Conspectus
The human innate immune system has evolved the means to reduce the bioavailability of first-row late d-block transition metal ions to invading microbial pathogens in a process termed “nutritional immunity”. Transition metals from Mn(II) to Zn(II) function as metalloenzyme cofactors in all living cells, and the successful pathogen is capable of mounting an adaptive response to mitigate the effects of host control of transition metal bioavailability. Emerging evidence suggests that Mn, Fe, and Zn are withheld from the pathogen in classically defined nutritional immunity, while Cu is used to kill invading microorganisms. This Account summarizes new molecular-level insights into copper trafficking across cell membranes from studies of a number of important bacterial pathogens and model organisms, including Escherichia coli, Salmonella species, Mycobacterium tuberculosis, and Streptococcus pneumoniae, to illustrate general principles of cellular copper resistance.
Recent highlights of copper chemistry at the host–microbial pathogen interface include the first high resolution structures and functional characterization of a Cu(I)-effluxing P1B-ATPase, a new class of bacterial copper chaperone, a fungal Cu-only superoxide dismutase SOD5, and the discovery of a small molecule Cu-bound SOD mimetic. Successful harnessing by the pathogen of host-derived bactericidal Cu to reduce the bacterial load of reactive oxygen species (ROS) is an emerging theme; in addition, recent studies continue to emphasize the importance of short lifetime protein–protein interactions that orchestrate the channeling of Cu(I) from donor to target without dissociation into bulk solution; this, in turn, mitigates the off-pathway effects of Cu(I) toxicity in both the periplasm in Gram negative organisms and in the bacterial cytoplasm. It is unclear as yet, outside of the photosynthetic bacteria, whether Cu(I) is trafficked to other cellular destinations, for example, to cuproenzymes or other intracellular storage sites, or the general degree to which copper chaperones vs copper efflux transporters are essential for bacterial pathogenesis in the vertebrate host.
Future studies will be directed toward the identification and structural characterization of other cellular targets of Cu(I) trafficking and resistance, the physical and mechanistic characterization of Cu(I)-transfer intermediates, and elucidation of the mutual dependence of Cu(I) trafficking and cellular redox status on thiol chemistry in the cytoplasm. Crippling bacterial control of Cu(I) sensing, trafficking, and efflux may represent a viable strategy for the development of new antibiotics.
1. Introduction
Copper is an essential transition metal found in all biological systems. All organisms have developed sophisticated copper homeostasis and resistance systems in order to maintain the normal cellular copper supply to essential cuproenzymes while detoxifying excess copper. Recent work suggests that host-derived copper is used as an antibacterial weapon; thus both host and bacterial pathogens actively engage cellular processes to manipulate copper levels at key sites during bacterial infections (for recent reviews, see refs (1)–3). In this Account, we follow the flow of copper from the extracellular space into human cellular cytosol to bacterial pathogens inside human cells, with emphasis on recent developments in our understanding of the molecular and mechanistic details that characterize the transport of copper across biological membranes at the host–bacterial pathogen interface.
Copper in biological systems can exist in either the reduced Cu(I) (3d10) state or the oxidized Cu(II) (3d9) state. The reduction potential of +150 mV of Cu(II)/Cu(I) ensures that Cu(I) is the major oxidation state of copper in the reducing environment of the cytosol (approximately −220 mV). Cu(I) is a soft acid, while Cu(II) is a borderline acid;4 as such, first coordination shell ligands in copper-sensing or transporting proteins are formed primarily by cysteine, methionine, and histidine residues. Cu(I) is relatively unique in biology in that it adopts thermodynamically stable complexes characterized by a low coordination number (n) of 2–4. In contrast, Cu(II) adopts higher coordination number complexes, with n = 4–6. The ability of copper to cycle between two oxidation states superimposed on the capacity to form thermodynamically stable yet ligand exchange labile coordination complexes establishes the foundation for understanding copper transport, sensing, trafficking, and utilization in biological systems.
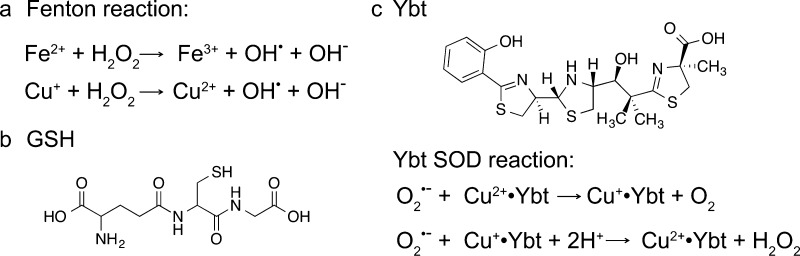
Excess copper negatively impacts bacterial cell viability. The original mechanistic proposal that Cu(I) catalyzes production of hydroxyl radical (OH•) from hydrogen peroxide (H2O2) via the Fenton reaction (Figure 1a), as firmly established for Fe(II) in the cytoplasm, seems increasingly unlikely since low molecular weight thiols (LMWTs) such as glutathione (GSH; Figure 1b) chelate free copper, rendering it incapable of cycling between Cu(I) and Cu(II) states.5 OH• damage is much more likely to occur in the periplasm of Gram-negative bacteria or in the extracellular space (Figure 2). More recent studies suggest that solvent exposed iron–sulfur clusters in metalloenzymes involved in branched-chain amino acid synthesis are primary targets of copper toxicity in Escherichia coli via Cu(I)-mediated displacement of Fe(II).6 Mis-metalation and iron–sulfur cluster disassembly by Cu(I) in enzymes involved in key metabolic processes including glucose catabolism and heme biosynthesis have also been shown to occur in other bacteria.7−9 More work needs to be done to establish the generality of this mechanism of Cu(I) toxicity, particularly in those organisms that lack iron–sulfur cluster proteins and in others, for example, lactic acid bacteria, that produce considerable endogenous hydrogen peroxide under aerobic conditions.10
Figure 1.
(a) Fenton reaction;, (b) chemical structure of reduced glutathione (GSH), and (c) chemical structure of yersiniabactin (Ybt) from uropathogenic E. coli (UPEC)22 and proposed reaction mechanism for the Cu-dependent superoxide dismutase (SOD) activity of Ybt.23
Figure 2.
