Abstract
Haplo-insufficiency of telomerase genes in humans leads to telomere syndromes such as dyskeratosis congenital and idiopathic pulmonary fibrosis. Generation of pluripotent stem cells from telomerase haplo-insufficient donor cells would provide unique opportunities towards the realization of patient-specific stem cell therapies. Recently, pluripotent human embryonic stem cells (ntESCs) have been efficiently achieved by somatic cell nuclear transfer (SCNT). We tested the hypothesis that SCNT could effectively elongate shortening telomeres of telomerase haplo-insufficient cells in the ntESCs using relevant mouse models. Indeed, telomeres of telomerase haplo-insufficient (Terc+/−) mouse cells are elongated in ntESCs. Moreover, ntESCs derived from Terc+/− cells exhibit naïve pluripotency as evidenced by generation of Terc+/−ntESC clone pups by tetraploid embryo complementation (TEC), the most stringent test of naïve pluripotency. These data suggest that SCNT could offer a powerful tool to reprogram telomeres and to discover the factors for robust restoration of telomeres and pluripotency of telomerase haplo-insufficient somatic cells.
Keywords: Embryonic stem cells, nuclear transfer, telomerase deficiency, telomere elongation
Introduction
Mammalian telomere lengths are primarily regulated by telomerase, consisting of a reverse transcriptase protein (TERT), a RNA subunit (TERC), and stabilizing proteins including dyskerin, in a temporally and spatially dependent manner (Gunes and Rudolph, 2013). Telomerase activity is high during early embryo development, but becomes undetectable in most adult cells in humans. Mutations of genes that encode telomerase components, TERT, TERC or DKC1, cause premature aging and age-related diseases, including aplastic anemia, dyskeratosis congenital (DC) and idiopathic pulmonary fibrosis (IPF), collectively referred to as “telomere syndromes” to reflect the short and dysfunctional telomeres commonly found in these patients’ cells (Armanios and Blackburn, 2012). Heterozygous mutations of TERT and TERC represent major mutations in genetically defined telomere syndromes. Mutations of telomerase components are also associated with decreased fertility in both animals and humans (Herrera et al., 1999; Yan et al., 2014).
Stem cell failure in highly proliferative tissues, such as hematopoietic stem cells, due to progressive telomere loss caused by insufficient telomerase activity is considered as an important therapeutic target for telomere syndromes (Armanios and Blackburn, 2012). Indeed, allogeneic stem cell transplantation can reverse the aplastic anemia phenotype (Armanios and Blackburn, 2012; Young et al., 2006). However, it has been a challenge to establish patient-specific induced pluripotent stem (iPS) cells with proper restoration of telomere defects using donor cells from telomere syndrome patients (Batista et al., 2011; Winkler et al., 2013).
Efficient generation of human ntESCs by SCNT using oocyte factors represents a major advancement towards the realization of patient-specific cell therapies (Chung et al., 2014; Tachibana et al., 2013; Yamada et al., 2014). Mouse ntESCs are transcriptionally and functionally indistinguishable from ESCs generated from fertilized embryos (Brambrink et al., 2006; Wakayama et al., 2001). Human ntESCs also closely resemble ESCs (Chung et al., 2014; Tachibana et al., 2013). Moreover, mice can be re-cloned by SCNT for more than 25 generations, without accumulation of errors, and show proper telomere maintenance (Wakayama et al., 2013). Earlier this year, Le et al. (2014) reported that telomeres in Terc null mouse cells were reprogrammed after SCNT; however, they did not investigate whether or not, and to what extent the telomeres of telomerase haplo-insufficient cells, which represents the majority of genotype-defined telomere syndrome patients, can be regenerated in ntESCs. The developmental pluripotency of telomerase insufficient ntESCs also remains to be determined. We performed experiments to test whether naïve pluripotent stem cells with robust telomere elongation can be derived via SCNT from telomerase defective donor cells, using donor cells from Terc+/− mice in comparison with G2 Terc−/− mice.
Results
Development of cloned embryos and derivation of ntESCs from telomerase deficient cells
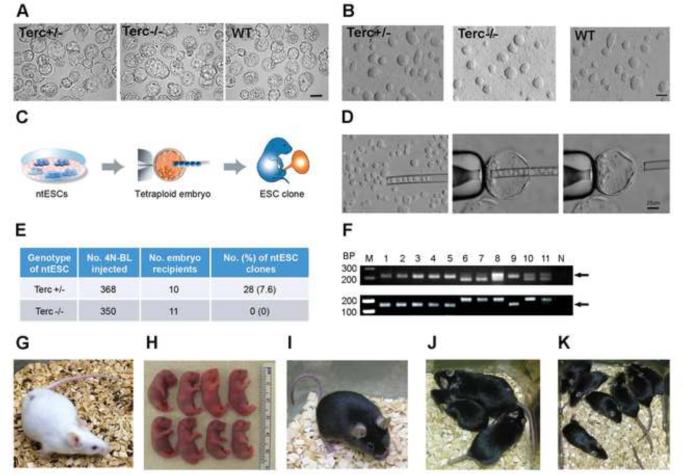
We performed SCNT using tail tip fibroblast (TTF) cells as donor cells isolated from heterozygous Terc (Terc+/−) C57BL/6 mice, in comparison with wild-type (WT) and G2 homozygous Terc KO (Terc−/−) mice. High rates of blastocyst (BL) development (Terc+/−: 43.7%; Terc−/−: 64.9% vs. WT: 73.7%) were achieved, despite telomerase deficiency (Fig. S1A). BLs of different Terc genotypes are morphologically indistinguishable (Fig. 1A).
