Abstract
The importance of Chromatin Immunoprecipitation (ChIP) technology has grown exponentially along with an increased interest in epigenetic regulation. The correlation of transcription factors with histone marks is now well established as the center of epigenetic studies; therefore, precise knowledge about histone marks is critical to unravel their molecular function and to understand their role in biological systems. This knowledge constantly accumulates and is provided openly in the expanding hubs of information such as the USCS Genome Browser. Nevertheless, as we gain more knowledge, we realize that the DNA-protein interactions are not driven by a “one size fits all” rule. Also, the diversity of interactions between DNA, histones, and transcriptional regulators is much bigger than previously considered. Besides a detailed protocol of sample preparation for the ChIP assay from primary human monocyte-derived macrophages (MDM)a, we show that differences between various types of cells exist. Furthermore, we can postulate that such variations exist between transformed macrophage-like cell lines and primary macrophages obtained from healthy volunteers. We found that the most efficient fixation time for MDM is 10 minutes. Finally, to perform multiple analytical assays, we showed that even with thorough methodology, the yield of material obtained from primary cells is the major challenge.
Keywords: Monocyte, macrophage, MDM, ChIP, histone modifications, UCSC Genome Browser
Introductionb
Epigenetics is the examination of functionally relevant changes to the living organism that do not result from changes in the nucleotide sequence in the genome. Cell differentiation is an insightful example of the importance of epigenetic regulation [1]. An example of epigenetic regulation is a fertilized egg that gives rise to an organism that consists of hundreds of different cell types, morphologically and functionally, while preserving genetic information contributed by both parents [2, 3]. The epigenetic regulation of gene expression is a highly complicated process; however, bringing to light the mechanisms that are responsible for the regulation of epigenetic modifications is imperative to advance personalized medicine and improve disease treatments and prevention.
Chromatin Immunoprecipitation (ChIP) is an epigenetic tool that compares transcription factors or modifications of DNA-bound proteins (e.g. histones) to specific genes across the whole genome [4]. An important feature of the ChIP assay is the fixation of protein-DNA interactions and histone post-translational modifications (PTM) in the cells, which reflect the influences on gene expression by capturing the state of regulation at a given point in time. Therefore, the importance of understanding epigenetic regulation extends beyond developmental biology into other biomedical fields, such as immune responses and toxicity. As such, in this study, we focus on the adoption and adaptation of methods to study epigenetic regulation of the primary, human monocyte-derived macrophages (MDM). This is a cell type that plays an integral role in the innate immunity system and also promotes crosstalk between innate immune cells and adaptive immune cells [5].
Macrophages originate from pro-monocytes and have high phenotypic plasticity to accommodate their need to rapidly respond to changes in the surrounding environment. For example, in response to inflammation, Ly-C6+ monocytes differentiate into inflammatory macrophages that can further polarize into specific phenotypes, such as M1 or M2 macrophages [6, 7]. A primary function of the macrophage is to survey the organism and respond quickly to pro-inflammatory signals, infection, or toxic insults to maintain homeostasis. Macrophages respond to such stimuli by secreting cytokines and producing reactive oxygen species, quinolinic acid, glutamate, arachidonic acid, and its metabolites, etc. contributing to the overall response of the innate immunity system [8, 9], all of which are regulated by epigenetic mechanisms. Therefore, abnormalities in epigenetic regulatory processes can be detrimental for their functions. Despite decades of research, we are still far from an in-depth understanding of the complex regulatory mechanisms by which pathogens and toxins contribute to a wide range of impairments of the innate immune system.
A more comprehensive understanding of the mechanisms controlling epigenetic regulation can be obtained by integrating multiple technologies, such as mass spectrometric analysis of histone PTM, profiling transcription regulatory expression and activity, investigating DNA methylation, and high-throughput sequencing of immunoprecipitated DNA fragments. Each of these technologies can provide unique information; however, the challenges that must be overcome can be daunting and not always obvious. In our research, we utilized both mass spectrometry-based quantification of histone PTMs as well as ChIP. We determined that the limited yield of DNA that can be efficiently isolated from primary cells is a severe impediment, but not impossible. Cell lines can provide abundant amounts of biological material as compared to primary cells obtained from human subjects.
Investigations into epigenetic regulation of macrophages have primarily used transformed macrophage-like cell lines, including THP-1, U937, and HL-60. Although beneficial, the findings of these studies have limited translatability to primary cells. While the UCSC Genome Browser provides the histone modifications for cells lines included in the Encyclopedia of DNA Coding Elements (ENCODE), it is also a great guideline for experimental design to determine the antibodies against PTMs for immunoprecipitation (IP). Nevertheless, we have identified differences between cell lines and primary MDM that need to be further characterized in future studies.
MDM are an excellent experimental model when looking at specific genes or globally to investigate specific pathways of regulation or to provide a much broader picture of intracellular mechanisms [10]. MDM from human donors reflect the in vivo situation more closely than transformed cell lines. Another important aspect of using primary cells from human donors is the ability to observe donor-to-donor variation and translate this information to broader conclusions about the function of specific mechanisms in diverse populations.
Despite numerous articles and book chapters describing methods used in biomedical research, ChIP techniques require optimization because of the experimental models and approaches, as well as the ever-increasing availability of new reagents and ready to use kits. Additionally, since most research employs cell lines, development of analytical methods to investigate primary cells is lagging. Therefore, we present detailed methodology to obtain DNA from MDM using ChIP techniques. Although this technique seems relatively straightforward and despite of continuous progress [11], the limited amount of immunoprecipitated DNA is the biggest constraint. In this study, we describe our approach for modifying the protocol from Active Motif, one of the kits currently available. Because our study is focused on ChIP-PCR, only a select number of genes were investigated.