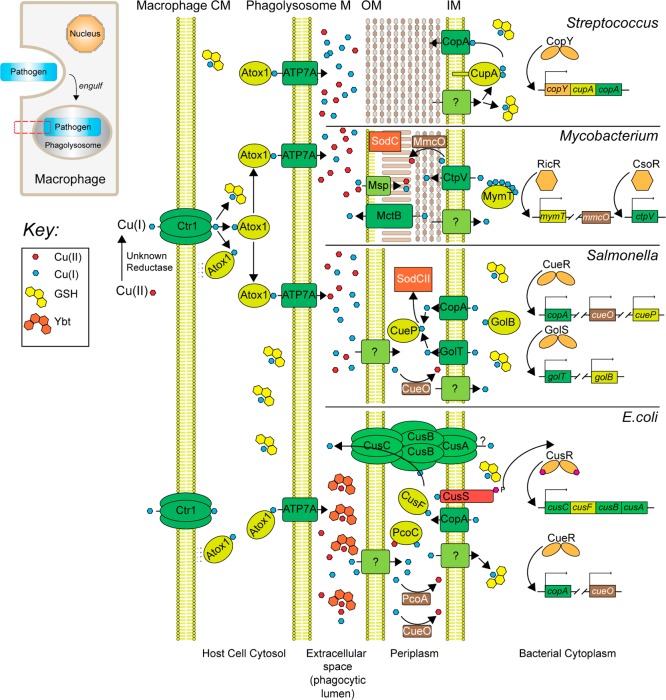
Pathways of copper transport, trafficking, sensing, and resistance in several well-studied bacterial pathogens, including the Gram positive pathogen S. pneumoniae, M. tuberculosis, and two similar Gram negative pathogens, E. coli and Salmonella spp., at the host–pathogen interface. Inset (upper left), cartoon representation of a host macrophage engulfing a bacterial pathogen, ultimately sequestered in an intracellular phagolysosomal compartment. The red box highlights the plasma, phagolysosomal, and outer/inner membranes of the bacterium (from left to right), expanded in the main body of the figure. E. coli CueO, plasmid-encoded PcoA, and mycobacterial MmcO60 are multicopper oxidases (MCOs).2 Both MmcO and an outer membrane channel MctB20 are required for mycobacterial copper resistance. CtpV is a copper exporting ATPase that is required for full virulence of M. turberculosis in murine models of infection,61 while Msp is a porin on the outer membrane of M. tuberculosis.27 Overexpression of Msp genes induces copper stress in M. turberculosis, consistent with a role in Cu uptake.27 Representative metalloregulatory proteins are also shown (right).1,30,43,62
2. Uptake and Trafficking of Copper from Host to Microbe
A cartoon representation of copper transport and trafficking from outside of the vertebrate host cell, for example, a macrophage, into an engulfed bacterial cell is shown in Figure 2. In eukaryotes from yeast to mammals, the acquisition of cellular copper requires a conserved plasma membrane-localized Cu(I) importer, Ctr1 (Figure 2, left).11 The copper trafficking hypothesis12 posits that free copper present in the cytosol is vanishingly low and that Cu(I) must be shuttled to distinct cellular targets by dedicated Cu(I) chaperones. The human Cu(I) chaperone Atox1 (Figure 2) is conserved from bacteria (denoted CopZ; vide infra) to man and is a small cytosolic protein with the ferredoxin-like fold harboring a solvent-exposed CXXC motif that forms a bisthiolate Cu(I) complex.13 Atox1 is proposed to acquire Cu(I) from the C-terminal domain of hCtr1 directly via a specific protein–protein interaction14 or via a glutathione–Cu(I) complex intermediate.15 It is unclear how cells ensure that Atox1 is capable of sequestering all Cu(I) imported through hCtr1. A recent study reveals that Atox1 is capable of binding to liposomes, a mimic of the plasma membrane, via weak electrostatic interactions in vitro thereby reducing the dimensionality of the search along the inner leaflet of the plasma membrane and facilitating Cu(I) loading.16 Likewise a recently characterized bacterial Cu(I) chaperone, CupA from Streptococcus pneumoniae, localizes to the plasma membrane via a single membrane-spanning helix (Figure 2, top).17
Under normal conditions, Atox1 transfers Cu(I) to the Cu(I)-transporting P1B-type ATPases, ATP7A and ATP7B, primarily localized in the trans-Golgi network (vide infra) to provide for systemic copper supply and maturation of specific cuproproteins. However, in phagocytic cells, Cu(I) accumulates in cytoplasmic vesicles that partially fuse with the phagolysosome that ultimately transitions into an antimicrobial compartment,3,18 and there is some evidence to suggest that this accumulation may be dependent upon the trafficking of ATP7A specifically to the membranes of these vesicles (Figure 2).18 Indeed, copper resistance has been shown to be required for virulence in two animal models of mycobacterial infection.19,20
3. Fate of Copper Accumulation in the Extracellular Space
Accumulation of copper in the lumen of the phagolysosomal compartment represents the host’s attempt to exploit the bactericidal power of free Cu as part of a toxic milieu characterized by an acidic pH, reactive nitrogen species (RNS), for example, nitric oxide, reactive oxygen species (ROS), including hydrogen peroxide and NADPH-oxidase derived superoxide anion O2–•, and reactive chlorine species (RCS), for example, hypochlorite (HOCl); in addition, pathogen-requiring Fe is mobilized outside of this compartment.21 Successful microbial pathogens exploit at least two recently characterized strategies to minimize the effects of copper toxicity in this compartment before Cu enters the bacterial cell (Figure 2).
Uropathogenic E. coli (UPEC) synthesizes a siderophore called yersiniabactin (Ybt) that binds Cu(II)22 and catalyzes the dismutation of superoxide to H2O2 and O2 (Figure 1c); recent work reveals that this activity bestows on UPEC a survival advantage in phagosomes.23 Similarly, the human fungal pathogen Candida albicans expresses a novel SOD, SOD5, that is now known to be representative of glycosylphosphatidylinositol (GPI)-anchored, extracellular, monomeric Cu-only SODs that are homologous to the classic cytosolic, dimeric Cu/Zn SOD1.24 Unlike Cu/Zn SOD, SOD5 is secreted as a disulfide-oxidized apoprotein that is readily metalated by the available extracellular copper and thus does not seem to require a chaperone. This presumably rapid metalation is facilitated by the relatively open and solvent exposed nature of the Cu site (Figure 3).24 Despite these structural differences, SOD5 catalyzes superoxide dismutation with turnover kinetics that approach the diffusion limit, like Cu/Zn SODs. Mycobacterium tuberculosis also encodes a Cu-only SOD5 (SodC) of known structure,25 and while there is some evidence that SodC is secreted into the extracellular space, it is also associated with the cell wall and membrane fractions where it functions extracytoplasmically (Figure 2).26
Figure 3.
