Abstract
Induction of cell autonomous apoptosis following oncogene-induced overproliferation is a major tumor-suppressive mechanism in vertebrates. However the detailed mechanism mediating this process remains enigmatic. In this study we demonstrate that dMyc-induced cell-autonomous apoptosis in the fruit fly Drosophila melanogaster relies on an intergenic sequence termed the IRER (Irradiation Responsive Enhancer Region). The IRER mediates expression of surrounding pro-apoptotic genes, and we use an in vivo reporter of the IRER chromatin state to gather evidence that epigenetic control of DNA accessibility within the IRER is an important determinant of the strength of this response to excess dMyc. In prior work we showed that the IRER also mediates P53-dependent induction of pro-apoptotic genes following DNA damage, and the chromatin conformation within IRER is regulated by Polycomb group-mediated histone modifications. dMyc-induced apoptosis and the P53-mediated DNA damage response thus overlap in a requirement for the IRER. The epigenetic mechanisms controlling IRER accessibility appear to set thresholds for the P53 and dMyc-induced expression of apoptotic genes in vivo and may have a profound impact on cellular sensitivity to oncogene-induced stress.
Keywords: Apoptosis, Overproliferation, DNA accessibility, reaper, Epigenetics
Introduction
Cell-autonomous apoptosis following Myc over-expression has long been regarded as a major tumor-suppression mechanism (1). Suppression of apoptosis, by means such as overexpression of Bcl-2, is required to unleash the tumorigenic potential of Myc in mammalian cells (2-4). The propensity of cells with elevated levels of Myc to undergo apoptosis depends on the availability of growth factors such as IGF (insulin-like growth factors) (5). It has been postulated that Myc-induced cell-autonomous apoptosis reflects a fundamental mechanism that maintains tissue homeostasis by inducing apoptosis when overproliferation is sensed. However, the mechanistic details of this apoptotic program remain enigmatic.
One extensively studied pathway implicated in Myc-induced cell autonomous apoptosis is the P53–mediated activation of pro-apoptotic genes and/or suppression of anti-apoptotic genes (6). Many important pro-apoptotic genes, such as apaf-1 and caspase-9, have been implicated in Myc-induced apoptosis in cell culture systems. However, study of animal models suggests that Myc-induced cell death can proceed in the absence of apaf-1 or caspase-9 (7). It is also clear that at least under some circumstances, Myc-induced cell death can proceed without the participation of P53 (8).
Despite its enigmatic nature, the mechanism of Myc-induced apoptosis appears to be highly conserved. Overexpression of the only Myc ortholog in the fruit fly Drosophila melanogaster, dMyc, also induces cell autonomous apoptosis (9, 10). Intriguingly, while the level of dP53 (Drosophila ortholog of mammalian tumor suppresser P53) mRNA is significantly increased following dMyc expression, the function of dP53 appears to be dispensable for dMyc-induced cell death (9). Ectopic expression of dMyc leads to increased cell size but fails to result in significant hyperplasia on its own (11). Moderate tissue overgrowth has only been observed after dMyc-induced apoptosis is blocked by co-expression of the viral caspase inhibitor P35 (9). This indicates that a blockade of apoptosis is required for dMyc-induced hyperplasia in Drosophila, which is similar to what is observed in mammalian c-Myc tumorigenesis models. In this study, we show that the induction of apoptosis following dMyc-induced overproliferation requires a highly conserved intergenic regulatory control region in the RHG (reaper, hid, and grim) genomic block.
A ~ 33kb intergenic region in the RHG block was defined as the IRER (irradiation responsive enhancer region) based on a requirement to mediate induction of reaper, hid, and sickle expression following ionizing irradiation in embryos (12). This region contains a previously identified response element for Drosophila P53 (13). Interestingly, the epigenetic status of this region undergoes a dramatic change at embryonic development stage 12, when most cells enter into post-mitotic differentiation. During this transition, the region becomes enriched for H3K27me3 and H3K9me3, and is bound by Polycomb group (PcG) proteins as well as Heterochromatin Protein 1 (HP1). Consequently, the DNA in this region becomes as inaccessible to DNase I as pericentromeric heterochromatin region. This epigenetic blocking of the IRER renders the three pro-apoptotic genes unresponsive to ionizing irradiation while other branches of the DNA damage response, such as the DNA repair pathway, remain active (12).
Here we report that the IRER is required to limit cell numbers of several organs during development, and that the functional significance of this regulatory control region in apoptosis is heightened in the context of oncogenic stress. While overexpression of dMyc in wild type animals failed to induce hyperplasia, significant overproliferation was observed in animals lacking the regulatory region IRER, indicating that the IRER is essential for the induction of apoptosis associated with oncogene-induced overproliferation. In addition, we found that cells with relatively open IRER are more sensitive to dMyc-induced cell autonomous apoptosis than those with relatively closed IRER, suggesting epigenetic regulation plays an important role in determining the cellular sensitivity to oncogenic stress.
Results
IRER mediates DNA-damage-induced pro-apoptotic gene expression in post-embryonic tissues
Our previous work revealed that IRER is strictly required for mediating irradiation-induced reaper, hid, and sickle expression in embryos before stage 12 (12) (Fig. 1A). In embryos deficient for this intergenic region (i.e. homozygotes of Df(IRER)), the transcriptional response of the three pro-apoptotic genes to DNA damage is fully blocked. While the irradiation responsiveness of the three pro-apoptotic genes is robust in stage 9-11 wild-type embryos, it is diminished in most cells in embryos post developmental stage 12. This sensitive-to-resistant transition is due to targeted epigenetic regulation of the IRER that requires the function of Polycomb group (PcG) proteins and histone deacetylase (HDAC) (12).
Figure 1. IRER mediates DNA damage –induced pro-apoptotic response in the developing wing discs.
(A) The four pro-apoptotic RHG genes (reaper, hid, grim, and sickle) are clustered in a ~ 280 kb genomic region. They are transcribed to the same direction. The two intergenic regions surrounding reaper are unusually long (99 kb and 40kb) and enriched for HCNEs (Highly conserved non-coding elements). Regulatory regions, such as NBRR (Neuroblast Regulatory Region) (26) or IRER (12), controls the expression of multiple RHG genes during development or in response to stress, respectively. The regions deleted in deficiency strains used in this study are indicated by dashed lines. The region cloned into the BAC-IRER rescue construct is indicated by the red bar on top of the DNA. The figure is not drawn to scale due to space limitation. Nucleotide coordinates are relative to the reaper TSS and based on Drosophila genome release 5.42. (B-E) Caspase 3 staining of wing discs without ionizing irradiation (IR) (B and D), or at 4 hours after 40Gy of IR (C and E). Irradiation induced apoptosis in wild-type wing discs (compare C with B) but not in Df(IRER) discs (compare E with D). Scale bar = 50μm. (F) Time course of irradiation-induced reaper expression from whole larvae. Early third instar larvae were irradiated with 40Gy of IR and collected for quantitative RTPCR (qRT-PCR) at various times after IR. The induction level of reaper, i.e. the ratio of IR vs. control (NT), appeared to be the highest between 1 and 2 hour after IR in both wild-type and Df(IRER) larvae. (G) Both reaper and hid transcripts were induced from wild-type wing discs at 1.5~2 hours following IR as shown by qRT-PCR. The magnitude of induction was significantly smaller in Df(IRER) wing discs. For all the qRT-PCR data, means + S.D. are shown (n=3).