Fig. 1. Production of all Terc+/− ntESC pups by tetraploid embryo complementation assay.
(A) Morphology of cloned embryos at blastocyst stage. Scale bar: 100 μm. (B) Morphology of ntESCs at Passage 5 under transmitted microscopy. Scale bar: 100 μm. (C) Illustration of the tetraploid embryo complementation assay. A tetraploid embryo at blastocyst stage (shown in orange) is used as the host embryo. The ntESCs (shown in blue) are injected into host embryos followed by embryo transfer. The tetraploid host embryo will only contribute to the placenta (orange), whereas all embryonic tissues are originated from ntESC (blue). (D) Microscopic images of ntESC injection in the TEC assay. Scale bar: 25 μm. (E) Success rates of ntESC pups by TEC assays. 4N-BL: tetraploid blastocyst. (F) Confirmation of specific origin of Terc+/− ntESC clones by microsatellite genotyping using D18Mit17 primers (top gel) and D12Mit136 primers (bottom gel). M: molecular marker. 1: Terc+/− donor fibroblast cells. 2: Terc+/−ntESC line#7. 3-5: pups derived from Terc+/− ntESC line#7. 6-8: placenta from Terc+/−ntESC clones. 9: WT B6 mouse. 10: ICR mouse. 11: BDF1 mouse. N: blank. Samples from Terc+/− fibroblast donor cells, Terc+/−ntESC line#7 and ntESC pups (lanes#1-5) show a single band (top arrow, ~213 bp) for D18Mit17, and a single band (bottom arrow, ~147 bp) for D12Mit136, the same pattern as WT C57BL/6 mouse (lane#9); whereas samples from placenta (lanes#6-8, ICR origin), surrogate mouse (lane#10, ICR), oocyte donor (lane#11, BDF1) show different patterns with two bands for D18Mit17 (~188 and 213 bp) and a band at ~191 bp for D12Mit136. (G) Surrogate mouse (ICR origin, albino coat and red eyes). (H) Newborn pups derived entirely from Terc+/− ntESCs (black eyes). (I) Adult Terc+/− ntESC Clone #3 (black coat and black eyes from C57BL/6 origin). (J) Three adult Terc+/−ntESC clones. (K) Pups fathered by Clone#3 after breeding with a WT C57BL/6 female. See also Fig. S1.
We next mechanically dissected the inner cell mass (ICM) from these cloned embryos and plated them on feeder cells to derive ntESCs. The efficiencies of ntESC derivation were similar among three groups (WT: 17.4%; Terc+/−: 19.1%; Terc−/−: 19.5%, Fig. S1B). These ntESC lines (Fig. 1B) showed characteristics typical of mouse ESC markers (Oct4, Nanog, Sox2 and SSEA1) by immunofluorescence microscopy (Fig. S1D), expressed pluripotent genes as measured by RT-PCR (Oct4, Sox2, Nanog and Rex1, Fig. S1E), and maintained normal karyotypes (66.6 to 86.7%), regardless of their genotypes (Fig. S1C). These results demonstrate that telomerase deficiency does not significantly compromise the in vitro development of cloned embryos and the derivation of ntESC lines.
Telomerase haplo-insufficient ntESCs show naïve pluripotency
Previously, we established WT mouse ntESCs and demonstrated that they support full-term development by tetraploid embryo complementation (TEC) (Sung et al., 2010), the most stringent test of naïve pluripotency (Jaenisch and Young, 2008). Here we tested naive pluripotency of Terc+/− and Terc−/− ntESC lines by TEC (Fig. 1C and D) to determine whether these cells are capable of supporting full-term development. We injected Terc+/− ntESCs with C57BL/6 genetic background to tetraploid embryos (n=368, ICR background) by micromanipulation and transferred the embryos to ten recipients (ICR background, Fig. 1G). Twenty-eight cloned Terc+/−ntESC pups were born, with a term-development rate of 7.6% (Fig. 1E and H), comparable to that of WT ntESCs (2%) (Sung et al., 2010), and that of Terc+/− ESCs derived from normally fertilized embryos (9%) (Huang et al., 2011). All Terc+/− pups showed C57BL/6 genetic background by microsatellite analysis, in contrast to the corresponding placentas with ICR background, confirming the clonal origin of the pups from the Terc+/− ntESCs (Fig. 1F). However, 350 tetraploid embryos injected with Terc−/− ntESCs failed to produce any pups (Fig. 1E), indicating significantly compromised in vivo pluripotency of Terc−/− ntESCs, resembling Terc−/− ESCs (Huang et al., 2011). These data show that telomerase haplo-insufficient ntESCs are capable of supporting full-term development, functionally validating their naïve pluripotent state.