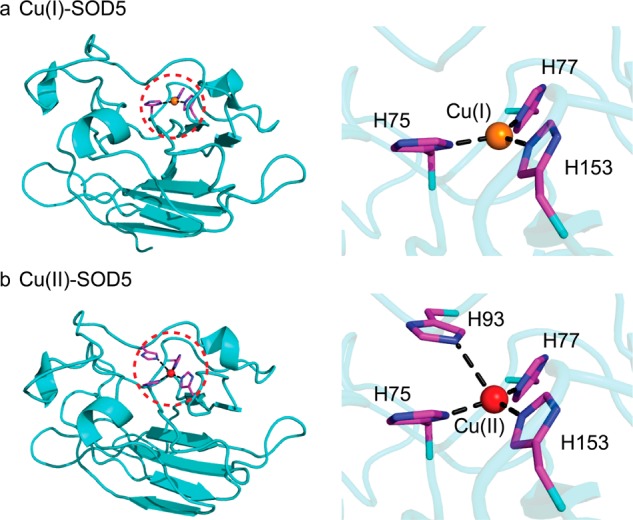
Molecular structures of (a) Cu(I)- and (b) Cu(II)-bound Candida albicans SOD5.24
Extracellular copper not utilized for these processes ultimately enters the bacterial cell, which for the Gram negative pathogens, for example, E. coli and Salmonella spp. and the acid-fast bacterium M. tuberculosis, is the periplasm, while for Gram positive bacteria, including Enterococcus ssp., S. pneumoniae, and Bacillus spp., Cu(I) must cross the bacterial inner or plasma membrane (Figure 2). Although a number of reports document a specific copper uptake system in pathogenic bacteria either into the periplasm27 or across the plasma membrane,28 there is generally a lack of specific uptake systems described for Cu(I)/Cu(II) outside of the photosynthetic bacteria. This is consistent with the fact that the predicted cuproproteome accounts for ≤0.3% of the proteome in all of bacteria29 and most cuproproteins either localize to the periplasm in Gram negative bacteria or are tethered to the plasma membrane, facing the extracellular space (Figure 2).
Since cytoplasmic Cu(I) is far more toxic than biologically useful, there is a robust program of Cu(I) detection in the periplasm, for example, by CusS, or sensing by Cu(I)-specific metalloregulatory proteins in the cytoplasm (Figure 2, right),30 examples of which are E. coli CueR and M. tuberculosis CsoR and RicR19 (Figure 4). These Cu(I) sensors increase the transcription of genes encoding for proteins that allow an organism to resist the effects of copper stress. Resistance proteins include cytoplasmic copper chelators like bacterial metallothionein (MymT) in M. tuberculosis, copper chaperones that operate in both the periplasm and the cytoplasm, multicopper oxidases (MCOs) that oxidize Cu(I) to less toxic Cu(II), and Cu(I) efflux pumps and outer membrane-localized channels that move copper across membranes (Figure 2).
Figure 4.
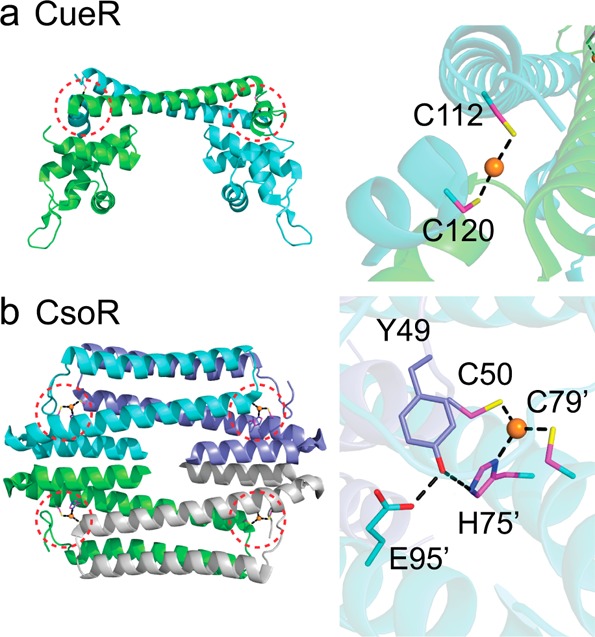
Molecular structures of two Cu(I)-specific metalloregulatory proteins: (a) Cu(I)-bound E. coli CueR;62 (b) Cu(I)-bound CsoR from Geobacillus thermodenitrificans.63 Subunits are differentially shaded with the Cu(I) binding sites circled and expanded (right).
4. Copper Resistance Determinants in the Periplasm
The periplasm in Gram negative bacteria can be considered a true subcellular compartment that minimizes the effects of cellular Cu(I) toxicity. Indeed, a number of copper-containing proteins in different organisms are found in this compartment, all of which have evolved the capacity to bind Cu(I), Cu(II), or both with high affinity and globally function as Cu trafficking proteins. These include E. coli CusF, Salmonella enterica serovar typhimurium CueP, and E. coli PcoC/Pseudomonas CopC (Figure 5).2 Each has a distinct role in Cu homeostasis, however, and there has been significant recent work on how these proteins acquire and traffic Cu.
Figure 5.