Exposure of Drosophila larvae to irradiation induces rapid and wide spread apoptosis in imaginal discs that is dependent on the function of P53 (14). To test whether IRER is required for DNA damage-induced cell death in post-embryonic tissues, we measured irradiation-induced caspase activation and pro-apoptotic gene expression in imaginal discs from Df(IRER) larvae. We subjected third instar larvae to similar treatment (i.e. 40Gy of γ-ray) and found that at 4 hours post irradiation, there was indeed a significant increase of apoptosis in the wild type wing discs, preferentially at the wing pouch (Fig. 1B vs. C). In sharp contrast, there was little detectable increase of caspase activation in discs from animals homozygous to Df(IRER) (Fig. 1D vs. E).
We then measured the mRNA levels of the RHG genes by quantitative PCR. In a time course analysis, we found that the induction of reaper and hid in wild type larvae was highest between 1 and 2 hours following irradiation (Fig. 1F). At this time point, irradiation-induced expression of reaper and hid was significantly lower in Df(IRER) wing discs but not absent as it is in Df(IRER) embryos (Fig. 1G). Levels of sickle and grim mRNAs in these wing discs remained barely detectable even after irradiation. The observation that irradiation-induced expression of reaper and hid is not completely blocked in Df(IRER) wing discs suggests that an alternate death mechanism exists in wing disc cells. It has been observed that a P53-indpendent mechanism also contributes to irradiation/DNA damage-induced cell death in the wing disc (15). In animals mutated for dp53 or chk2, such a mechanism could eventually compensate for the loss of dP53 or Chk2 function and induce apoptosis at a much later (12-16hr post irradiation) time point (15). It is possible that the incomplete block of reaper/hid induction in the wing discs reflects the existence of such a mechanism in wing disc cells. However, it is clear that the suppression of γ-ray-induced reaper/hid expression in Df(IRER) wing discs was sufficient to result in a significant decrease in the frequency of apoptotic cells (i.e. cells with activated caspase) at 4 hours post irradiation. Indeed, previous data from Brodsky's lab showed that heterozygosity at the hid locus was sufficient to suppress irradiation-induced death (14). Therefore, we conclude that the IRER is also required to mediate DNA damage-induced pro-apoptotic gene expression and cell death in post-embryonic tissue.
The cis regulatory function of IRER is required for tissue homeostasis and organ size control
While analyzing γ-ray-induced cell death in developing tissues, we noticed that discs from homozygous Df(IRER) larvae were consistently larger than those with wild-type IRER or Df(IRER) heterozygotes (Supplementary Fig. 1A-B & D-E). Statistical analysis revealed that both eye and wing imaginal discs from homozygous Df(IRER) larvae were significantly larger than those from heterozygous siblings (Supplementary Fig. 1C&F). Close inspection of these discs indicated that the cell size in the Df(IRER) discs, reflected both as the distance between DAPI-stained nuclei as well as density of cells within a given area, was not significantly different from cells in the heterozygous discs. Thus it appears that developing Df(IRER) imaginal discs contain more cells, and are consequently larger than normal discs.
To confirm that the observed increase in imaginal disc size was not due to changes in the timing of proliferation or developmental cell death, we evaluated the final size of adult wings that grow from these discs. About 10-20% homozygous Df(IRER) animals survive to adulthood. The wings of these Df(IRER) flies are significantly larger than those of wild type animals raised in parallel (Fig. 2A-A’). Measurement of the anterior and posterior compartments separately revealed that while both overgrew, the growth was relatively more pronounced in the posterior compartment (Supplementary Fig. 2A-B).
Figure2. The cis regulatory function of IRER is required for controlling organ size.
(A) A representative image of the wild-type wing, with five longitudinal veins labeled as L1 through L5. The squares indicate the areas sampled to measure hair number. (A’) Wings from IRER−/− adult flies (red) are larger than those from wild-type flies (black). Bar=100μm in (A) and (A’). (B) The density of cells was estimated by counting the number of trichomes from fixed areas on the wing blade. Four 75px2 - areas (squares in A) were selected for trichome counting (L1-L2, L2-L3, L4-L5 and L5). In the posterior compartment, i.e. L4-5 and L5, the cell density is higher in IRER−/− wings (p= 0.0022 and 0.0002, respectively; n=12). There is no significant difference in cell density between the two genotypes in the anterior compartment. (p= 0.0566 and 0.0783, for L1-2 and L2-3, respectively; n=12). (C) Quantification of adult wing sizes. As shown in A’, wings of Df(IRER) animals are larger than the wild-type controls (p = 2.6e-5). A similar wing size increase is observed with the transheterozygous Df(ED225)/Df(IRER) (p<0.0001 between ED225−/IRER-(n=20) and +/+ (n=25)), p=0.0581 between ED225−/IRER− and IRER−/− (n=26)). In contrast, the wing overgrown phenotype was partially rescued by Df(ED224) (p<0.0001 between ED224-/IRER− (n=27) and +/+, p=0.0236 between ED224−/IRER− and IRER−/−). The deletion regions in ED224 and ED225 are indicated in Fig 1A. (D) Introducing the BAC-IRER to the second chromosome failed to rescue the increased wing size of IRER−/− (p< 0.0001 between Bac-IRER, IRER−/− (n=30) and +/+, p= 0.2669 between Bac-IRER, IRER−/− and IRER−/−. (E) The basal levels expression of reaper and sickle are significantly reduced in IRER−/− larvae as measured by qRTPCR. (F) The expression of reaper is also decreased in the IRER−/− wing discs. The unpaired student t-test was used to calculate p-value.
Measurement of trichome (wing hair) densities provides additional evidence that the increased size of Df(IRER) adult wings is not due to increased cell size but rather to increased numbers of cells. Trichome density is slightly higher in enlarged Df(IRER) wings than in wild type wings, suggesting that Df(IRER) adult wing cells are in fact slightly smaller than wild type cells. The difference was statistically significant for the posterior compartments (reflected by increased hair density in L4-L5 and L5 regions marked in Fig. 2A-B). When corrected for this difference in cell density, wings from homozygous Df(IRER) animals have about 10% more cells than the wings of wild type animals.
To rule out the possibility that the cell number and organ size phenotype was due to potential background mutation(s) on the Df(IRER) chromosome, complementation tests were performed with well-defined deficiencies of the RHG region (Fig. 1A & 2C). The Df(ED224) and Df(ED225) deficiency chromosomes were generated by the DrosDel project with defined break points and in an isogenic background (16). Df(ED225) removes the IRER region, while Df(ED224) retains the IRER region and sickle but lacks the transcribed regions of reaper, grim and hid. Both deficiencies are homozygous lethal but viable in trans to Df(IRER), which allows assessment of adult wing phenotypes. The wing size of Df(IRER)/Df(ED225) animals is enlarged to a similar degree as Df(IRER) homozygotes (Fig. 2C). In contrast, the overgrown phenotype of the Df(IRER) allele was partially complemented by Df(ED224) (Fig. 2C). This result suggests that expression of sickle, controlled by IRER, may contribute to limiting wing size independent of reaper/hid. Alternatively, the normal copy of the IRER on the Df(ED224) chromosome may act in trans to regulate the expression of reaper and/or hid on the homologous chromosome. Since the genetic background of Df(ED225) is independent to that of Df(IRER), we conclude that the wing overgrowth phenotype is due to the lack of the genomic region containing IRER.