1. Methodology
ChIP is a multi-step procedure that requires optimization for each cell type or tissue. In this study, the ChIP-IT Express Enzymatic Kit (Active Motif) was used as the basis for development of our own protocol aimed at using human MDM. The experimental scheme including the original manufacturer’s protocol are provided in Fig. 1. Specific modifications are described in the text.
Figure 1. Flow Chart of Methodology.
General steps for ChIP that can be adapted for multiple experimental models.
2. Materials
2.1 Solution and Buffer Formulations
-
-
Macrophage Serum Free Media (MSFM 1x, Life Technologies, Grand Island, NY) supplemented with 1x HEPES buffer (Invitrogen), 1x Nutridoma (Roche), and 10 ng/ ml MCSF (macrophage colony stimulating factor) (Preprotech, Inc.)
-
-
Lysis Buffer (LBuf; in-house): Final concentration of the following will be 25 mM Hepes (pH 7.8), 10 mM KCl, 1.5 mM MgCl2, 0.1% Igepal CA-630, 5 mM Sodium Butyrate and 5 µl 20X PIC plus 5 µl 100 mM PMSF (phenylmethanesulfonyl fluoride) for the total volume 100 µl.
-
-
5X shearing buffer (SBuf; in-house): 0.5% Sodium Dodecyl Sulfate (SDS), 0.05 M EDTA, 0.25 M Tris and 5 µl 20X PIC (0.2X final) plus 5 µl 100 mM PMSF (0.1 mM final), pH 8.0 to reach final volume of 500 µl.
-
-
In-house Buffer 1: 100 mM NaHCO3 and 0.5% SDS
-
-
In-house Buffer 2: 0.3125 M NaCl
2.2 Cell Culture
Human monocytes were collected by leukopheresis and purified by elutriation from healthy donors, who are sera negative for HIV-1, −2, and Hepatitis B [12]. All cells were collected from individuals that had provided written informed consent on research protocols approved by the UNMC Institutional Review Board. Monocytes were plated in MSFM, at a final density of 1 × 106 cells per mL in 100-mm tissue culture treated dishes (BD Falcon, San Jose, CA). In our experience, using the aforementioned density of monocytes for plating prevents overcrowding, which as a result of lack of space can lead to significant cell loss. Cells were maintained at 37 °C and 5% CO2 and left undisturbed for three days to promote cell adherence. Fresh media was added on day three post-plating, and a half-media exchange was performed on day five post-plating. Using these methods, within seven days of plating, differentiated cells were phenotypically similar to macrophages (e.g. double in size, irregular shape). Differentiation of monocytes to macrophages was monitored by morphological changes as previously described by other studies [13] and us [14].
2.3 Cell Fixation and Collection
Protein-DNA interactions were preserved by fixing cells using a 1% formaldehyde solution. In order to establish the optimal fixation time for primary MDM, this fixing step was performed using a time course over 10 min. For fixation, media was removed, and cells were treated with a 1% methanol-free formaldehyde solution diluted in phosphate buffered saline (PBS). Cells were then washed using 1x PBS, and the fixation was quenched using 0.3125 M glycine for 5 min. The cells were once again washed using 1x PBS, and cells were collected in 7 mL of PBS by manual cell scraping.
2.4 Lysis
Following DNA-protein cross-linking, cells were pelleted by centrifugation at 1,250 × g for 15 min at 4 °C. At this point, for future studies, the cells can be frozen at −80 °C following an addition of 5 µl 20X protease inhibitor cocktail (PIC; 0.2X) and 5 µl 100 mM PMSF (0.1 mM) to the pellet as recommended by Active Motif. Pelleted cells were re-suspended in 5 ml of Lysis buffer (LBuf), briefly vortexed, and incubated for 30 min at 4 °C to complete cell lysis. Nuclei and cell debris were pelleted by centrifugation at 1,250 × g for 15 min at 4 °C and then the supernatant was removed. The pellet was re-suspended in 730 µl Milli-q™ H2O and 182 µl of 5X shearing buffer (SBuf), producing 910 µl 1X SBuf, pH 8.0. The chromatin fragments were released from the nucleus by incubating on ice for 10 min.
2.5 Sonication
Released chromatin fragments were then subjected to sonication to generate the DNA fragments of 200–500 base pairs (bp). This step needs to be optimized for different cell types. We used four times: 5, 10, 15 and 20 min. At minimum, the sonicator should have: an intensity of 3 watts and 2% duty cycle at 4 °C. The S-series SonoLAB Single sonicator (Covaris, Woburn, MA) was used, which utilized sonication tubes that are limited to 130 µl and were most efficient with 3–5 × 106 cells per tube. Therefore, for sonication each microtube was used seven times for each sample of 20 × 106 cells. The combined samples were then centrifuged for 10 min at 13,000 × g at 4 °C, and the supernatants were transferred to a new tube. This is called the stock solution and is comprised of soluble chromatin and was stored at −20 °C.