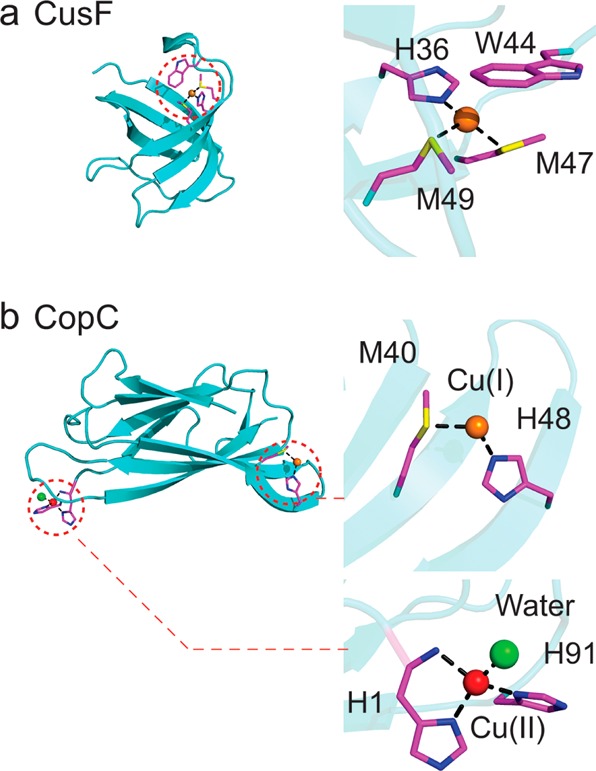
Molecular structures of selected periplasmic copper homeostasis proteins: (a) Cu(I)-bound E. coli CusF;34 (b) Cu(I)/Cu(II) bound CopC.64
CusCBA is representative of large proton antiporters derived from the resistance–nodulation–cell division (RND)/heavy metal efflux (HME) superfamily. It consists of the transmembrane efflux pump CusA, a periplasmic membrane fusion protein CusB, and an outer membrane β-barrel CusC, conforming to a 3:6:3 CusA/CusB/CusC complex with pseudo- 3-fold rotational symmetry.31 The crystal structures of all components of the CusCBA complex are now known, and its structure and mechanism have recently been reviewed in detail.31,32
E. coli CusF is a Cu(I) binding protein that adopts an oligonucleotide/oligosaccharide-binding (OB)-fold (Figure 5a) whose genetic deletion results in a copper sensitivity phenotype.33 Structural and spectroscopic studies on Cu(I)–CusF reveal a methionine-rich Met2-His Cu(I) coordination complex that is capped by a cation−π interaction with a nearby tryptophan.34 CusF is a bona fide periplasmic metallochaperone as documented by X-ray absorption spectroscopy (XAS), where the distinct Cu(I) EXAFS signatures of the Met2-His site of CusF and Met3 site of the N-terminal region of the CusB were exploited to uncover an equimolar distribution of Cu(I) between CusF and CusB, suggesting that these two sites have similar Cu(I) affinities.35 These two proteins interact, and direct Cu(I) transfer is proposed to be dependent on an interaction between the N-terminal region of CusB and CusF.36 Metalated CusF has also been shown to deliver copper to CusB in the context of CusCBA through a specific protein–protein interaction.36 Although in vitro measurements suggest that Cu(I) transfer or equilibration is bidirectional, this equilibrium would likely be shifted by active translocation of Cu(I) by CusCBA antiporter with the net flow of Cu(I) toward the extracellular space (Figure 2).35 A more recent trielement XAS study exploiting protein-specific selenomethione labeling and Se, Ag, and Cu XAS further clarifies the role of CusB and suggests that Cu(I)–CusB activates Cu(I) transfer from CusF to CusA into a novel S2(O/N) Cu(I) site formed by two methionines and one glutamate.37 CusA, CusB, and CusF continue the theme of Met-rich Cu(I) coordination complexes in the more oxidizing periplasm because they are less susceptible to oxidation than the cysteine-rich coordination sites that dominate in the cytoplasm.38
Since CusF is a soluble periplasmic protein and cells may want to restrict the concentrations of toxic Cu(I) in this compartment, how CusF is metalated for delivery to the CusCBA antiporter is an important question. Recent experiments in E. coli suggest that CusF is efficiently Cu(I) loaded through a physical interaction with the cytoplasmic membrane copper exporting P1B-type ATPase CopA. Cu(I) transfer from CopA to CusF has been shown to be unidirectional and dependent upon ATP hydrolysis by the transporter39 and appears to require a specific, cognate CopA–CusF interaction from the same bacterial species. Immunoprecipitation experiments suggest an interaction between CopA in the E1·Cu(I) state (vide infra) and apo-CusF that is fully consistent with a model of Cu(I) trafficking to CusF during a productive CopA ATPase cycle.39 Although many questions remain, for example, what drives dissociation of Cu(I)–CusF from the transporter, as well as a full accounting of the relative rates at which Cu(I) is loaded onto CusF by CopA vs “free” Cu(I), this is an exciting finding that suggests that periplasmic domains of P1B-type ATPases may have evolved specific interaction surfaces to allow for “channeling” of copper once it is removed from the cytoplasm (Figure 2).
Salmonella enterica serovar typhimurium (S. typhimurium), a Gram negative rod-shaped bacterium, has evolved to survive in the phagolysosomes of macrophages (Figure 2) and has served as a model organism alongside the closely related E. coli for understanding copper sensing and resistance in this cellular compartment. However, S. typhimurium differs from E. coli in that it does not encode a CusCBFA system but instead encodes CueP as a major copper resistance protein; in fact, this dichotomy of Cu(I)-resistance function is preserved in all Gram negative bacteria: they harbor either the cus system under the control of CusRS40 or a CueR-regulated cueP locus.41 Although Cavet and co-workers originally identified CueP as a major Cu binding protein in copper-stressed cells,42 recent work defines the Cu stoichiometry as 1:1 and provides genetic and biochemical evidence that Cu–CueP, like CusF in E. coli, obtains Cu(I) directly from one of the two encoded P1B ATPases (CopA, GolT) and functions as a Cu(I) chaperone for the periplasmic superoxide dismutase SodCII at low [Cu]43 (Figure 2). Although many details of this exciting finding remain to be elucidated, it provides additional evidence that Cu(I) is trafficked from a cytoplasmic Cu(I) effluxer to a specific destination in the periplasm, not only mitigating the effects of cytoplasmic Cu(I) toxicity but also using the copper to reduce superoxide loads in this compartment, with clear parallels to Ybt in UPEC.
5. Copper Resistance Determinants in the Cytoplasm
Copper chaperones are found in all kingdoms of life and function as high affinity Cu(I) binding proteins that deliver their cargo to specific molecular targets. Solution NMR structures of two bacterial Cu(I) chaperones, Enterococcus CopZ and Bacillus subtilis CopZ, and crystallographic studies of S. cerevisiae Atx1 and human Atox1 in either apo-reduced, apo-oxidized or variously metallated states all reveal the same mixed α/β ferrodoxin-like fold with a solvent-exposed CXXC Cu(I) binding motif (Figure 6a).13 CopZ and related proteins, for example, cyanobacterial Atx1 (scAtx1),13 form two- or three-coordinate Cu(I) complexes that exploit the low coordination number and ligand exchange properties of Cu(I) to transiently form metal-cross-linked intermediates, facilitating Cu(I) transfer without dissociation into bulk solvent. However, recent work reveals that an obligatory requirement for a CopZ-like chaperone to deliver Cu(I) to its major target in bacteria, the Cu(I)-exporting P1B-type ATPases,44 is not fully consistent with the data,45 a finding compatible with the idea that not all bacteria encode a known Cu(I) chaperone, for example, E. coli and M. tuberculosis (Figure 2), and genetic deletion of the chaperone often has little impact on cellular copper resistance or copper allocation.45−47 Rather, bacterial CopZ-like Cu(I) chaperones play principal roles in buffering low levels of Cu(I) thereby minimizing the deleterious off-pathway impact of free Cu(I) on other metal homeostasis systems in the cell.45
Figure 6.