The effect of the IRER loss on organ size control is very likely due to altered regulation of proximal genes. There is no predicted coding or non-coding sequence in the IRER genomic region (Flybase.org), and no transcript with coding potential has been detected in this region by any RNA sequencing projects (based on data collected by Fly base or ModENCODE). To rule out the possibility that the phenotypes associated with the Df(IRER) genomic allele are due to a non-coding transcript within the IRER, we cloned the genomic region encompassing the IRER into a rescue construct (Fig. 1A). A BAC clone containing the genomic region Chromosome 3L:18391335–18432601, corresponding to the span from just before the start site of reaper ORF to the stop codon of sickle and including the complete IRER sequence (Fig. 1A), was used to make a transgenic fly via phiC31 integrase-mediated transformation. Animals carrying the reaper-sickle interval fragment appear to be normal without any noticeable change in viability or organ size and were referred to as “Bac-IRER” hereafter. When this chromosome was combined with the Df(IRER) chromosome, it failed to rescue the suppression of irradiation-induced reaper expression (Supplementary Fig. 3) and increased wing size associated with homozygosity for Df(IRER) (Fig. 2D). From this evidence, we conclude that the IRER does not encode any essential trans acting factors and that the cis regulatory function of IRER thus underlies the requirement for this region in tissue homeostasis.
To monitor how the lack of IRER cis regulatory function affects pro-apoptotic gene expression in development, we compared the expression of the four pro-apoptotic genes, reaper, hid, grim, and sickle, between wild-type and homozygous Df(IRER) larvae. We found levels of reaper and sickle mRNA to be significantly lower in larvae homozygous for the IRER deletion, while the levels of hid and grim were not significantly changed (Fig. 2E). When measuring RHG transcripts in third-instar larval wing discs, a similar decrease of reaper mRNA was observed in homozygous Df(IRER) wing discs, whereas the level of hid mRNA was not significantly changed (Fig. 2F). The transcripts of sickle and grim in wing discs were too low to be reliably measured with qRT-PCR (Fig. 2F). Interestingly, a loss-of-function mutation of reaper does not increase wing size (Supplementary Fig. 4), suggesting that blocking reaper expression alone cannot account for the overgrowth phenotype observed for the Df(IRER) animal. Deregulation of other RHG genes such as sickle or grim, or other undefined IRER targets, may also contribute to the observed overgrowth phenotype. More sensitive assay needs to be performed to measure the expression of these genes in wing discs in future study. Based on above observations, we conclude that IRER is required for mediating appropriate expression of several pro-apoptotic genes in larval cells.
To examine whether IRER is also responsible for mediating developmental cell death in other tissues, we monitored developmental cell death in the embryonic central nervous system (CNS). We noted that about 10% of the homozygous Df(IRER) embryos have an enlarged CNS (Supplementary Fig.5 A-B). This enlarged CNS phenotype is not as strong as that in Df(H99) mutant embryos, which lack coding sequences for three pro-apoptotic genes reaper, hid and grim (17, 18). This likely indicates that some, but not all RHG-mediated developmental cell death in the CNS requires a functional IRER. To examine cell numbers during the formation of the Drosophila central nervous system, we used the slit1.0-lacZ reporter to label the pool of ensheathing midline glia. In stage 16-17 embryos homozygous for Df(IRER), extra slit1.0-lacZ-positive cells are present in the abdominal segments (Supplementary Fig. 5 C-D), suggesting that the IRER is also required to limit cell numbers in this group of nervous system cells.
The pattern of cells expressing reaper and sickle in developing tissues such as the wing disc is dynamic and transient, which is partly due to the fact that cells expressing these genes are quickly removed via apoptosis. To bypass this difficulty, we turned to the embryo to examine the effect of IRER on gene expression. We noted that in contexts where the reaper expression is cell lineage-specific, such as neuroblasts in post stage 14/15 embryos, there is no detectable change of reaper expression in Df(IRER) homozygous embryos. However, the segmentally repeated expression of reaper in the epidermal cells was significantly weakened in animals lacking IRER (Supplementary Fig. 6).
IRER is required for the induction of apoptosis following dMyc-induced overproliferation
While the wings of Df(IRER) animals consistently contain more cells than that of wild type control wings, the magnitude of this developmental effect is relatively moderate. In light of this observation, we next sought to determine whether this requirement for IRER in tissue homeostasis was heightened under the pathologic condition of oncogene overexpression. Similar to what has been observed for c-Myc in mammalian systems, overexpression of dMyc in Drosophila tissues leads to increased ribosome biogenesis, increased cell size, and overproliferation (reviewed in (19)). The dMyc-induced overproliferation is quickly followed by induction of apoptosis, which largely cancels out any impact on final cell number. For instance, dMyc expression in retinal cells with the GMR-Gal4 driver leads to a significant increase of cell proliferation and enlarged eyes and ommatidia in the resulting adults (9). However, this overproliferation is accompanied by increased TUNEL-positive staining, such that there is little overall effect on the final number of cells in GMR-dMyc eyes. These reported observations were reconfirmed when we established the system with GMR-Gal4 and UAS-dMyc (Fig. 3A).
Figure 3. The regulatory function of IRER is required for dMyc-induced cell death.
(A-C) Female adult eyes from indicated genotypes. GMR-Gal4 driving expression of UAS-dMyc transgene (GMM: GMR-Gal4/UAS-dMyc) induced both overproliferation and apoptosis and had little impact on the size of the adult eye (A). There was no significant overgrowth of eyes in IRER−/− adults (B). Expression of dMyc in the homozygous Df(IRER) animals resulted in significant overgrown phenotype (C), with some eyes containing protruding cell mass (black arrow). (D-F) Scanning electron micrographs (SEM) from adult eyes of indicated genotypes. The hyperplasia of GMM,IRER−/− was evident with SEM. Note that the ommatidia from GMM,IRER−/− were disorganized. (G-I) Fluorescent in situ hybridization (FISH) was performed to the third-instar eye discs to detect reaper transcripts. reaper was significantly induced by GMM in the GMR-domain in wild-type eye discs (H), and this induction was largely blocked in IRER−/− eye discs (I). The white bars mark the expression domain of GMR-Gal4 (H & K). (J-L) Cleaved caspase3 staining of third-instar larval eye imaginal discs. The yellow arrow marks a group of apoptotic cells in eye discs expressing dMyc in the GMR-Gal4 domain. (M) Merged layers of the representative wing images of tub>dmyc (black) and tub>dmyc,IRER−/− (red). (N) Histogram shows quantification of the effects on wing size. Either lack of IRER (IRER−/−) or overexpression of dMyc (tub>dmyc) alone was sufficient to increase wing size (p < 0.0001 between +/+ (n=30) and IRER−/− (n=26), p< 0.0001 between +/+ and tub>dmyc (n=20), p=0.8818 between IRER−/-and tub>dmyc). When combined together (tub>dmyc,IRER−/−)(n=19), the wing size was increased further, suggesting the synergy between Df(IRER) and dMyc-overexpression (p=0.0012 between IRER−/− and tub>dmyc,IRER−/−, p=0.0027 between tub>dmyc and tub>dmyc,IRER−/−).
