduction the cells were exposed to puromycin 5ug/mL (Sigma Aldrich). Cells that continued to grow in puromycin containing medium were passaged to generate clonal cell populations by serial dilution. Once confluent, cells were harvested and RNA isolated as described below.
RT-PCR
Total RNA was isolated from cells using Trizol (Invitrogen) according to manufacturer's instructions and DNase treated (Qiagen, Valencia, CA) before being reverse transcribed with First Strand Superscript Synthesis kit (Invitrogen). Quantitative PCR analysis was performed using the following primers: (1) Mouse HPRT – Forward 5′- TGCCGAGGATTTGGAAAAAG-3′; Reverse 5′-CCCCCCTTGAGCACACAG; (2) MouseS100A8 – Forward 5′-CCATGCCCTCTACAAGAATG-3′; Reverse 5′- ATCACCATCGCAAGGAACTC-3′; (3) Mouse Serpinb3a – Forward 5′-CAGATGATGAAACAAAACATCG-3′; Reverse 5′-AGACCTTGAGTGCTGCTCATA-3′; (4) Human GAPDH – Forward 5′-GGGGAAGGTGAAGGTCGGAGTCA-3′; Reverse 5′-AGCCTTGACGGTGCCATGGAAT-3′; (5) Human S100A8 – Forward 5′-ATGTTGACCGAGCTGGAGAAAG-3′; Reverse 5′-TTCAGGTCATCCCTGTAGACGG-3′; (6) Human SERPINB4 – Forward 5′-GTCGATTTACACTTACCTCGG-3′; Reverse 5′-GCCTTGTGTAGGACTTTAGATACT-3′.
Human Expression Data
Human expression data from Guttman-Yassky et al (Guttman-Yassky et al., 2009) (GSE16161) were imported into GeneSpring GX software (Agilent Technologies, Santa Clara, CA), and the intensities for each chip were normalized relative to the median of the control samples.
RNA-Seq Expression Analysis
RNA was extracted from mouse skin using the Trizol reagent. 2.5μg RNA from three mice from each treatment group (WT-SAL, WT-ASP, KO-SAL, KO-ASP) were used for RNA-Seq analysis. RNA-Seq was performed by Genomics Sequencing Core in the University of Cincinnati. The RNA-Seq library was constructed by using PrepX SPIA RNA-Seq kit (IntegenX, Pleasanton, CA) and Apollo 324 NGS Library Prep System (IntegenX), 10 ng of total RNA was converted into cDNA suitable for mRNA sequencing. The cDNA was then sheared by Covaris S2 (Covaris, Woburn, MA) under the condition recommended by IntegenX, follow by Bioanalyzer assay of the size distribution with Agilent High Sensitivity DNA kit (Agilent, Santa Clara, CA). The properly sheared cDNA fragments were purified by Agencourt AMPure XP meganetic beads (Beckman Coulter, Brea CA). Using IntegenX PrepX ILM DNA library kit for Illumina and Apollo 324 NGS Library Prep System, 500 ng purified cDNA fragments were then go through end repair, addition of a single ‘A’ base, ligation of adapters and indexed individually. The products were purified and enriched by PCR to create the final cDNA library targeting mRNAs. The size of the generated library was validated by Bioanalyzer, and the library was quantified by using Kapa Library Quantification kit (Kapabiosystem, Woburn, MA). Twelve individually indexed cDNA libraries were equal amount pooled for clustering in cBot system (Illumina). Libraries at the concentration of 6.5 pM were clustered onto a flow cell using Illumina's TruSeq SR Cluster Kit v3, and sequenced for 50 cycles using TruSeq SBS kit on Illumina HiSeq system.
Sequence reads were aligned to the reference mouse genome (mm10) using TopHat aligner (Trapnell et al., 2009). The counts of reads aligning to each gene's coding region were summarized using ShortRead (Morgan et al., 2009) Bioconductor package and custom-written R programs (Ihaka and Gentleman, 1996). Statistical analysis to identify genes differentially expressed between ASP and SAL treated WT and KO mice was performed using the negative-binomial model of read counts as implemented in DESeq Biocondoctor package(Anders and Huber, 2010). Transcripts with 2.5 fold change and p-value<0.05 were considered differentially expressed and were further analyzed by Ingenuity Pathways Analysis (IPA). The GEO accession number is GSE54640.