2.6 Chromatin Reverse Cross-linking and Efficiency of Sonication
A 50 µl aliquot of stock solution prepared as described above (see 2.5) was used to clean up chromatin by first reverse cross-linking the proteins from DNA. To begin with, 150 µl Milli-q™ H2O and 10 µl 5 M NaCl were added to the sample and heated overnight (16 hrs) at 65 °C. Then, 1 µl RNase A was added for 15 min at 37 °C. The proteins were digested by adding 10 µl of 0.5 µg/µl Proteinase K (ThermoFischer Scientific, City, State) at 42 °C for 1.5 hours. The PCR Purification Column (Qiagen, Hilden, Germany) was used to purify the DNA instead of the phenol:chloroform method recommended by Active Motif. To evade the toxic carry-over that can occur with this reagent, phenol:chloroform is avoided The protocol provided for the columns was followed with some minor alterations, which will be highlighted as the method is discussed. The Qiagen Buffer PB (PB) with pH indicator was added to the sample in a 5:1 ratio. Sodium acetate (3 M, pH 5.0) was added for a final pH of ≤ 7.5. The DNA was bound to the spin column by centrifuging at 10,000 rpm for 30 sec at room temperature. The flow-through was discarded and 750 µl Qiagen Buffer PE (PE) was added and incubated for 2 min before centrifuging at 12,000 rpm for 1 min. The flow-through was discarded, and the tubes were centrifuged again to ensure the ethanol had passed through the column. The filter tube was placed on a new 1.7 ml tube that was used to collect the eluted DNA; the filter was air dried to evaporate the ethanol from the filter. The Qiagen Buffer EB (EB) was pre-warmed to 65 °C to increase efficiency of the pull down. Although the instructions recommended 50 µl of EB, we added 20 µl to the column as one of modifications and incubated at room temperature for 5 min with shaking. The columns were centrifuged for 1 min at 18,000 × g. An additional 20 µl of EB was added to get the maximum amount of DNA extracted from the filter. The Nanodrop 8000 (ThermoFisher Scientific, Waltham, MA) was used to quantify the DNA, and approximately 400 ng was run on an agarose gel at 80 V for 70 min to evaluate shearing efficiency.
2.7 Chromatin Immunoprecipitation and Clean Up
IP was performed by using the structure of Active Motif ChIP-IT Express Enzymatic manufacturer’s protocols with modifications as described below. Each sample was divided into five aliquots for IP and for clean-up, after which samples were then recombined. In siliconized tubes, the IP reaction was set up with 130 µl of stock solution, as described in Table 1. The reaction was incubated overnight at 4 °C on an end-to-end rotator. The highest recommended amount of antibody (3 µg) was applied to each reaction (Active Motif).
Table 1.
Composition of reagents for IP setup.c
| Active Motif recommendations | |||
|---|---|---|---|
| Compontents | < 60 µl of chromatin |
> 60 µl of chromatin |
Actual |
| Protein G Magnetic Beads (µl) | 25 | 25 | 25 |
| ChIP Buffer 1 (µl) | 10 | 20 | 20 |
| Sheared Chromatin (µl) | 20–60 | 61–150 | 130 |
| 20X PIC (µl) | 1 | 2 | 2 |
| dH2O (µl) | variable | variable | 20 |
| Antibody (µg) | 1–3 | 1–3 | 3 |
| Total Volume (µl) | 100 | 200 | 200 |
Active Motif recommends these parameters for IP; we outlined the actual amounts that we used for our protocol.
After incubation, the supernatants were collected and stored in −20 °C for later studies. For the wash steps, the manufacturer’s protocol was followed. Note that the beads should not be vortexed, but inverted to wash them. While the tubes were in the magnetic stand, the beads were washed as follows: one wash of 800 µl ChIP Buffer 1 and two washes of 800 µl ChIP Buffer 2.
The chromatin was eluted from the Dynabeads® protein G beads (Life Technologies) by using 50 µl Active Motif Elution buffer AM2 and following the manufacturer’s guidelines. To test the efficacy in chromatin elution, the beads were re-suspended in 50 µl in-house buffer 1 for 15 min at room temperature with intermittent agitation. The Active Motif Reverse Cross-Link Buffer was also used to unbind the DNA to proteins as the manufacturer recommended. To compare the efficiency, 50 µl in-house buffer 2 was added to the beads to reverse cross-link the DNA and proteins. A 10 µl “input” aliquot from the stock solution is prepared by adding 88 µl ChIP Buffer 2 and 2 µl 5 M NaCl in a 0.2 ml PCR tube. All of these samples were incubated at 95 °C for 15 min in a thermocycler.
After the above step, the tubes were returned to room temperature and transferred to a 1.7 ml siliconized tube or the samples stored at −20 °C. The proteins were degraded by adding 2 µl Proteinase K to each tube and incubating at 37 °C for 1 hour.
The samples can be stored at −20 °C or can be cleaned up using the PCR Purification Column (Qiagen) as described above in section 2.6. Note, five columns were used for a 20 × 106 cell sample. The DNA is eluted for each IP individually; but once this is done, they are combined into one siliconized tube. The samples were desiccated until dried, then re-suspended in 12 µl of Milli-q™ H2O and quantified by the Qubit High Sensitivity Assay (Life Technologies).
3. Results and Discussion
3.1 Fixation Time Optimization
The fixation time is a very important step and needs to be specifically optimized to the tissue or cell and can range from 10 min for THP-1 cells [15] to as long as overnight for zebrafish adult heart cells [16]. An optimized fixation time is important to promote strong DNA-protein interactions that can prevent low DNA yield. Alternatively, if excess fixation occurs, the antigens can be shielded from the antibody by the aggregates (Diagenode, Denville, NJ). Protein-DNA cross-linking was optimized by fixing for 1, 2.5, and 10 min. We found that the different fixation times yield the same amount of DNA for the in-house buffers (Table 2). There is a significant difference between the DNA yield of Active Motif buffers used in the elution steps and the in-house buffers as outlined above in section 2.7. The Student’s t-test was used to demonstrate that there is a difference between the buffers for the 10 min fixation. Our parameters were: two tailed test with our samples as equal variance. The average values for the two different buffers are shown in Table 2; these are from three different donors for each buffer: #1021, 1018, 1010 for the Active Motif buffers and #1010, 1022, 1004 for the in-house buffers. For the Active Motif Buffers, the average DNA concentration was 0.434, 0.522, and 0.406 ng/µl for the fixation time of 1, 2.5, and 10 min respectively. The in-house buffers are more effective at obtaining more DNA after IP. Again, the 10 min fixation time was significant between the two kinds of buffers. The average DNA concentration was 1.663, 1.28, and 1.47 ng/µl for the fixation times of 1, 2.5, and 10 min respectively. The PCR results indicate that 2.5 and 10 min have the same or better amplification than 1 min for the cytokine genes, and 10 min had lower standard deviation of yield between donors. Therefore, we concluded that 10 min of fixation should be used for MDM. DNA amounts needed for our downstream experiments were sufficient for downstream assays such as PCR and ChIP-Sequencing (ChIP-Seq).