The cis regulatory function of IRER is required for overproliferation-induced apoptosis in the GMR-dMyc (GMM) model. Under normal conditions, there is no detectable defect in the eyes of homozygous Df(IRER) adults (Fig. 3B), suggesting that most developmentally programmed cell deaths in the eye proceed normally in the absence of IRER. When dMyc was over-expressed in eye discs lacking IRER, the resulting adult eyes are larger than GMM alone (Fig. 3C) and contain superfluous ommatidia (Fig. 3D-F).
It has been shown that cell autonomous apoptosis following dMyc-induced overproliferation is accompanied by the induction of reaper and sickle (9). Using fluorescent in situ hybridization (FISH), we found that reaper mRNA is significantly elevated from cells behind morphogenetic furrow in GMM larval eye discs (Fig. 3G-H), which is consistent with previously reported observation of apoptotic cells in this model. In addition to the elevated reaper mRNA, we also observed reproducible increase in activated caspase-3 signal in the GMR-Gal4 expression domain posterior to the morphogenetic furrow in GMM eye discs (Fig. 3K) relative to GMR-Gal4 alone (Fig. 3J). Critically, an intact IRER is required for both of these dMyc-induced events. Cell autonomous expression of reaper was almost absent in GMM discs homozygous for Df(IRER) (Fig. 3I), and the GMM active caspase-3 was also lowered in the IRER-deficient background (Fig. 3L). In order to rule out the possibility that the overgrowth of GMM,IRER−/− eyes is due to an increased proliferation, we performed BrdU incorporation in GMM eye discs in both wild-type and Df(IRER) background, and detected no difference (Supplementary Fig. 7). The observations demonstrate that the IRER region is required for ectopic apoptosis induced by overexpressed dMyc in the developing eye.
It has been documented that constitutively elevated level of dMyc introduced by tubulin>dMyc (tub-dMyc) leads to an increased size of the wing, due mainly to increased cell size rather than cell number (10). When tub-dMyc was crossed into the Df(IRER) background, we found that the wing sizes were significantly larger than either Df(IRER) alone or tub-dMyc alone (Fig. 3M,N), suggesting that more cells survived when the IRER is absent in dMyc-overexpressing wings cells.
Cells with relatively open IRER are more sensitive to dMyc-induced cell death
Our previous analyses have shown that the accessibility of IRER in developing embryos is subject to epigenetic regulation (12). In cells with an ‘open’ IRER chromatin state, reaper, sickle, and hid are rapidly induced within 15-30 minutes following ionizing irradiation. In contrast, none of these three genes can be induced when IRER forms a heterochromatin-like structure enriched with both H3K27me3 and H3K9me3. In order to monitor the accessibility of IRER in individual cells, an ubiquitin-DsRed reporter was inserted into IRER through homologous recombination, with DsRed driven by a 2.2kb upstream regulatory sequence of the ubiquitin-63E gene (Fig. 4A). Our analysis indicated that the reporter expression reflects the DNA accessibility of the IRER locus (manuscript in preparation). The expression level of IRER{ubi-DsRed} is senstive to the epigenetic status of IRER and negatively correlates with suppressive histone modifications in IRER (Supplementary Fig. 8). Thus the IRER{ubi-DsRed} reporter allows us to monitor the epigenetic status of the local chromatin environment in individual cells during development.
Figure 4. Cells with open IRER are more sensitive to dMyc -induced cell death.
(A) An ubiquitin-DsRed reporter was inserted, via homologous recombination, into the middle of IRER, a locus that showed strongest resistance to DNase I in late stage embryos (12). The strain was referred to as “IRER{ubi-DsRed}” and served as a reporter for the epigenetic status of IRER. The IRER{ubi-DsRed} reporter signal is variegated in the developing wing discs. (B’). Expression of GFP (green) in the dpp domain does not change the variegated expression pattern of IRER{ubi-DsRed} (B”). (C-C”) When dMyc was co-expressed with GFP, there was a conspicuous lack of DsRed- positive cells in the dpp zone (yellow arrows). (D-D”) When cell death in the dpp zone was blocked by co-expressing UAS-diap1, many DsRed -positive cells were rescued (yellow arrows). (E) DsRed intensity quantification from dpp-expression domain of indicated genotypes in B, C and D. Mean+S.E.M. are shown, n=6 for each genotype. (F-F”) dMyc-overexpression clones, marked by GFP (dashed lines), were generated by the Act>y>Gal4 flip-out in conjugation with UAS-GFP and UAS-dMyc transgene expression. Note cells in the dMyc-overexpressing clones have lower levels of IRER{ubi-DsRed} signal comparing to neighboring cells.
To examine whether cells with an open IRER are more sensitive to dMyc-induced apoptosis, we expressed dMyc in wing discs using the UAS/Gal4 system and monitored the IRER{ubi-DsRed} activity. dMyc driven by dpp-Gal4 at the anterior/posterior boundary of the wing discs induces apoptosis in both the dpp expression domain and in the zone immediately anterior to it through cell autonomous and non-autonomous mechanisms (10). In the background of IRER{ubi-DsRed}, the sizes of cells in the dpp>dMyc expression domain (marked in green, GFP in Fig. 4) are noticeably larger than the neighboring cells, which is reflected by the diluted DAPI signal intensity and verified by anti-Dlg staining (Fig. 4C vs. B, Supplementary Fig. 9). Moreover, there is a conspicuous lack of DsRed-positive cells in the dMyc expression zone as well as the zone immediately anterior to it, where apoptosis is reported to occur in dpp-dMyc discs (10); in contrast, the variegated patterns of DsRed in other regions of the dpp>dMyc wing discs are indistinguishable from those in discs without dMyc overexpression (Fig. 4C’ vs. B’, Fig. 4E). The lack of IRER{ubi-DsRed} expression was not specific to the dpp-Gal4 system. A similar phenomenon was observed when dMyc was expressed in Act>y+>Gal4 ‘flip-out’ clones (Fig. 4FF”).
The reduction of IRER{ubi-DsRed} expression following dMyc expression could reflect rapid elimination of cells with highly accessible IRER. To assess whether cells with higher levels of DsRed expression and thus a more accessible chromatin state on the IRER, are preferentially eliminated by dMyc-induced cell death, we co-expressed the anti-apoptotic factor Diap1 (Fig. 4D-D”) or the caspase inhibitor P35 (Supplementary Fig. 10) in the dpp-dMyc system to block apoptosis. These anti-apoptotic transgenes led to the appearance of brightly DsRed-positive cells in the dpp zone (Fig. 4 D’, 4E, supplementary Fig. 10B). Moreover, the restoration of DsRed signal was accompanied by an accumulation of cleaved caspse-3 in ‘undead’ cells in which dMyc-induced apoptosis was blocked by P35 (Supplementary Fig. 10B).
Taken together, our experiments indicate that DsRed-positive cells, i.e. those with relatively open IRER, are preferentially eliminated in response to dMyc overexpression. One prediction from this observation is that cells with epigenetically blocked IRER will have a higher threshold for oncogenic stress-induced apoptosis. Although the Polycomb group protein (PcG) is required for the formation of suppressive histone modifications in IRER (12), the mechanism that targets PcG to the IRER remains elusive. Thus as a surrogate for reducing IRER activity by epigenetic means, we utilized a chromosome with an IRER deletion to determine if cells without IRER are more resistant to oncogenic stress.