IPA Analysis
Data were analyzed through the use of QIAGEN's Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity).The biological functions and/or diseases that were most significant were identified, using the list of differentially regulated genes and the Ingenuity Pathways Knowledge Base. Fisher's exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to the list is due to chance alone. The list of 53 genes was overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base and networks then algorithmically generated based on connectivity.
Statistical analysis
Reported values are expressed as mean ± SD. All experiments were done with at least 6-8 mice per group. Each dot on the graphs represents an independent mouse. Data shown are representative of three independent replicates. All statistical analysis was done using PRISM software (GraphPad Software Inc., La Jolla, CA). For all studies statistical significance was assessed using one-way ANOVA followed by a Tukey-Kramer posttest (for statistical significance between groups). Significance was set at a p value of 0.05. Correlation analysis was performed using a Spearman correlation test. For the human expression data, statistical significance was determined using the Mann-Whitney test.
Acknowledgments
The authors would like to thank all contributing authors to “Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis” (Guttman-Yassky, E., et al, 2009) for making the data publicly available.
Supported by NIH 2U19AI70235 (G.K.K.H); P30ES06096 and UL1 RR026314 (M.M)
Abbreviations
- AD
Atopic Dermatitis
- TEWL
Transepidermal Water Loss
- WT
Wild Type
- KO
Knockout
- SAL
Saline
- ASP
Aspergillus fumigatus extract
Footnotes
Conflict of Interest: All authors declare they have no conflict of interest.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando M, Zou X, Hiroshima Y, et al. Mechanism of interleukin-1alpha transcriptional regulation of S100A9 in a human epidermal keratinocyte cell line. Biochimica et biophysica acta. 2013;1829:954–62. doi: 10.1016/j.bbagrm.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt EB, Kovacic MB, Lee GB, et al. Diesel exhaust particle induction of IL-17A contributes to severe asthma. The Journal of allergy and clinical immunology. 2013;132:1194–204 e2. doi: 10.1016/j.jaci.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt EB, Sivaprasad U. Th2 Cytokines and Atopic Dermatitis. Journal of clinical & cellular immunology. 2011;2 doi: 10.4172/2155-9899.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccardo CJ, Mahaffey SB, Strand M, et al. Peeling off the layers: skin taping and a novel proteomics approach to study atopic dermatitis. The Journal of allergy and clinical immunology. 2009;124:1113-5 e1–11. doi: 10.1016/j.jaci.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YH, Jin GY, Li LC, et al. Inhibition of protein kinase C delta attenuates allergic airway inflammation through suppression of PI3K/Akt/mTOR/HIF-1 alpha/VEGF pathway. PloS one. 2013;8:e81773. doi: 10.1371/journal.pone.0081773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cork MJ, Danby SG, Vasilopoulos Y, et al. Epidermal barrier dysfunction in atopic dermatitis. The Journal of investigative dermatology. 2009;129:1892–908. doi: 10.1038/jid.2009.133. [DOI] [PubMed] [Google Scholar]
- de Koning PJ, Kummer JA, de Poot SA, et al. Intracellular serine protease inhibitor SERPINB4 inhibits granzyme M-induced cell death. PloS one. 2011;6:e22645. doi: 10.1371/journal.pone.0022645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming SA, Koch M, Krieger A, et al. Differential proteomic analysis distinguishes tissue repair biomarker signatures in wound exudates obtained from normal healing and chronic wounds. Journal of proteome research. 2010;9:4758–66. doi: 10.1021/pr100456d. [DOI] [PubMed] [Google Scholar]