Table 2.
Comparison of DNA yield (ng/µl) for commercial and in-house buffersa.
| Time Min. |
Active Motif Buffer (average) |
St. Dev. | In-House Buffer (average) |
St. Dev. (+/−) |
|---|---|---|---|---|
| 1 | 0.434 | 0.195 | 1.663 | 0.878 |
| 2.5 | 0.522 | 0.315 | 1.280 | 0.485 |
| 10 | 0.406 | 0.054 | 1.470 | 0.339 |
- Two buffers, one provided by Active Motif, Elution BufferAM2 and Reverse Cross-linking Buffer and one made in-house 1: 100 mM NaHCO3 and 2: 0.5% SDS, and in-house buffer 2: 0.3125 M NaCl were compared. Each set of time points is from the same donor, but three different donors were used to get multiple biological replicates for each buffer.
3.2 DAPI visualization for Lysis
The lysis of primary cells needs to be optimized because MDM require more SDS to break open the cell membrane than transformed cell lines. Our LBuf is efficient at releasing the DNA from the nucleus as visualized by lack of DAPI staining in Fig. 2B.
Figure 2. DAPI Staining before and after lysis.
Panel A: DAPI staining of intact MDM at a 40X magnification before adding lysis buffer. Panel B: The released DNA after lysis at 63X magnification. DAPI stains the A-T rich regions in the DNA.
3.3 Sonication
The shearing of chromatin into fragments is assessed by the smear pattern between 200 and 500 bp on an agarose gel. The shearing time point that gives the best smear between 200 and 500 bp on an agarose gel was between 10 and 15 min (Fig. 3). For future experiments, 12 min was used to shear chromatin. DNA must be sheared into fragments in order to have an efficient ChIP-Seq. The fragments need to be one to two nucleosomes long because then they are short enough to have good enrichment resolution and specificity, and long enough to have an efficient alignment for sequencing.
Figure 3. Agarose Gel for Varying Sonication Times.
The sonication time needs to be optimized to get the majority of DNA sheared between 200–500 bp long. The sonication times are 5, 10, 15, and 20 min. The optimal time is within 10 and 15 min; therefore, we chose 12 min to use for future samples. This time is best because the fragments of DNA are the correct length to make downstream application the most efficient. The 200–500 bp length is especially important for ChIP-Seq because of the conditions needed to effectively map and specify the sequence that is associated with this histone modification.
3.4 Immunoprecipitation
Different trials of IP were completed with antibodies of H3K9ac (ab10812), H3K14ac (ac46984), RNA pol II (Active Motif 39097), and negative control (Active Motif ChIP-IT Control q-PCR cat. 53026). The DNA yield is compared between the input or clean-up, IP, and the supernatants as well as with each of the antibodies in Table 3. The reaction continued overnight (16 hours) on an end-to-end rotator at 4 °C. We also experimented with other adjustments to our protocol. The antibody and sample were pre-blocked for 4 hours before the beads were applied for an overnight incubation. Additionally, the chromatin was diluted to see if that would help obtain a higher DNA yield after IP. Initially, 2 µg of H3K9ac antibody was used for IP; however, only 1–3 ng of DNA could be obtained for one sample of 20 × 106 MDM. We then increased the amount of H3K9ac antibody to 3 µg, which is well within the capacity of the Dynabeads® and within the parameters set by Active Motif. With this increase, we determined that there was more DNA being bound. For downstream projects, such as ChIP-Seq, the Epigenetic Core at University of Nebraska Medical Center only requires samples with 12 ng of DNA to begin the process. The different fixation times yielded a total of 14.7–16.6 ng of DNA per 20 × 106 MDM.
Table 3.
DNA yield and antibody efficiency. d
| Antibody | H3K9ac (10 min fixation) | H3K14ac | RNA pol II | Negative | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Clean Up (50 ul) | IP | Sup* | Clean Up (50 ul) | IP | Sup | Input (10 ul) | IP | Sup | Input (10 ul) | IP | Sup |
| DNA (ng/ul) | 47.3 | 1.23 | 16.6 | 48.6 | 0.057 | 44.55 | 24.4 | 1.475 | 55.72 | 24.4 | 0.84 | 50.68 |
| DNA (ng) | 1892 | 12.3 | 664 | 1944 | 0.57 | 1782 | 976 | 14.75 | 2229 | 976 | 8.4 | 2027 |
| Theoretical per IP ug | 5.12 | 1.80 | 5.27 | 4.83 | 6.04 | 5.49 | ||||||
Four different antibodies have been utilized for IP. H3K9ac, (ab10812) H3K14ac (ac46984), RNA pol II (Active Motif 39097), and a negative antibody (Active Motif 53026) were used to pull down DNA from MDM. The samples are from the clean-up step (Section 2.6), or input (Section 2.7), the IP, and the supernatant. The clean-up and input samples are directly from the stock solution of chromatin; they are simply different proportions of the stock solution. The concentrations for the input and IP samples were measured by the Qubit®, while the clean-up and supernatant samples were measured by the NanoDrop. Theoretical per IP is the amount of DNA that could be obtained from each IP tube; there are six tubes per 20 ×106 MDM.