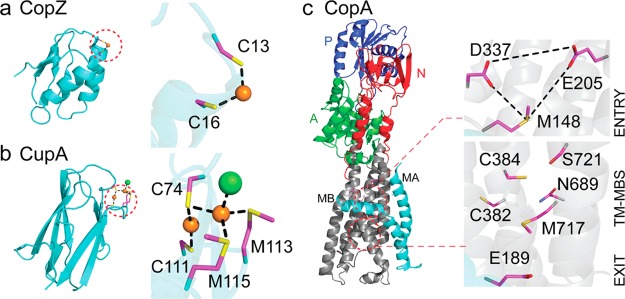
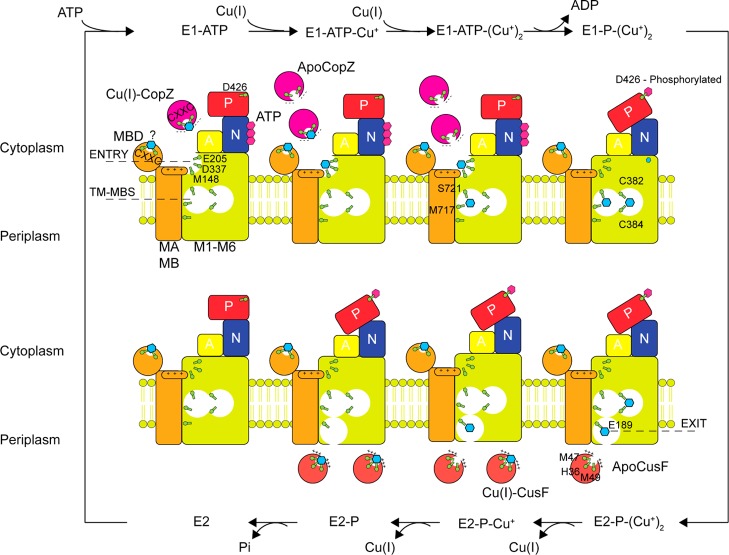
Molecular structures of two cytoplasmic Cu(I) chaperones and the apo-structure of the Cu(I)-effluxing P1B-type ATPase CopA from L. pneumophila. (a) B. subtilis CopZ, representative of the Atox1-like ferredoxin-like fold metallochaperones. The S–Cu–S bond angle of 120° suggests a third ligand from solvent to complete a trigonal coordination structure.13 (b) The Cu(I) chaperone CupA from S. pneumoniae that harbors a binuclear Cu(I) center.17 (c) L. pneumophila CopA with the proposed copper entry, transmembrane (TM-MBS), and exit sites indicated.50,51 The MA and MB helices are shaded cyan, with the actuator (A, green), nucleotide-binding (N, red) and phosphorylated (P, blue) domains also highlighted.50
A new structural class of bacterial Cu(I) chaperones, exemplified by S. pneumoniae CupA (Figure 6b), is distributed widely among lactobacilli and streptococci that lack a CopZ-like metallochaperone.17 CupA expression is induced in the lungs and nasopharynx of intranasally infected mice48 and, unlike other CopZs, is essential for cellular Cu resistance under conditions of Cu stress in liquid culture. The core domain of CupA is homologous to the N-terminal metal binding domain (MBD) of S. pneumoniae CopA, a conserved feature of other cognate chaperone–CopAMBD pairs.13 The crystal structures of the soluble Cu(I)-bound CupA (sCupA) and CopAMBD reveal isostructural cupredoxin-like folds, each harboring a binuclear Cu(I) cluster unprecedented in bacterial copper trafficking. NMR studies are consistent with unidirectional Cu(I) transfer from the low-affinity site (S2) on sCupA to the high-affinity site (S1) of CopAMBD down a thermodynamic gradient (Figure 6b). CupA localizes to the plasma membrane via a single-pass N-terminal transmembrane (TM) helix, and a Cu(I)-binding competent, membrane-localized CupA is obligatory for cellular copper resistance.17
Although much of the structural work in bacterial systems has focused on the molecular details of Cu(I) transfer from the chaperone to the CopAMBD,49 recent evidence, at least for Archaeoglobus fulgidus (Af) CopA, suggests that this transfer is not on pathway for Cu(I) efflux, but instead plays a regulatory role in allosteric activation of CopA. The first crystallographic structure of a bacterial CopA, from Legionella pneumophila (Lp), in the metal-free apo state50 reveals a core structure of six transmembrane helices, M1–M6, flanked by two CopA-unique N-terminal helices, MA and MB, and the typical P-type ATPase actuator (A), nucleotide binding (N), and phosphorylation (P) domains (Figure 6c). The N-terminal MBD, adjacent to the MA helix, was not resolved in this structure. The general mechanism for the P-type ATPase catalytic cycle is based on the Albers–Post model (Figure 7).51 Briefly, the E1·ATP state binds substrate Cu(I) to the transmembrane metal binding sites harboring the core CPC motif in TM4 (TM-MBS; Figure 6c), activating ATP hydrolysis to generate an Asp–phosphoryl intermediate E1·P (D426 in Lp CopA). This drives a conformational change that allows TM-MBS-bound Cu(I) ions to move to an “exit” site to form the E2·P state, from which Cu(I) is released into the extracellular space. Hydrolysis and release of inorganic phosphate forms the E2 state, which is recycled to the E1·ATP state upon ATP binding.51
Figure 7.
Cartoon of a mechanistic model of Cu(I) efflux across the bacterial inner membrane that summarizes recent structural and biochemical studies on CopA from E. coli, A. fulgidus and L. pneumophila. CopA is postulated to transport two Cu(I) ions per ATPase cycle. E. coli CusF has been shown to be metalated upon Cu(I) release from CopA through a protein–protein interaction.39
The structure of Lp CopA coupled with biochemical and molecular dynamics experiments in Af and Lp CopAs provide support for a continuous Cu(I) translocation pathway through the membrane involving three spatially resolved Cu(I) binding sites, termed entry, transmembrane (TM-MBS), and exit (Figure 6c). The entry site (M148, E205, and D337; Figure 6c) is buttressed by a cluster of positively charged residues, which constitute a “platform” helix as part of the kinked MB helix. The integrity of the entry site and the positive charge of the platform helix has now been experimentally validated in Af CopA,52 but only if Cu(I) bound to Af CopZ and not free Cu(I) was used to measure Cu(I)-stimulated CopA ATPase activity. This finding is consistent, albeit not confirmatory, of a transient and direct physical interaction between the cognate chaperone and the “throat” of the pump (Figure 6c) via electrostatic complementarity. Given the high Cu(I) binding affinity of CopZ, Cu(I) is postulated to move from the chaperone to the entry site via ligand exchange.13 However, a Cu(I)-trapped CopZ–CopA complex, as a presumed intermediate on the Cu(I)-transfer pathway, has not yet been characterized; in addition, given two major classes of bacterial copper chaperones, it will be interesting to assess the molecular specificity of this entry-site mediated Cu(I)-transfer process.