- Gittler JK, Shemer A, Suarez-Farinas M, et al. Progressive activation of T(H)2/T(H)22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. The Journal of allergy and clinical immunology. 2012;130:1344–54. doi: 10.1016/j.jaci.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guajardo JR, Schleifer KW, Daines MO, et al. Altered gene expression profiles in nasal respiratory epithelium reflect stable versus acute childhood asthma. The Journal of allergy and clinical immunology. 2005;115:243–51. doi: 10.1016/j.jaci.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Gupta J, Grube E, Ericksen MB, et al. Intrinsically defective skin barrier function in children with atopic dermatitis correlates with disease severity. The Journal of allergy and clinical immunology. 2008;121:725–30 e2. doi: 10.1016/j.jaci.2007.12.1161. [DOI] [PubMed] [Google Scholar]
- Guttman-Yassky E, Suarez-Farinas M, Chiricozzi A, et al. Broad defects in epidermal cornification in atopic dermatitis identified through genomic analysis. The Journal of allergy and clinical immunology. 2009;124:1235–44 e58. doi: 10.1016/j.jaci.2009.09.031. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Kiyoshima T, Matsuo K, et al. Effect of SCCA1 and SCCA2 on the suppression of TNF-alpha-induced cell death by impeding the release of mitochondrial cytochrome c in an oral squamous cell carcinoma cell line. Tumor Biol. 2005;26:165–72. doi: 10.1159/000086949. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: A language for data analysis and graphics. Journal of computational and graphical statistics. 1996;5:299–314. [Google Scholar]
- Karaaslan C, Birben E, Keskin O, et al. The role of SCCA1 in asthma related physiological events in the airway epithelium and the effect of promoter variants on asthma and gene function. Respiratory medicine. 2013;107:368–79. doi: 10.1016/j.rmed.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Katagiri C, Iida T, Nakanishi J, et al. Up-regulation of serpin SCCA1 is associated with epidermal barrier disruption. Journal of dermatological science. 2010;57:95–101. doi: 10.1016/j.jdermsci.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Katagiri C, Nakanishi J, Kadoya K, et al. Serpin squamous cell carcinoma antigen inhibits UV-induced apoptosis via suppression of c-JUN NH2-terminal kinase. The Journal of cell biology. 2006;172:983–90. doi: 10.1083/jcb.200508064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima H, Nishimata S, Kashiwagi Y, et al. Squamous cell carcinoma-related antigen in children with atopic dermatitis. Pediatrics International. 2000;42:448–50. doi: 10.1046/j.1442-200x.2000.01253.x. [DOI] [PubMed] [Google Scholar]
- Kumari S, Bonnet MC, Ulvmar MH, et al. Tumor necrosis factor receptor signaling in keratinocytes triggers interleukin-24-dependent psoriasis-like skin inflammation in mice. Immunity. 2013;39:899–911. doi: 10.1016/j.immuni.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Lu ZR, Park D, Lee KA, et al. Profiling the dysregulated genes of keratinocytes in atopic dermatitis patients: cDNA microarray and interactomic analyses. Journal of dermatological science. 2009a;54:126–9. doi: 10.1016/j.jdermsci.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Lu ZR, Park TH, Lee ES, et al. Dysregulated genes of extrinsic type of atopic dermatitis: 34K microarray and interactomic analyses. Journal of dermatological science. 2009b;53:146–50. doi: 10.1016/j.jdermsci.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Mitsuishi K, Nakamura T, Sakata Y, et al. The squamous cell carcinoma antigens as relevant biomarkers of atopic dermatitis. Clin Exp Allergy. 2005;35:1327–33. doi: 10.1111/j.1365-2222.2005.02353.x. [DOI] [PubMed] [Google Scholar]
- Morgan M, Anders S, Lawrence M, et al. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–8. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami A, Suminami Y, Hirakawa H, et al. Squamous cell carcinoma antigen suppresses radiation-induced cell death. British J Cancer. 2001;84:851–58. doi: 10.1054/bjoc.2000.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva M, He Z, Kallberg E, et al. Human S100A9 protein is stabilized by inflammatory stimuli via the formation of proteolytically-resistant homodimers. PloS one. 2013;8:e61832. doi: 10.1371/journal.pone.0061832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saaf AM, Tengvall-Linder M, Chang HY, et al. Global expression profiling in atopic eczema reveals reciprocal expression of inflammatory and lipid genes. PloS one. 2008;3:e4017. doi: 10.1371/journal.pone.0004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schakel K, Dobel T, Bosselmann I. Future treatment options for atopic dermatitis - Small molecules and beyond. Journal of dermatological science. 2014;73:91–100. doi: 10.1016/j.jdermsci.2013.11.009. [DOI] [PubMed] [Google Scholar]