Sup- Supernatant
3.5 Polymerase Chain Reaction (PCR)
PCR was performed on IP samples. The flow-through from the IP and what did not bind to the antibody of choice will be called the supernatant. We chose to look at the UCSC Genome Browser and selected the genes from secreted cytokines of macrophages. In UCSC, we found the gene promoter and used a span of ~1000 bp long to input into Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/). We also selected areas that were non-repeating sequences to create the most effective primer set. Other relevant settings for Primer3 include: a PCR product size of about 200 bp, primer size of 23 bp, melting temperature of 60 °C, and the nucleotide GC percentage of 35–40%. There are several options that are given from this database and those can be put into Integrated DNA Technologies (IDT, Inc.) in order to look at the quality of the primer set. IDT can provide the maximum delta G for a given set of primers; we wanted this parameter to be less than −42 kcal/mole. Also, the delta G of a given primer dimer was assessed; the goal was to minimize this number. The average delta G was −5.5 kcal/mole; as the number gets closer to zero, it is less likely that the primer dimer will form. A stock solution of 100 µM of each primer was made, and the NanoDrop microvolume UV/VIS instrument was used to quantify them. The best method to determine the concentration was to use the absorbance and then find the concentration from the specification sheet provided by IDT. We then diluted a portion of the stock sample to 5 µM. We ran a gradient PCR for each set of primers at 60 °C ± 8 °C in order to determine the best annealing temperature. Also, we used the Roche FastStart Kit (Roche, Indianapolis, IN) for our PCR experiments, and our sample was human genomic DNA. The conditions for PCR were 35 cycles at the determined annealing temperature, using a total of 25 µl for each sample as seen in Table 3.
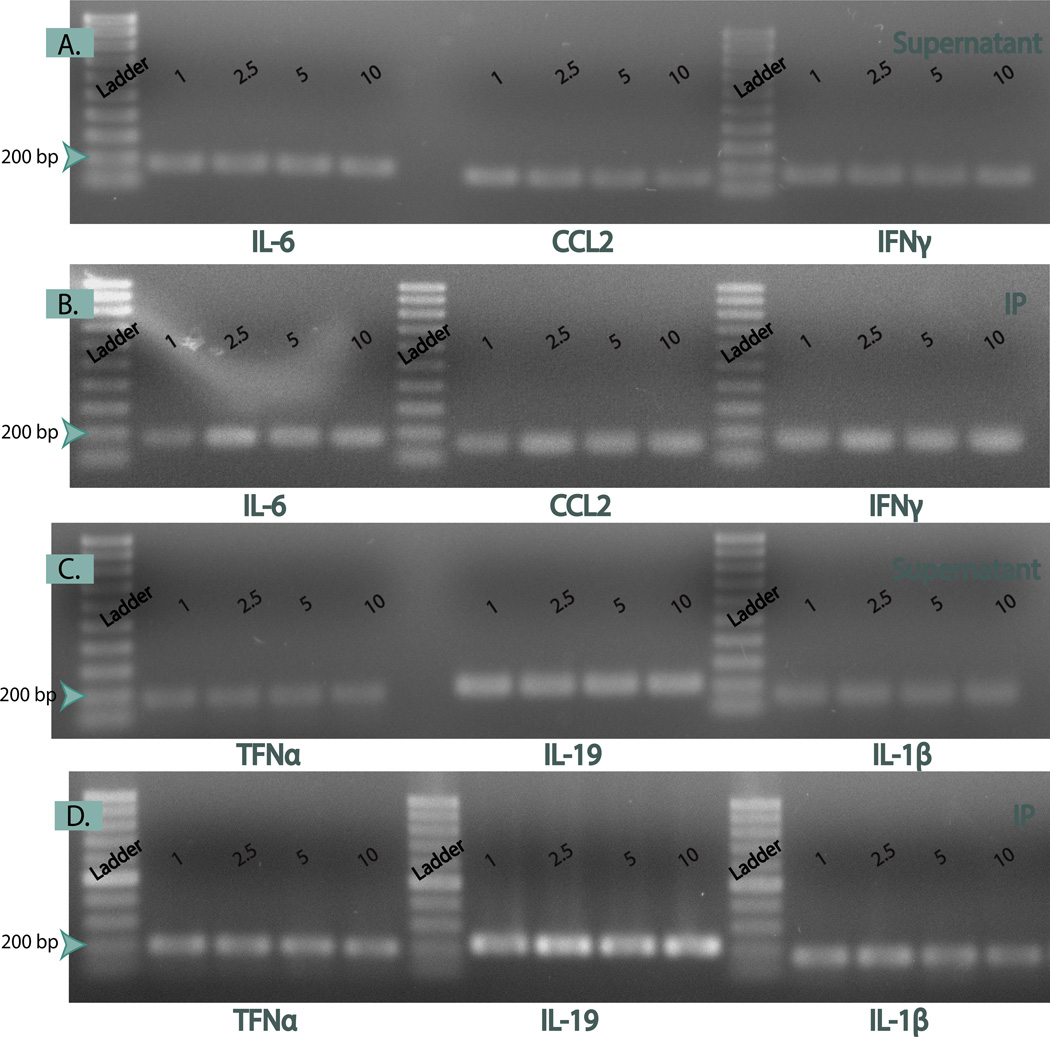
As expected, in ChIP-PCR there was incomplete binding; and DNA remained in the supernatants after IP [17]. However, the higher intensities of the PCR bands indicate that more DNA was pulled down in the IP than the supernatant (Fig. 4 B & D). For the agarose gel presented in Fig. 4, the same donor (#1010) was used to eliminate donor-to-donor variability. The same amount of DNA was loaded from the bound (IP) and unbound (supernatants); thus, they do make a direct comparison.
Figure 4. PCR results for IP and Supernatants.