Cells lacking the cis regulatory function of IRER have the propensity to overgrow and are resistant to stress-induced cell death
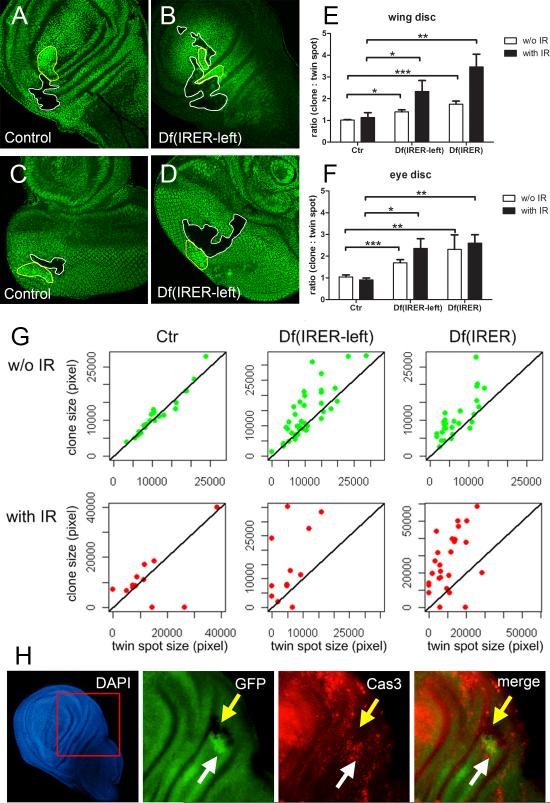
To study the behavior of IRER-deficient cells in proliferating larval tissues, we generated mosaic clones bearing IRER deletions in imaginal discs. The heat shock-induced FLP (hsFLP) system was titrated to generat~1 clone per disc, which facilitated identification and measurement of the size of each clone and its paired twin spot. As shown in Figure 5, control (wild type) clones and their twin spots grew to similar size in either the wing or the eye discs (Fig. 5 A & C). In contrast, clones lacking either the left section of IRER (Df(IRER_left)) (12), or the entire IRER (Df(IRER)), showed a propensity to overgrow relative to their respective twin spots (Fig. 5 B & D). Although there was variation in the extent of this overgrowth, the overall phenotype was robust and statistically significant (Fig. 5 E & F, open bars). A similar overgrown phenotype was observed with clones induced by ey-FLP in adult eyes (Supplementary Fig. 11 B-D).
Figure 5. Cells lacking IRER have the propensity to overgrown and are resistant to stress –induced cell death.
(A-D) Clones were induced using the FLP/FRT-mediated mitotic recombination and labeled by the absence of GFP (white lines). The sibling wild-type twin spots are marked by 2XGFP (yellow lines). While neutral clones generally grow to similar size as the simultaneously induced twin spots, homozygous IRER-deficient clones tend to overgrow (A & B, wing disc; C & D, eye disc, 72hr following clone induction). (E & F) While there is a considerable variation in how much the Df(IRER) clone overgrows, the overgrown phenotype is significant (open bars in E & F. wing disc: p=0.017 between Ctr and Df(IRER-left), p<0.001 between Ctr and Df(IRER); eye disc: p<0.001 between Ctr and Df(IRER-left), p=0.002 between Ctr and Df(IRER)). Wilcoxon rank-sum test was used to for p-value calculation. When animals are subject to a sub-lethal dose (10Gy) of x-ray at 24 hours following clone induction, the size difference between Df(IRER) clones and the twin spots is significantly increased (closed bars in E & F. wing disc: p=0.021 between Ctr and Df(IRER-left), p=0.002 between Ctr and Df(IRER); eye disc: p=0.027 between Ctr and Df(IRER-left), p=0.002 between Ctr and Df(IRER)). Size was measured at 72~96hr after clone induction and the ratio of clone size vs. twin spot size was calculated. (G) Scatter plot of clone size and twin spot size measured at 72~96hr after clone induction. Red dots and green dots represent the clones induced with or without IR, respectively. (H) Animals were irradiated (40Gy) at 48 hr post clone induction. Apoptosis was measured by Caspase 3 staining at 4hr following IR. There is a lack (or reduced level) of apoptotic cells in the Df(IRER) clones. The size difference between the mutant clone and the twin spot in this setting is not significant due to the relatively short time following clone induction and x-ray treatment.
Importantly, the propensity of IRER mutant clones to outgrow their wild-type counterparts was significantly heightened when a cytotoxic stress was introduced to the system. When growing animals were subjected to a non-lethal dose of irradiation (10Gy) at 24 hours following clone induction, the ratio between the size of the IRER-deficient clones and their respective twin spots was greatly increased, both in the wing discs (Fig. 5 E & G) and the eye discs (Fig. 5F & Supplementary Fig. 11A). Indeed, under this heightened stress condition, wild-type twin spots were eliminated in about 15% of the cases, leaving only the IRER mutant clones at the end of larval development. Even when these ‘missing clone’ instances were omitted from calculation of mutant: twin spots size ratios, the overgrowth phenotype of both Df(IRER_left) and Df(IRER) clones remained statistically significant (Fig. 5E-F, closed bars).
The propensity to overgrowth displayed by cells lacking the cis regulatory function of IRER was accompanied by resistance to stress-induced cell death. When irradiation-induced cell death was monitored in discs containing Df(IRER) clones at 4 hours post γ-ray treatment (40Gy), there was a conspicuous lack of activated caspase-3 signal in the clones deficient for IRER despite abundant apoptotic cells in the twin spots as well as the non-recombinant heterozygous tissues (Fig. 5H). In line with this observation, it has been well documented that imaginal disc clones mutant for the Drosophila initiator caspase dronc have a similar growth advantage (20).
Synergy between Df(IRER) and dMyc in inducing hyperplasia
To study the behavior of cells with elevated dMyc and blocked (deleted) IRER, we took advantage of the MARCM strategy (21) to generate clones of cells overexpressing dMyc and simultaneously deficient for IRER (Fig. 6 A). Clones of cells that overexpress dMyc alone (dMyc+) (Fig. 6 B-B’, C-C’) or have elevated dMyc and are deficient for IRER (dMyc+,IRER−/−) (Fig. 6 D-D’, E-E’) were generated in parallel at the same developmental stage. Critically, dMyc+,IRER−/− clones grow significantly larger than dMyc+ clones. When clone size was measured as the percentage of the whole disc, dMyc+,IRER−/− clones are about 3 times larger than dMyc+ clones in the eye disc (Fig. 6F-G). The size ratio between dMyc+, IRER−/− and dMyc+ clones is also significantly higher than the ratio between IRER−/− and IRER+/+ (i.e. wild type) clones (Fig. 5 E, F). Thus it is apparent that there is significant synergy between elevated dMyc and Df(IRER) in the induction of tissue overproliferation and hyperplasia.
Figure 6. Synergy between Df(IRER) and dMyc in driving overproliferation and hyperplasia.