- Sehra S, Yao Y, Howell MD, et al. IL-4 regulates skin homeostasis and the predisposition toward allergic skin inflammation. Journal of immunology. 2010;184:3186–90. doi: 10.4049/jimmunol.0901860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon H, Amar D, Levdansky E, et al. PrtT-regulated proteins secreted by Aspergillus fumigatus activate MAPK signaling in exposed A549 lung cells leading to necrotic cell death. PloS one. 2011;6:e17509. doi: 10.1371/journal.pone.0017509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaprasad U, Askew DJ, Ericksen MB, et al. A nonredundant role for mouse Serpinb3a in the induction of mucus production in asthma. The Journal of allergy and clinical immunology. 2011;127:254–61. 61 e1–6. doi: 10.1016/j.jaci.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaprasad U, Warrier MR, Gibson AM, et al. IL-13Ralpha2 has a protective role in a mouse model of cutaneous inflammation. Journal of immunology. 2010;185:6802–8. doi: 10.4049/jimmunol.1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song KJ, Ahn HJ, Nam HW. Anti-apoptotic effects of SERPIN B3 and B4 via STAT6 activation in macrophages after infection with Toxoplasma gondii. The Korean journal of parasitology. 2012;50:1–6. doi: 10.3347/kjp.2012.50.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Farinas M, Lowes MA, Zaba LC, et al. Evaluation of the psoriasis transcriptome across different studies by gene set enrichment analysis (GSEA) PloS one. 2010;5:e10247. doi: 10.1371/journal.pone.0010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suminami Y, Nagashima S, Vujanovic NL, et al. Inhibition of apoptosis in human tumour cells by the tumour-associated serpin, SCC antigen-1. British journal of cancer. 2000;82:981–9. doi: 10.1054/bjoc.1999.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Higuchi D, Takahashi T, et al. Overexpression of serpin squamous cell carcinoma antigens in psoriatic skin. The Journal of investigative dermatology. 2002;118:147–54. doi: 10.1046/j.0022-202x.2001.01610.x. [DOI] [PubMed] [Google Scholar]
- Tian S, Krueger JG, Li K, et al. Meta-analysis derived (MAD) transcriptome of psoriasis defines the “core” pathogenesis of disease. PloS one. 2012;7:e44274. doi: 10.1371/journal.pone.0044274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titapiwatanakun B, Miyahira Y, Mayuzumi N, et al. SCCA2-transfected human keratinocytes show increased secretion of IL-1alpha and IL-6, but not of TNF-alpha. Archives of dermatological research. 2005;297:274–7. doi: 10.1007/s00403-005-0609-1. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turato C, Ruvoletto MG, Biasiolo A, et al. Squamous cell carcinoma antigen-1 (SERPINB3) polymorphism in chronic liver disease. Digestive and liver disease : official journal of the Italian Society of Gastroenterology and the Italian Association for the Study of the Liver. 2009;41:212–6. doi: 10.1016/j.dld.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Ullman E, Pan JA, Zong WX. Squamous cell carcinoma antigen 1 promotes caspase-8-mediated apoptosis in response to endoplasmic reticulum stress while inhibiting necrosis induced by lysosomal injury. Molecular and cellular biology. 2011;31:2902–19. doi: 10.1128/MCB.05452-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CG, Fu J, Xue P, et al. Dose-dependent transitions in Nrf2-mediated adaptive response and related stress responses to hypochlorous acid in mouse macrophages. Toxicology and applied pharmacology. 2009;238:27–36. doi: 10.1016/j.taap.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane Y, Moriyama K, Yasuda C, et al. New horny layer marker proteins for evaluating skin condition in atopic dermatitis. International archives of allergy and immunology. 2009;150:89–101. doi: 10.1159/000210385. [DOI] [PubMed] [Google Scholar]