Detection of cytokines by immunoprecipitation for: IL-6, CCL2, IFNγ, TFNα, IL19, IL-1β at each of the time points: 1, 2.5, 5, and 10 min. Panel A & C. Cytokine detection in supernatants. Panel B & D. Cytokine detection from IP. The IP bands have a higher intensity than the Supernatant counterparts, meaning more of that gene is amplified in that sample. For 2.5, 5, and 10 min, the bands are more intense than the 1 min fixation. We conclude that 1 min is not long enough to fix the DNA-protein interactions. Because this is the same donor (#1010) for each sample and the same amount of DNA (1 ng) was used for each PCR reaction, a direct comparison can be made.
In conclusion, the PCR results demonstrated there are differences between primary MDM and cells lines in whether or not genes have histone H3K9ac marks associated with them. We selected six cytokines to analyze: Interleukin 6 (IL-6), chemokine ligand 2 (CCL-2), interferon-gamma (IFN-γ), tumor necrosis factor- alpha (TNFα), IL-19, and IL-β; the sequences can be seen in Table 4. The genome browser shows there is a strong positive correlation of the H3K9ac histone mark with IL-6 and CCL-2 promoter region. Our PCR results confirmed this association and validated our methodology. Also, the UCSC data demonstrates a positive correlation for H3K9ac modification and TNFα and IL-1β promoter regions, but the signal was weak for cell lines. Nevertheless, our results determined that both the TNFα and IL-1β promoters positively corresponded to H3K9ac in MDM. Finally, the browser data shows that there is a negative relationship between the IFNγ and IL-19 promoter regions, but our data illustrates that both have a positive correlation with the H3K9ac modification. In order to validate our results, we chose the NDNL2 gene, which is unrelated to cytokines coding genes. For NDNL2, a set of primers was selected where the promoter binds strongly to H3K9ac, according to UCSC. Another set of primers was selected from a region where the NDNL2 was not interacting with this histone mark (negative control). IP of H3K9ac associated DNA as expected showed positive amplification when primers for interacting region of NDNL2 were used and did notproduce signal when primers for non-interacting region of this gene were used (Fig. 5). PCR amplification showed positive signal for both regions in this DNA sample. This shows that if an interaction occurs, it can be visualized by PCR (Fig. 5). Instead of simple gel, qPCR can be used if the degree of interaction of specific DNA region and histone mark is necessary. In this study, we were interested in the positive v. negative result.
Table 4.
Composition of reagents for PCR.e
| Components, per reaction | Gradient PCR | ChIP-PCR |
|---|---|---|
| 10X PCR Reaction Buffer (µl) | 2.5 | 2.5 |
| 10 mM dNTPs (µl) | 0.5 | 0.5 |
| 25 mM MgCl2 (µl) | 3.5 | 3.5 |
| 5 U/µl FastStart Taq (µl) | 0.20 | 0.20 |
| DNA (ng/ul) | 11.0 | 0.33 |
| 5 µM Forward Primer (µl) | 1.5 | 1.5 |
| 5 µM Reverse Primer (µl) | 1.5 | 1.5 |
| H2O | 12.3 | 12.3 |
| Total Volume (µl) | 25.0 | 25.0 |
The Roche FastStart Kit components were used to run PCR for each of the cytokines selected. In order to find the optimal annealing temperature for each primer set, a gradient PCR reaction was completed. The gradient was 60 °C ± 8 °C using genomic DNA. The best temperature was selected by analyzing the bands on an agarose gel. The gradient PCR reactions used a significant amount of DNA (33 ng) making it easily visualized on the gel. The optimized temperature was used for subsequent ChIP-PCR reactions performed with 1 ng of DNA.
Figure 5. PCR amplification of NDNL2 gene immunoprecipitated using H3K9ac antibody.
Lane 1 shows positive amplification of the promoter region of NDNL2 after IP with anti-H3K9ac modification while lane 2 shows no amplification of NDNL2 downstream to the promoter region in the same sample. Lanes 3 and 4 show the amplification, thus the presence of both regions in samples containing isolated DNA (Sup Pos and Sup Neg). Altogether it supports specificity of binding and detection of NDNL2 promoter but no downstream region to H3K9ac histone mark.
3.6 UCSC Genome Browser
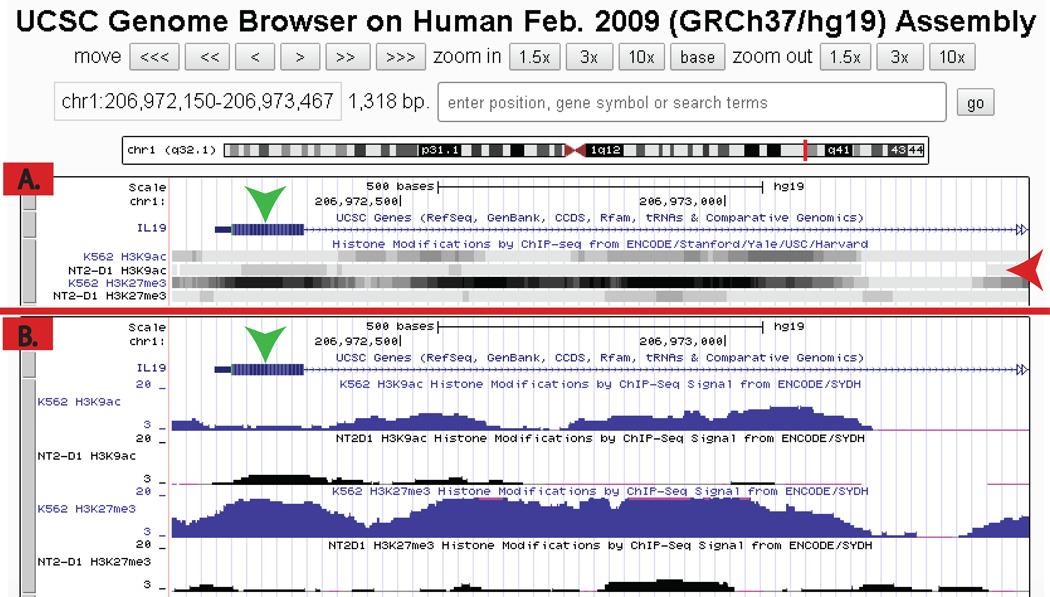