Cu(I) bound to the entry site is proposed to be transient50,51 and must move to the TM-MBS sites. The TM-MBSs in Af CopA characterized by XAS revealed two spectroscopically distinct Cu(I) sites involving six ligands (Figure 6c); these findings played a crucial role in identifying candidate Cu(I) ligands in the apo-Lp CopA structure.53 Molecular dynamics simulations of Lp CopA suggest that TM-MBS site I, formed by the two cysteines of the CPC motif and a tyrosine from the conserved IYNV motif in M5 (Figure 6c), is likely more important in transporting Cu(I).51 Finally, the femtomolar affinity of the TM-MBSs for Cu(I) poses an energetic barrier for Cu(I) release into the extracellular bulk solvent;53 this led to the proposal of a Cu(I) exit site involving conserved glutamate and methionine residues (Figure 6c). Simulations reveal that solvent can enter the TM region from the periplasmic side and reach Glu189 in the proposed exit site; furthermore, this solvent filled pathway is predicted to be wide enough for Cu(I) to be shuttled from the TM-MBS to the exit site with particularly important roles proposed for Cys382 and Met717 in this process (Figure 6c). Mutagenesis experiments in Lp CopA are consistent with the functional importance of the exit site, in terms of both ATP hydrolysis and cellular copper resistance.
6. Conclusion and Perspectives
This Account summarizes our increasingly sophisticated understanding of copper transport and trafficking in bacteria and how human macrophages exploit excess copper to poison invading pathogens. While this process of Cu-mediated killing is opposite to that of “nutritional immunity” in which the host attempts to withhold essential transition metal ions from the pathogen,54 both strategies reflect host efforts to manipulate metal bioavailability to eliminate bacterial infections. An ongoing area of investigation is the degree to which Cu(I) toxicity is linked to Cu(I)-induced dissociation of cognate metals ions from metalloenzymes harboring cofactors composed of poorly competitive metals, for example, Mn(II) and Fe(II), as well as the interplay between copper homeostasis and cellular redox maintenance systems, which are intrinsically linked by sulfur chemistry.55,56 Glutathione and perhaps other related LMWTs in bacterial pathogens that lack glutathione, for example, bacillithiol and mycothiol in S. aureus and M. tuberculosis, respectively, must help to buffer “free” Cu(I) to very low levels given its high affinity for Cu(I),57 since in some45 but not all58 cases, crippling glutathione biosynthesis is synergistic with the deletion of the copper chaperone with respect to Cu(I) stress resistance. Likewise, in S. pneumoniae, abrogation of the ability to uptake glutathione (Spn does not biosynthesize GSH) gives rise to a severe growth defect under copper stress.59 These studies thus generally link the redox status of the cytoplasmic compartment as defined by the GSH/GSSG couple to copper homeostasis and resistance.56
Further, because the reduced cysteines in Atox1 are responsible for coordinating Cu(I), the redox state of those cysteines is also maintained through equilibrium with the GSH/GSSG pair in human cells.55 Atox1, in turn, may functionally replace GSH under conditions of low total glutathione, consistent with the original functional description of Atox1 as an antioxidant protein.55 Further, human glutaredoxin 1 (hGrx1) has been shown to catalyze the interchange between disulfide-oxidized and reduced Atox1 as a function of the GSH/GSSG ratio.56In vitro experiments also demonstrate that hGrx1 binds Cu(I) with high affinity, indicating a potential role in Cu(I) buffering.56 The dual roles of GSH, Atox1, and Grx1 in cellular redox and copper homeostasis strongly suggest a functional linkage between these two systems. Future studies that define the chemical and physical origins of this linkage and, more generally, how copper stress impacts other kinds of thiol chemistry that might be manifest in a typical antimicrobial host response, promises new surprises at the host–pathogen interface.
Biographies
Yue Fu obtained his B.S. in Biological Sciences from the University of Science and Technology of China in 2009. He is currently a graduate student in Professor Giedroc’s group where he studies copper resistance in Streptococcus pneumoniae.
Feng-Ming James Chang obtained his B.Sc. in Chemistry from National Taiwan University in 2004 and in 2014 earned his Ph.D. with Professor Giedroc for structural studies of CsoR family copper sensors.
David P. Giedroc is a Pennsylvania State University graduate and earned his Ph.D. in Biochemistry at Vanderbilt University School of Medicine in 1984. Following postdoctoral studies at Yale University, he joined the faculty at Texas A&M University in 1988. In 2007, he joined the Department of Chemistry at Indiana University, Bloomington, where he is currently Professor and Chair of Department. He has general interests in the biophysical chemistry of infectious disease, with current projects in bacterial transition metal homeostasis, sulfur homeostasis in Staphylococcus aureus, and the replication of human coronaviruses. He was elected a Fellow of the American Association for the Advancement of Science in 2013.