(A) Schematic representation of the MARCM strategy used to induce dMyc expression in IRER-mutant clones. When compared to wild-type clones with dMyc-expression (B-B’ in eye disc, C-C’ in wing disc), IRER-deficient clones (D-D’ & E-E’) with dMyc-expression are significantly larger in size. Clones are monitored at 72hours after heat-shock and individual channels or merged channels are shown as indicated. The percentage occupancy of clones is examined by calculating the ratio between the GFP-positive area and the entire disc area. The dMyc+,IRER−/− clones occupy a much larger area than the dMyc+ clones in both eye discs (F) and wing discs (G). Mean + SE. are shown (n=5).
Discussion
Induction of apoptosis following oncogene-induced overproliferation is a key tumor-suppressor mechanism. Previous studies have shown that, similar to mammalian systems, overexpression of dMyc led to overproliferation as well as cell autonomous induction of apoptosis in Drosophila (9, 22). These earlier works opened the door for deciphering the molecular mechanisms of overproliferation-induced apoptosis in this genetic model organism. In this study, we show that an intergenic regulatory region, previously identified for its role in mediating DNA damage-induced apoptosis, is required to mediate dMyc-induced apoptosis. More importantly, since the accessibility of this intergenic region is epigenetically controlled through histone modifications, the findings provide a mechanistic link between epigenetic regulation and oncogenic stress-induced apoptosis and hyperplasia (Figure 7).
Figure 7. Diagram summarizing the working mechanism of IRER.
Ectopic expression of proto-oncogene dMyc leads to overproliferation, which is balanced/corrected by reaper/sickle-dependent apoptosis. IRER functions as the pivotal regulator for the induction of apoptosis following overproliferation. Epigenetic regulation of IRER defines cellular sensitivity to developmental constraints and oncogenic stress. In general, cells with an open IRER are more sensitive to stress-induced apoptosis and have less potential to over-proliferate; whereas cells with a closed IRER are more resistant to stress and exhibit higher proliferation potential in the context of Myc over-expression.
IRER serves as the gatekeeper for overproliferation-induced apoptosis in Drosophila
In mammalian systems, transcriptional regulation of BH3-only pro-apoptotic genes, such as noxa and Bim (Bcl-2 interacting mediator of cell death), plays an important role in mediating c-Myc-induced apoptosis (reviewed by Hoffman and Liebermann (6)). In fact, the increased oncogenic potential of tumor-derived c-Myc mutants have been attributed to their failure to induce Bim (and apoptosis) while retaining the activity to stimulate proliferation (23). Consistent with this hypothesis, co-expression of the anti-apoptotic factor Bcl-2 renders wild type c-Myc as oncogenic as tumor-derived c-Myc mutants. Our current work and previous work by the Gallant group (9) both implicate reaper (and likely sickle) as the upstream pro-apoptotic genes responsible for mediating dMyc-induced apoptosis in Drosophila. One striking similarity between the mammalian genes responsible for mediating c-Myc-induced cell death and the two Drosophila pro-apoptotic genes, is that they are all potential transcriptional targets of P53 and are involved in mediating DNA damage-induced cell death. Montero et al. found that dMyc overexpression significantly induces reaper and sickle, and that a chromosome deletion that removes a large region including both reaper and sickle dominantly suppresses dMyc-induced apoptosis (9). In this study, we showed that the intergenic regulatory region between reaper and sickle, i.e. IRER, is required for mediating dMyc-induced reaper (and sickle) expression and apoptosis.
The IRER is required for mediating P53-depdendent induction of multiple pro-apoptotic genes in the RHG regulatory block following irradiation of early stage embryos (12, 24). Transcriptional activation of the pro-apoptotic RHG genes plays a pivotal role mediating apoptosis during development as well as in response to a variety of stresses (25). With the exception of hid, the other RHG genes (reaper, grim, and sickle) seem to be exclusively expressed in cells that are eliminated shortly after one or multiple of these genes are expressed. Centralized regulation, in which one regulatory region coordinates the expression of multiple RHG genes, appears to be the general theme in transcriptional regulation of RHG genes. In addition to our finding that IRER is responsible for mediating radiation-induced expression of reaper, sickle and hid, studies from the White laboratory have shown that a ~22kb intergenic region between grim and reaper termed the NBRR (neuroblast regulatory region) is required for cell lineage specific developmental expression of reaper, grim, and sickle in embryonic neuroblasts (26). The IRER and NBRR are situated upstream and downstream of reaper, respectively. Both are highly enriched for HCNEs (Highly Conserved Non-coding Elements) and appear to fulfill important regulatory roles in gene expression in vivo (27, 28).
In the mammalian system, the role of P53 in Myc-induced cell death seems to vary depending on the cell type and experimental context. In Drosophila, although P53 mRNA is induced by overexpression of dMyc, its function is dispensable for dMyc-induced cell death (9). The IRER is required for P53-dependent induction of RHG genes following irradiation (12). There is a consensus P53-response element within the IRER, and reporter transgenes containing the P53RE is DNA damage-inducible (13). Although there is currently no evidence that dMyc directly binds to IRER sequences to induce RHG gene expression, the IRER contains multiple sequences that match the consensus Myc binding site. Significant technical challenges must be overcome to test this model and to investigate whether these elements support Myc-induced transcription of reaper (and sickle). Given the central role that the IRER plays in mediating P53 and dMyc-induced pro-apoptotic gene expression, an improved knowledge of transcription factor binding sites within this region may shed light on interactions between Myc and P53 in the developmental and homeostatic control of cell death.
Epigenetic control of oncogenic stress-induced apoptosis
Abnormal histone modifications have been postulated to be a major oncogenic mechanism in vertebrates (reviewed in (29). A wide spectrum of histone modification abnormalities has been observed in different cancer cells. However, it is necessary to discern epigenetic changes responsible for driving tumor formation from those that are byproducts of the oncogenic process. Epigenetic silencing of pro-apoptotic genes, by blocking the pro-apoptotic tumor suppressor mechanism, could potentially serve as a key event in early stages of tumorigenesis. For instance, noxa, a c-Myc target gene that encodes a BH3-only pro-apoptotic protein, is subject to Bmi-mediated suppressive histone modification (H3K27me3) (30). Murine Bmi, the ortholog of Drosophila PcG protein Psc (Posterior sex combs), collaborates with c-Myc in promoting tumorigenesis by suppressing apoptosis (31). Similarly, PcG proteins, such as Psc, are functionally required for suppressive histone formation in the IRER (12). Our prior work showed that formation of suppressive histone modifications on the IRER renders this region as inaccessible to DNase I as the centromeric heterochromatin, suggestive of a very condensed, transcriptionally silent chromatin environment. Accordingly, RHG pro-apoptotic genes are no longer responsive to DNA damage under this heterochromatic condition. As IRER deletions used in this study do not remove any transcribed region or defined promoter, the behavior of cells without the IRER (i.e. Df/Df) can be viewed as a reflection of cells with complete epigenetic silencing of the IRER. As shown here, the lack of IRER blocks dMyc-induced apoptosis and leads to significant hyperplasia. In summary, these findings suggest that epigenetic control of oncogenic stress-induced apoptosis is a fundamental tumor-suppressor mechanism that is conserved from insects to mammals. Abnormal epigenetic regulation of P53-targeted proapoptotic genes could potentially serve as a driving event for tumorigenesis.