The UCSC Genome Browser is a tool that displays a comprehensive but working draft compilation of genomes and reference sequences developed and maintained by the Genome Bioinformatics Group and the Center for Biomolecular Science and Engineering (CBSE) at the University of California Santa Cruz (UCSC) (http://genome.ucsc.edu/). We used the UCSC Genome Browsers as a primary source to investigate interactions between regions of particular genes of interest with specific histone marks. In this study, we compiled a list of six cytokines (IL-6, CCL-2, IFN-γ, TNFα, IL-19, and IL-β) secreted by macrophages to search their potential interactions with histone modifications. The credentials for a search were the following: mammal, human, and Feb. 2009 (GRCh37/hg19) for the assembly. The gene of interest can be found by inputting the Uniprot ID or the protein name. This will yield the gene locus of transcript and genomic sequences. To display histone modifications, go to “Regulation” and select “show” in the ENCODE Histone Modification drop-down menu. On the “Regulation” bar, click the refresh button to display the cell lines listed by the histone modification. A right click on the corresponding bar and selecting “full” will expand to show the signal as is related to the gene. In Fig. 6 we show an example of how IL-19 is related to H3K9ac. If the modifications correlate with the promoter, then that alteration can play a part in controlling transcription. The promoter is indicated by the green arrows in Fig. 6. For example, IL-19 was used to show that there is a negative correlation between H3K9ac and the promoter. A signal that is not strong (negative) is defined in this paper as the following: either one cell line had a strong signal and the other had a weak (less than half) signal for the modification or both had a weak signal. Fig. 6 shows the difference between a strong and weak signal. For IL-19, there is a weak signal of H3K9ac for both cell lines; but for H3K27me3, the signals are not equivalent between cell lines.
Figure 6. UCSC genome data for IL-19.
K562 and NT2-D1 cells lines are compared to each other with the same histone modifications, H3K9ac and H3K27m3. Panel A. The bars for each cell line are displayed. Panel B. Displays the “full” version; the ChIP-Seq signals are visible. The data can be analyzed to see if these modifications correlate positively or negatively with the promoter. In this example, neither of the cell lines shows an association with H3K9ac. However, H3K27me3 can be seen in Panel B. It shows a strong signal for K562 cells but a weak signal for NT2-D1.
UCSC Genome Browser is primarily based on common cell types selected by ENCODE, which seeks to identify functional elements in the human genome. For a detailed description of cell lines and rationale for their selection, we refer readers to the ENCODE site (http://www.genome.gov/26524238). Briefly, ENCODE lists three tiers of cells, including more than a dozen different cell lines. Tier 1 includes a lymphoblastoid cell line, GM12878; an immortalized chronic myelogenous leukemia (CML) cell line, K562; and H1 human embryonic stem cell line. Tier 2 includes HeLa-S3, an immortalized cell line that was derived from a cervical cancer patient; HepG2, a cell line derived from a male patient with liver carcinoma; and HUVEC (human umbilical vein endothelial cells). While the rationale for including easy growing cells such as HeLa-S3 is obvious, the quantity of primary cells obtained from human subjects will be a highly limiting factor. Tier 2.5 includes an additional nine cell types: SK-N-SH, IMR90 (ATCC CCL-186), A549 (ATCC CCL-185), MCF7 (ATCC HTB-22), HMEC or LHCM, CD14+, CD20+, Primary heart or liver cells, and differentiated H1 cells. Nevertheless, most of the information pertaining to DNA interactions with modified histones is provided for K562, GM78, and NT2-D1 cells. This has to be carefully accounted for in the interpretation of experimental data obtained from other cells.
While a higher yield of DNA allows researchers to perform global (ChIP-Seq) or targeted (ChIP-PCR) tests, data interpretation of the IP step is the most challenging. As much as UCSC serves as a great guideline for determining antibodies against PTMs for immunoprecipitation, we observed differences as compared to MDM. For instance, the UCSC Genome has compiled data from seven different cells lines that look at the H3K27ac showing the differences between cell lines used for establishing this database. Not only do the cell types make a difference, but also the conditions used to prepare the chromatin alter the pull-down of proteins and needs to be considered [4].
Each of the cytokines or chemokines was evaluated in the database. These six (IL-6, CCL-2, IFN-γ, TNFα, IL-19, and IL-β) were chosen to determine the relationship of the information available through the UCSC and the data we obtained with macrophages. ChIP-PCR was carried out on the IP samples and their corresponding supernatants with these six cytokines. All of the genes were found in the supernatants and the IP (Fig. 4). The samples were run on the gel as follows: IL-6, CCl-2, IFN- γ, TNFα, IL-19, and IL- β for each fixation time of: 1, 2.5, 5 and 10 min.
Based on multiple experiments and additional quality of beads tests (data not shown), we agree with previously published observations that the quality of antibodies plays a role in the efficiency of DNA yield, which is critical factor for the success of ChIP [18]. An availability of many antibodies from various sources and their ability to efficiently IP modified histones needs to be addressed on an individual bases.
Table 5.