We are grateful to the US National Institutes of Health (Grant GM042569) for support of our work on transition metal homeostasis.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Braymer J. J.; Giedroc D. P. Recent developments in copper and zinc homeostasis in bacterial pathogens. Curr. Opin. Chem. Biol. 2014, 19, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi K. S.; Henderson J. P. Pathogenic adaptations to host-derived antibacterial copper. Front. Cell. Infect. Microbiol. 2014, 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanovic M. I.; Ding C.; Thiele D. J.; Darwin K. H. Copper in microbial pathogenesis: Meddling with the metal. Cell Host Microbe 2012, 11, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino J. T.; Franz K. J. Coordination chemistry of copper proteins: How nature handles a toxic cargo for essential function. J. Inorg. Biochem. 2012, 107, 129–143. [DOI] [PubMed] [Google Scholar]
- Macomber L.; Rensing C.; Imlay J. A. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 2007, 189, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L.; Imlay J. A. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 8344–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillappagari S.; Seubert A.; Trip H.; Kuipers O. P.; Marahiel M. A.; Miethke M. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J. Bacteriol. 2010, 192, 2512–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouzi A.; Steunou A. S.; Durand A.; Khalfaoui-Hassani B.; Bourbon M. L.; Astier C.; Bollivar D. W.; Ouchane S. Coproporphyrin III excretion identifies the anaerobic coproporphyrinogen III oxidase HemN as a copper target in the Cu(+)-ATPase mutant copA(−) of Rubrivivax gelatinosus. Mol. Microbiol. 2013, 88, 339–351. [DOI] [PubMed] [Google Scholar]
- Djoko K. Y.; McEwan A. G. Antimicrobial action of copper is amplified via inhibition of heme biosynthesis. ACS Chem. Biol. 2013, 8, 2217–2223. [DOI] [PubMed] [Google Scholar]
- Ramos-Montanez S.; Kazmierczak K. M.; Hentchel K. L.; Winkler M. E. Instability of ackA (acetate kinase) mutations and their effects on acetyl phosphate and ATP amounts in Streptococcus pneumoniae D39. J. Bacteriol. 2010, 192, 6390–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutsenko S. Human copper homeostasis: A network of interconnected pathways. Curr. .Opin. Chem. Biol. 2010, 14, 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae T. D.; Schmidt P. J.; Pufahl R. A.; Culotta V. C.; V. O’Halloran T. Undetectable intracellular free copper: The Requirement of a copper chaperone for superoxide dismutase. Science 1999, 284, 805–808. [DOI] [PubMed] [Google Scholar]
- Boal A. K.; Rosenzweig A. C. Structural biology of copper trafficking. Chem. Rev. 2009, 109, 4760–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z.; Loughlin F.; George G. N.; Howlett G. J.; Wedd A. G. C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: Sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking. J. Am. Chem. Soc. 2004, 126, 3081–3090. [DOI] [PubMed] [Google Scholar]
- Maryon E. B.; Malloy S. A.; Kaplan J. H. Cellular glutathione plays a key role in copper uptake mediated by human copper transporter 1. Am. J. Physiol.: Cell Physiol. 2013, 304, C768–C779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A.; Unger V. Atox1 contains positive residues that mediate membrane association and aid subsequent copper loading. J. Membr. Biol. 2013, 246, 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.; Tsui H.-C. T.; Bruce K. E.; Sham L.-T.; Higgins K. A.; Lisher J. P.; Kazmierczak K. M.; Maroney M. J.; Dann C. E.; Winkler M. E.; Giedroc D. P. A new structural paradigm in copper resistance in Streptococcus pneumoniae. Nat. Chem. Biol. 2013, 9, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C.; Lee J.; Kambe T.; Fritsche K.; Petris M. J. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 2009, 284, 33949–33956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X.; Festa R. A.; Ioerger T. R.; Butler-Wu S.; Sacchettini J. C.; Darwin K. H.; Samanovic M. I. The copper-responsive RicR regulon contributes to Mycobacterium tuberculosis virulence. mBio 2014, 5, e00876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschendorf F.; Ackart D.; Shrestha T. B.; Hascall-Dove L.; Nolan S.; Lamichhane G.; Wang Y.; Bossmann S. H.; Basaraba R. J.; Niederweis M. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 1621–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz M.; Fritsche G.; Brunner P.; Talasz H.; Hantke K.; Weiss G. Interferon-gamma limits the availability of iron for intramacrophage Salmonellta typhimurium. Eur. J. Immunol. 2008, 38, 1923–1936. [DOI] [PubMed] [Google Scholar]
- Chaturvedi K. S.; Hung C. S.; Crowley J. R.; Stapleton A. E.; Henderson J. P. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 2012, 8, 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi K. S.; Hung C. S.; Giblin D. E.; Urushidani S.; Austin A. M.; Dinauer M. C.; Henderson J. P. Cupric yersiniabactin is a virulence-associated superoxide dismutase mimic. ACS Chem. Biol. 2014, 9, 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason J. E.; Galaleldeen A.; Peterson R. L.; Taylor A. B.; Holloway S. P.; Waninger-Saroni J.; Cormack B. P.; Cabelli D. E.; Hart P. J.; Culotta V. C. Candida albicans SOD5 represents the prototype of an unprecedented class of Cu-only superoxide dismutases required for pathogen defense. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 5866–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo L.; Toro I.; D’Orazio M.; O’Neill P.; Pedersen J. Z.; Carugo O.; Rotilio G.; Battistoni A.; Djinovic-Carugo K. Unique features of the sodC-encoded superoxide dismutase from Mycobacterium tuberculosis, a fully functional copper-containing enzyme lacking zinc in the active site. J. Biol. Chem. 2004, 279, 33447–33455. [DOI] [PubMed] [Google Scholar]
- Sartain M. J.; Belisle J. T. N-Terminal clustering of the O-glycosylation sites in the Mycobacterium tuberculosis lipoprotein SodC. Glycobiology 2009, 19, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer A.; Rowland J. L.; Haeili M.; Niederweis M.; Wolschendorf F. Porins increase copper susceptibility of Mycobacterium tuberculosis. J. Bacteriol. 2013, 195, 5133–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillappagari S.; Miethke M.; Trip H.; Kuipers O. P.; Marahiel M. A. Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. J. Bacteriol. 2009, 191, 2362–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C. L.; Grass G.; Rensing C. Copper toxicity and the origin of bacterial resistance-new insights and applications. Metallomics 2011, 3, 1109–1118. [DOI] [PubMed] [Google Scholar]
- Ma Z.; Jacobsen F. E.; Giedroc D. P. Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 2009, 109, 4644–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F.; Su C. C.; Lei H. T.; Bolla J. R.; Do S. V.; Yu E. W. Structure and mechanism of the tripartite CusCBA heavy-metal efflux complex. Philos. Trans. R. Soc. London, Ser. B: Biol. Sci. 2012, 367, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealman T. D.; Blackburn N. J.; McEvoy M. M. Metal export by CusCFBA, the periplasmic Cu(I)/Ag(I) transport system of Escherichia coli. Curr. Top. Membr. 2012, 69, 163–196. [DOI] [PubMed] [Google Scholar]