Unlike mammalian systems, key upstream pro-apoptotic genes in Drosophila are clustered in the RHG genomic region and are coordinately regulated by IRER. Understanding mechanisms that control the epigenetic status of the IRER could reveal how suppressive histone modifications can target oncogenic stress-responsive tumor suppressor genes. There is no discernible PRE (Polycomb response element) in the IRER, and the formation of heterochromatin in IRER is independent of anterior/posterior cell positioning. Monitoring DNA accessibility in the IRER with the innovative IRER{ubi-DsRed} reporter reveals that IRER epigenetic status is dynamically regulated during development, stochastically regulated in tissue compartments, and highly responsive to environmental stresses. The IRER thus represents a unique system to explore the relationship between chromatin regulation and oncogenic stress-induced apoptosis.
Materials and methods
Drosophila strains and culture
Flies were maintained on a standard corn-agar medium at 25°C except otherwise mentioned. The strains used in this study were described in supplementary table I. Irradiation was performed as described (12).
The IRER{ubi-DsRed} strain was generated via “Ends-out” gene targeting method (32). We first generated a donor strain carrying a pEnds-out2[LA-ubi-DsRed-RA] construct on the × chromosome. The ubi-DsRed is a florescence reporter consisting a full length DsRed coding sequence driven by a 2.2kb fragment from the ubiquitin-63E gene. LA and RA represent two 3kb consecutive segments within IRER separated by an Nde I site. The LA and RA homologous sequences were enzyme digested from a BAC clone (BACR35F04, DGRC) by XhoI & BamHI, gel purified, and sub-cloned to the pEnds-out2 vector to make the donor construct. The recombination event was first recovered based on autosomal inheritance of ubi-DsRed and subsequently verified with PCR and southern hybridization. The correlation of DeRed expression with the epigenetic status of IRER was verified with DNase sensitivity assay, ChIP assay, as well as disruption of IRER epigenetic status through Su(var)3-9 knocking down (Supplementary Fig. 8).
The Bac-IRER strain was generated via Recombineering mediated gap-repair (33). The DNA fragment spanning reaper promoter/enhancer-IRER was retrieved from the BACR35F04 clone (DGRC), and was cloned to the P[acman] vector (33). The 40kb P[acman]-reaper promoter-IRER construct was integrated to the genomic locus 51D on the 2nd chromosome through ΦC31 integrase-mediated transgenesis.
Clone induction
Clones were generated with either the standard FLP/FRT system or the MARCM system. Hspflp and ey-flp were used to induce clones from imaginal discs and adult eyes, respectively. Clone size was determined by measuring the 2-dimensional area of clones at 200X magnification.
Act>dMyc clones with or without IRER: Larvae of the genotype hsp70-flp22; Act-Gal4, UASGFP/UAS-dMyc; Df(IRER), FRT80B (or: FRT80B)/tub-Gal80, FRT80B were subjected to a heat shock of 30min at 37°C at 60hr after egg laying, and were dissected at 72hr after heat-shock.
IRER-deficient clones in the imaginal discs: larvae of the genotype hsp70-flp1; Df(IRER), FRT80B (or: FRT80B)/ubi-GFP, FRT80B were heat shocked for 1.5hr at 37°C 48-52hr after egg laying. The imaginal discs were dissected at 72-96hr after heat-shock.
For all the clone induction experiments, control clones are always induced and analyzed in parallel with the mutant clones.
Immunohistochemistry and microscopy
Imaginal disc dissection and antibody staining were performed following the standard protocol (34). Primary antibodies used in this study included: anti-cleaved Caspase 3 (Cell signaling, 1:200), anti-Dlg (DSHB, 1:50), anti-BrdU (BD Biosciences, 1:100). Secondary antibodies (anti-rabbit conjugated Cy5 or anti-mouse conjugated Cy3) were used as recommended (Jackson ImmunoResearch). In situ hybridization was carried out as described (12). FISH signal was detected by HRP-conjugated anti-DIG antibody (1:500, Roche) and subsequently amplified by the Tyramid Signal Amplification Kit (PerkinElmer, Waltham, MA, USA).
Fluorescent images were taken with a Zeiss Axioplan imaging 2 Microscope or a Leica SP5 Confocal Microscope. The Open lab software was used to acquire the images. Clone size was measured at 200X magnification with Image J (NIH) or Photoshop (Adobe). Fluorescence intensity in wing discs was quantified with Image J (NIH).
Scanning electron microscopy (SEM) was performed as described (34).
Wing size measurement
Flies were raised at a standard density (~20 flies per vial) and three replicate vials were established for each genotype. Wings were dissected and mounted onto slides in Permount mounting medium. Wing images were acquired using the Leica DMLB light microscope and wing area was determined as described (35).
Gene expression analysis
RNA was extracted from ~20 wing discs or ~10 larvae of the desired genotypes using the RNeasy Minikit (Qiagen). qRT-PCR was performed as described (12).
Statistics
Wilcoxon rank-sum test was used to compare the size difference between clones and twin spots. In other cases, data were verified to be normally distributed and student's t-tests were used to determine the statistical significance.
Supplementary Material
Acknowledgements
We are grateful for Dr. Laura Johnston in Columbia University, Dr. Kristin White in Massachusetts General Hospital, the Bloomington stock center and Vienna Drosophila RNAi Center for providing us fly strains.
Footnotes
Author Contributions: LZ and CZ formulated the overall design of the experiments. SCT and EM designed and carried out the Act>(y)>dMyc experiment. The majority of experiments were carried out by CZ, with some experiments performed together with, or alone by, NL, MC, and LZ. GL performed statistical and bioinformatics analysis. LZ and CZ wrote the manuscript with input from SCT and KHM.
Conflict of interest.
The authors declare no conflict of interest.