Sequences of PCR primers.f
| Primer | Forward primer sequence (5′-3′) | Reverse primer sequence (5′-3′) |
|---|---|---|
| IL-6 | TAGGTCTGTGAAGCTCCTTTTTG | ATTTATGATTTGGCACTTTGGTG |
| CCL-2 | CTGTGCTTCATTCACCTTAGCTT | CATTAAGACGGACCAATAATCCA |
| IFN-γ | GAGTCATTTTCAACCACAAACAA | TAATTCTCTCGGAAACGATGAAA |
| TNFα | GGGGAAGAAACAAGTTGATATGGA | TGAGGTGTCTGGTTTTCTCTCTC |
| IL-19 | CAACAAGAGTATTCACCAAGGTT | AAGCAGAAAGGAGTGACTACGTG |
| IL-1β | ATCTGGTCTCCAAGCACAGATAA | GGGAAGTCACTCATTTTCTCCTT |
| NDNL-2 Pos | CACCAAGAAGCATTTAATTTTCG | GGCCACAAACTTAAGAACTTTCA |
| NDNL-2 Neg | GGGCTTCATCTTGTTATTGTCTG | GCTGGCCAAGTTATGAATACAAG |
PCR primers were selected for cytokines from IDT. The cytokines that we tested for include the following: IL-6, CCL-2 IFN-γ, TNFα, IL-19, and IL-β. NDNL-2 was used as positive and negative primer sets to test the interaction of the H3K9ac antibody with its target area.
Acknowledgments
Financial support was provided by National Institutes of Health grants R01 DA030962 and P30 MH062261. The authors would like to thank Dr. David Klinkebiel from Epigenetics Core Facility at the UNMC for his assistance in experiments performed for this publication. We would like to thank Ms. Robin Taylor for help in editing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
MDM- Monocyte Derived Macrophages; an acceptable in-vitro model for primary, human macrophage cells.
Abbreviations: Center for Biomolecular Science and Engineering (CBSE); Chemokine ligand 2 (CCL-2); Chromatin Immunoprecipitation (ChIP); Chronic myelogenous leukemia (CML); Encyclopedia of DNA Coding Elements (ENCODE); Human umbilical vein endothelial cells (HUVEC); Immunoprecipitation (IP); Integrated DNA Technologies (IDT); Interferon-gamma (IFN-γ); Interleukin (IL); Lysis Buffer (LBuf); Macrophage Serum Free Media (MSFM); MCSF (macrophage colony stimulating factor); Monocyte-derived macrophages (MDM); Nucleotide GC (Guanine-Cytosine); Phenylmethanesulfonyl fluoride (PMSF); Phosphate buffered saline (PBS); Protease inhibitor cocktail (PIC); Qiagen Buffer EB (EB); Qiagen Buffer PB (PB); Shearing buffer (SBuf); Sodium Dodecyl Sulfate (SDS); Tumor necrosis factor- alpha (TNFα); base pairs (bp);
Literature citations
- 1.Perdigoto CN, Valdes VJ, Bardot ES, Ezhkova E. Cold Spring Harbor perspectives in medicine. 2014;4 doi: 10.1101/cshperspect.a015263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill ME, Erkek S, Peters AH. Current opinion in cell biology. 2012;24:387–396. doi: 10.1016/j.ceb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Feil R. The International journal of developmental biology. 2009;53:191–201. doi: 10.1387/ijdb.082654rf. [DOI] [PubMed] [Google Scholar]
- 4.Ku CS, Naidoo N, Wu M, Soong R. Journal of medical genetics. 2011;48:721–730. doi: 10.1136/jmedgenet-2011-100242. [DOI] [PubMed] [Google Scholar]
- 5.Mills CD. Critical reviews in immunology. 2012;32:463–488. doi: 10.1615/critrevimmunol.v32.i6.10. [DOI] [PubMed] [Google Scholar]
- 6.Lawrence T, Natoli G. Nature reviews. Immunology. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 7.Nakata K, Yamamoto M, Inagawa H, Soma G. Anticancer research. 2013;33:2849–2853. [PubMed] [Google Scholar]
- 8.McCusker RH, Kelley KW. The Journal of experimental biology. 2013;216:84–98. doi: 10.1242/jeb.073411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michallet MC, Rota G, Maslowski K, Guarda G. Current opinion in microbiology. 2013;16:296–302. doi: 10.1016/j.mib.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz M, Kipnis J, Rivest S, Prat A. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:17587–17596. doi: 10.1523/JNEUROSCI.3241-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adli M, Bernstein BE. Nature protocols. 2011;6:1656–1668. doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gendelman HE, Orenstein JM, Martin MA, Ferrua C, Mitra R, Phipps T, Wahl LA, Lane HC, Fauci AS, Burke DS, et al. The Journal of experimental medicine. 1988;167:1428–1441. doi: 10.1084/jem.167.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinet F, Dupont A, Bencherif N, Guihot AL, Quatannens B, Amouyel P. Cellular and molecular biology. 2003;49:899–905. [PubMed] [Google Scholar]
- 14.Ciborowski P, Kadiu I, Rozek W, Smith L, Bernhardt K, Fladseth M, Ricardo-Dukelow M, Gendelman HE. Virology. 2007;363:198–209. doi: 10.1016/j.virol.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Curry HM, Zwilling BS, Lafuse WP. Journal of immunology. 2005;174:5687–5694. doi: 10.4049/jimmunol.174.9.5687. [DOI] [PubMed] [Google Scholar]
- 16.Chablais F, Jazwinska A. Journal of visualized experiments : JoVE. 2012 doi: 10.3791/3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haverland N, Pottiez G, Wiederin J, Ciborowski P. Journal of translational medicine. 2010;8:137. doi: 10.1186/1479-5876-8-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, Cheung MS, Day DS, Gadel S, Gorchakov AA, Gu T, Kharchenko PV, Kuan S, Latorre I, Linder-Basso D, Luu Y, Ngo Q, Perry M, Rechtsteiner A, Riddle NC, Schwartz YB, Shanower GA, Vielle A, Ahringer J, Elgin SC, Kuroda MI, Pirrotta V, Ren B, Strome S, Park PJ, Karpen GH, Hawkins RD, Lieb JD. Nature structural & molecular biology. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]