- Franke S.; Grass G.; Rensing C.; Nies D. H. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 2003, 185, 3804–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y.; Davis A. V.; Balakrishnan G.; Stasser J. P.; Staehlin B. M.; Focia P.; Spiro T. G.; Penner-Hahn J. E.; O’Halloran T. V. Cu(I) recognition via cation-π and methionine interactions in CusF. Nat. Chem. Biol. 2008, 4, 107–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagai I.; Rensing C.; Blackburn N. J.; McEvoy M. M. Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry 2008, 47, 11408–11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealman T. D.; Bagai I.; Singh P.; Goodlett D. R.; Rensing C.; Zhou H.; Wysocki V. H.; McEvoy M. M. Interactions between CusF and CusB identified by NMR spectroscopy and chemical cross-linking coupled to mass spectrometry. Biochemistry 2011, 50, 2559–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon K. N.; Mealman T. D.; McEvoy M. M.; Blackburn N. J.. Tracking metal ions through a Cu/Ag efflux pump assigns the functional roles of the periplasmic proteins. Proc. Natl. Acad. Sci. U. S. A. 2014, in press [DOI] [PMC free article] [PubMed]
- Davis A. V.; O’Halloran T. V. A place for thioether chemistry in cellular copper ion recognition and trafficking. Nat. Chem. Biol. 2008, 4, 148–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Benavides T.; George Thompson A. M.; McEvoy M. M.; Arguello J. M. Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: The interaction of Escherichia coli CopA and CusF. J. Biol. Chem. 2014, 289, 20492–20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipaty S. A.; Larsen A. S.; Rensing C.; McEvoy M. M. Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol. Lett. 2012, 330, 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontel L. B.; Soncini F. C. Alternative periplasmic copper-resistance mechanisms in Gram negative bacteria. Mol. Microbiol. 2009, 73, 212–225. [DOI] [PubMed] [Google Scholar]
- Osman D.; Waldron K. J.; Denton H.; Taylor C. M.; Grant A. J.; Mastroeni P.; Robinson N. J.; Cavet J. S. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J. Biol. Chem. 2010, 285, 25259–25268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman D.; Patterson C. J.; Bailey K.; Fisher K.; Robinson N. J.; Rigby S. E. J.; Cavet J. S. The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P1B-type ATPase copper efflux and periplasmic CueP. Mol. Microbiol. 2013, 87, 466–477. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guerrero M.; Arguello J. M. Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 5992–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey S.; Patterson C. J.; Banci L.; Bertini I.; Felli I. C.; Pavelkova A.; Dainty S. J.; Pernil R.; Waldron K. J.; Foster A. W.; Robinson N. J. Cyanobacterial metallochaperone inhibits deleterious side reactions of copper. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 95–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford D. S.; Kihlken M. A.; Borrelly G. P.; Harwood C. R.; Le Brun N. E.; Cavet J. S. CopZ from Bacillus subtilis interacts in vivo with a copper exporting CPx-type ATPase CopA. FEMS Microbiol. Lett. 2003, 220, 105–112. [DOI] [PubMed] [Google Scholar]
- Corbett D.; Schuler S.; Glenn S.; Andrew P. W.; Cavet J. S.; Roberts I. S. The combined actions of the copper-responsive repressor CsoR and copper-metallochaperone CopZ modulate CopA-mediated copper efflux in the intracellular pathogen Listeria monocytogenes. Mol. Microbiol. 2011, 81, 457–472. [DOI] [PubMed] [Google Scholar]
- Shafeeq S.; Yesilkaya H.; Kloosterman T. G.; Narayanan G.; Wandel M.; Andrew P. W.; Kuipers O. P.; Morrissey J. A. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 2011, 81, 1255–1270. [DOI] [PubMed] [Google Scholar]
- Banci L.; Bertini I.; McGreevy K. S.; Rosato A. Molecular recognition in copper trafficking. Nat. Prod. Rep. 2010, 27, 695–710. [DOI] [PubMed] [Google Scholar]
- Gourdon P.; Liu X. Y.; Skjorringe T.; Morth J. P.; Moller L. B.; Pedersen B. P.; Nissen P. Crystal structure of a copper-transporting PIB-type ATPase. Nature 2011, 475, 59–64. [DOI] [PubMed] [Google Scholar]
- Andersson M.; Mattle D.; Sitsel O.; Klymchuk T.; Nielsen A. M.; Moller L. B.; White S. H.; Nissen P.; Gourdon P. Copper-transporting P-type ATPases use a unique ion-release pathway. Nat. Struct. Mol. Biol. 2014, 21, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Benavides T.; McCann C. J.; Argüello J. M. The mechanism of Cu+ transport ATPases: Interaction with Cu+ chaperones and the role of transient metal-binding sites. J. Biol. Chem. 2013, 288, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guerrero M.; Eren E.; Rawat S.; Stemmler T. L.; Arguello J. M. Structure of the two transmembrane Cu+ transport sites of the Cu+-ATPases. J. Biol. Chem. 2008, 283, 29753–29759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood M. I.; Skaar E. P. Nutritional immunity: Transition metals at the pathogen–host interface. Nat. Rev. Microbiol. 2012, 10, 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori Y.; Clasen S.; Hasan N. M.; Barry A. N.; Lutsenko S. Functional partnership of the copper export machinery and glutathione balance in human cells. J. Biol. Chem. 2012, 287, 26678–26687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose J.; La Fontaine S.; Wedd A. G.; Xiao Z. Redox sulfur chemistry of the copper chaperone Atox1 is regulated by the enzyme glutaredoxin 1, the reduction potential of the glutathione couple GSSG/2GSH and the availability of Cu(I). Metallomics 2014, 6, 793–808. [DOI] [PubMed] [Google Scholar]
- Banci L.; Bertini I.; Ciofi-Baffoni S.; Kozyreva T.; Zovo K.; Palumaa P. Affinity gradients drive copper to cellular destinations. Nature 2010, 465, 645–648. [DOI] [PubMed] [Google Scholar]
- Helbig K.; Bleuel C.; Krauss G. J.; Nies D. H. Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol. 2008, 190, 5431–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter A. J.; Trappetti C.; Paton J. C. Streptococcus pneumoniae uses glutathione to defend against oxidative stress and metal ion toxicity. J. Bacteriol. 2012, 194, 6248–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland J. L.; Niederweis M. A multicopper oxidase is required for copper resistance in Mycobacterium tuberculosis. J. Bacteriol. 2013, 195, 3724–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S. K.; Abomoelak B.; Hoye E. A.; Steinberg H.; Talaat A. M. CtpV: A putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol. Microbiol. 2010, 77, 1096–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changela A.; Chen K.; Xue Y.; Holschen J.; Outten C. E.; O’Halloran T. V.; Mondragon A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 2003, 301, 1383–1387. [DOI] [PubMed] [Google Scholar]
- Chang F. M.; Coyne H. J.; Cubillas C.; Vinuesa P.; Fang X.; Ma Z.; Ma D.; Helmann J. D.; Garcia-de Los Santos A.; Wang Y. X.; Dann C. E. 3rd; Giedroc D. P. Cu(I)-mediated allosteric switching in a copper-sensing operon repressor (CsoR). J. Biol. Chem. 2014, 289, 19204–19217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Koay M.; Maher M. J.; Xiao Z.; Wedd A. G. Intermolecular transfer of copper ions from the CopC protein of Pseudomonas syringae. Crystal structures of fully loaded Cu(I)Cu(II) forms. J. Am. Chem. Soc. 2006, 128, 5834–5850. [DOI] [PubMed] [Google Scholar]