References
- 1.Meyer N, Kim SS, Penn LZ. The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol. 2006 Aug;16(4):275–87. doi: 10.1016/j.semcancer.2006.07.011. PubMed PMID: 16945552. Epub 2006/09/02. eng. [DOI] [PubMed] [Google Scholar]
- 2.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002 May 3;109(3):321–34. doi: 10.1016/s0092-8674(02)00738-9. PubMed PMID: 12015982. Epub 2002/05/23. eng. [DOI] [PubMed] [Google Scholar]
- 3.Letai A, Sorcinelli MD, Beard C, Korsmeyer SJ. Antiapoptotic BCL-2 is required for maintenance of a model leukemia. Cancer Cell. 2004 Sep;6(3):241–9. doi: 10.1016/j.ccr.2004.07.011. PubMed PMID: 15380515. Epub 2004/09/24. eng. [DOI] [PubMed] [Google Scholar]
- 4.Strasser A, Harris AW, Bath ML, Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990 Nov 22;348(6299):331–3. doi: 10.1038/348331a0. PubMed PMID: 2250704 Epub 1990/11/22. eng. [DOI] [PubMed] [Google Scholar]
- 5.Harrington EA, Bennett MR, Fanidi A, Evan GI. c-Myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 1994 Jul 15;13(14):3286–95. doi: 10.1002/j.1460-2075.1994.tb06630.x. PubMed PMID: 8045259. Pubmed Central PMCID: 395225. Epub 1994/07/15. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008 Oct 27;27(50):6462–72. doi: 10.1038/onc.2008.312. PubMed PMID: 18955973. Epub 2008/10/29. eng. [DOI] [PubMed] [Google Scholar]
- 7.Scott CL, Schuler M, Marsden VS, Egle A, Pellegrini M, Nesic D, et al. Apaf-1 and caspase-9 do not act as tumor suppressors in myc-induced lymphomagenesis or mouse embryo fibroblast transformation. J Cell Biol. 2004 Jan 5;164(1):89–96. doi: 10.1083/jcb.200310041. PubMed PMID: 14709542. Pubmed Central PMCID: 2171953. Epub 2004/01/08. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu B, Marin MC, Elnaggar AK, Stephens LC, Brisbay S, Mcdonnell TJ. Evidence That C-Myc Mediated Apoptosis Does Not Require Wild-Type P53 during Lymphomagenesis. Oncogene. 1995 Jul 6;11(1):175–9. PubMed PMID: ISI:A1995RJ29500020. English. [PubMed] [Google Scholar]
- 9.Montero L, Muller N, Gallant P. Induction of apoptosis by Drosophila Myc. Genesis. 2008 Feb;46(2):104–11. doi: 10.1002/dvg.20373. PubMed PMID: 18257071. Epub 2008/02/08. eng. [DOI] [PubMed] [Google Scholar]
- 10.de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004 Apr 2;117(1):107–16. doi: 10.1016/s0092-8674(04)00214-4. PubMed PMID: 15066286. Epub 2004/04/07. eng. [DOI] [PubMed] [Google Scholar]
- 11.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999 Sep 17;98(6):779–90. doi: 10.1016/s0092-8674(00)81512-3. PubMed PMID: 10499795. Epub 1999/09/28. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Lin N, Carroll PM, Chan G, Guan B, Xiao H, et al. Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos. Dev Cell. 2008;14(4):481–93. doi: 10.1016/j.devcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000 Mar 31;101(1):103–13. doi: 10.1016/S0092-8674(00)80627-3. PubMed PMID: 10778860 Epub 2000/04/25. eng. [DOI] [PubMed] [Google Scholar]
- 14.Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, et al. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol. 2004 Feb;24(3):1219–31. doi: 10.1128/MCB.24.3.1219-1231.2004. PubMed PMID: 14729967. Pubmed Central PMCID: 321428 Epub 2004/01/20. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wichmann A, Jaklevic B, Su TT. Ionizing radiation induces caspase-dependent but Chk2- and p53-independent cell death in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006 Jun 27;103(26):9952–7. doi: 10.1073/pnas.0510528103. PubMed PMID: 16785441. Pubmed Central PMCID: 1502560. Epub 2006/06/21. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004 Jun;167(2):797–813. doi: 10.1534/genetics.104.026658. PubMed PMID: 15238529. Pubmed Central PMCID: 1470913 Epub 2004/07/09. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou L, Hashimi H, Schwartz LM, Nambu JR. Programmed cell death in the Drosophila central nervous system midline. Curr Biol. 1995;5(7):784–90. doi: 10.1016/s0960-9822(95)00155-2. [DOI] [PubMed] [Google Scholar]
- 18.Gambis A, Dourlen P, Steller H, Mollereau B. Two-color in vivo imaging of photoreceptor apoptosis and development in Drosophila. Dev Biol. 2011 Mar 1;351(1):128–34. doi: 10.1016/j.ydbio.2010.12.040. PubMed PMID: 21215264 Pubmed Central PMCID: 3051417. Epub 2011/01/11. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallant P. Drosophila Myc. Adv Cancer Res. 2009;103:111–44. doi: 10.1016/S0065-230X(09)03005-X. PubMed PMID: 19854354. Epub 2009/10/27. eng. [DOI] [PubMed] [Google Scholar]
- 20.Verghese S, Bedi S, Kango-Singh M. Hippo signalling controls Dronc activity to regulate organ size in Drosophila. Cell Death Differ. 2012 Oct;19(10):1664–76. doi: 10.1038/cdd.2012.48. PubMed PMID: 22555454. Pubmed Central PMCID: 3438497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001 May;24(5):251–4. doi: 10.1016/s0166-2236(00)01791-4. PubMed PMID: 11311363. Epub 2001/04/20. eng. [DOI] [PubMed] [Google Scholar]
- 22.Bellosta P, Gallant P. Myc Function in Drosophila. Genes Cancer. 2010 Jun 1;1(6):542–6. doi: 10.1177/1947601910377490. PubMed PMID: 21072325. Pubmed Central PMCID: 2976539. Epub 2010/11/13. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005 Aug 11;436(7052):807–11. doi: 10.1038/nature03845. PubMed PMID: 16094360. Epub 2005/08/12. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin N, Zhang C, Pang J, Zhou L. By design or by chance: cell death during Drosophila embryogenesis. Apoptosis. 2009 Aug;14(8):935–42. doi: 10.1007/s10495-009-0360-8. PubMed PMID: 19466551. Pubmed Central PMCID: 2901921 Epub 2009/05/26. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008 Jul;15(7):1132–8. doi: 10.1038/cdd.2008.50. PubMed PMID: 18437164. Epub 2008/04/26. eng. [DOI] [PubMed] [Google Scholar]
- 26.Tan Y, Yamada-Mabuchi M, Arya R, St Pierre S, Tang W, Tosa M, et al. Coordinated expression of cell death genes regulates neuroblast apoptosis. Development. 2011;138(11):2197–206. doi: 10.1242/dev.058826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006 Apr 21;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. PubMed PMID: 16630819. Epub 2006/04/25. eng. [DOI] [PubMed] [Google Scholar]
- 28.Kikuta H, Laplante M, Navratilova P, Komisarczuk AZ, Engstrom PG, Fredman D, et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007 May;17(5):545–55. doi: 10.1101/gr.6086307. PubMed PMID: 17387144. Pubmed Central PMCID: PMC1855176. Epub 2007/03/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011 Oct;11(10):726–34. doi: 10.1038/nrc3130. PubMed PMID: 21941284. Epub 2011/09/24. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamashita M, Kuwahara M, Suzuki A, Hirahara K, Shinnaksu R, Hosokawa H, et al. Bmi1 regulates memory CD4 T cell survival via repression of the Noxa gene. J Exp Med. 2008 May 12;205(5):1109–20. doi: 10.1084/jem.20072000. PubMed PMID: 18411339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999 Oct 15;13(20):2678–90. doi: 10.1101/gad.13.20.2678. PubMed PMID: 10541554. Pubmed Central PMCID: 317101. Epub 1999/10/29. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc Natl Acad Sci U S A. 2003 Mar 4;100(5):2556–61. doi: 10.1073/pnas.0535280100. PubMed PMID: 12589026. Pubmed Central PMCID: 151379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006 Dec 15;314(5806):1747–51. doi: 10.1126/science.1134426. PubMed PMID: 17138868. [DOI] [PubMed] [Google Scholar]
- 34.Sullivan W, Ashburner M, Hawley RS. Drosophila Protocols. 1 ed Vol. 15. Cold Spring Harbor Laboratory Press; Jan, 2000. p. 2000. [Google Scholar]
- 35.Gilchrist AS, Partridge L. A comparison of the genetic basis of wing size divergence in three parallel body size clines of Drosophila melanogaster. Genetics. 1999 Dec;153(4):1775–87. doi: 10.1093/genetics/153.4.1775. PubMed PMID: 10581284 Pubmed Central PMCID: 1460863. Epub 1999/12/03. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.