Robust telomere elongation in telomerase haplo-insufficient ntESCs
Telomere lengths highly correlate with the developmental pluripotency of ESCs/iPSCs (Huang et al., 2011). To determine whether telomere lengths are elongated in ntESCs with high developmental pluripotency, we measured relative telomere content/length (RTL) by qPCR, shown as relative telomere to single copy gene (T/S) ratio (Callicott and Womack, 2006), of donor fibroblast cells and ntESC lines.
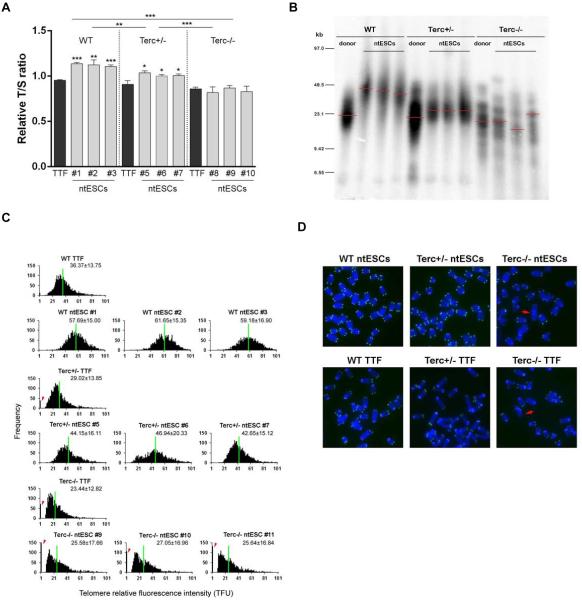
SCNT effectively restored telomeres in all WT ntESCs (RTL 1.11-1.14), compared to those of donor TTFs (RTL 0.95, Fig. 2A). In contrast, Terc−/− ntESCs had shorter RTLs in all lines examined (0.82-0.87), similar to those of donor TTFs (0.86) (Fig. 2A), suggesting failure of telomere elongation due to lack of telomerase. Interestingly, RTLs of Terc−/− ntESCs were maintained at similar level to those of donor cells, rather than shortening without telomerase, after ntESC derivation and in vitro culture, suggesting that telomerase independent mechanisms may be activated to slow down telomere attrition in these cells.
Fig. 2. Telomere lengths in ntESCs.
(A) Relative T/S ratio (mean ±SEM) in ntESCs of different genotypes. TTF: tail-tip fibroblast. Comparisons within the same genotype were made with TTF. (B) Telomere restriction fragment (TRF) analysis of ntESCs. (C) Histogram shows distribution of relative telomere length expressed as fluorescence intensity (TFU, telomere fluorescence unit). Heavy black bars on the Y-axis indicated by arrows show number of telomere signal-free ends. Average telomere length is shown as mean TFU±SD. TTF: tail-tip fibroblast donor cells. Numbers following genotype indicate ntESC line number. (D) Representative telomere Q-FISH images of ntESCs at P6. Telomeres are labeled with telomere PNA probes (green), and chromosomes labeled with DAPI (blue). Red arrows: telomere free ends, indicative of telomere loss. *P<0.05; **P<0.01; ***P<0.001. See also Fig. S2 and S3.
Notably, telomere lengths of all Terc+/− ntESCs (1.00-1.04) were robustly elongated to reach levels significantly longer than those of donor TTFs (RTL 0.91) and Terc−/− ntESCs (RTL 0.82-0.87) (Fig. 2A). Differences in telomere lengths between Terc+/− and Terc−/− ntESCs coincided with the outcomes of TEC experiments.
To validate the findings obtained by qPCR, we measured telomere lengths using Southern blot-based telomere restriction fragment (TRF) analysis (Fig. 2B) (Blasco et al., 1997). Consistent with the qPCR findings, telomeres were elongated robustly in WT, and also in Terc+/−, but not in Terc−/− ntESCs, compared with their donor cells. We also measured telomere length and function (telomere integrity and chromosome stability) of ntESCs by telomere quantitative fluorescent in situ hybridization (Q-FISH) (Fig. 2C). Relative telomere lengths shown as telomere fluorescence intensities (TFU) were shorter in TTFs of all three genotypes, but a correlation of the TFU with telomerase sufficiency was found with the TFU highest in WT (36.37±13.75), followed by Terc+/− (29.02±13.85), and lowest in Terc−/− TTF cells (23.44±12.82). Consistent with qPCR data, Telomeres shown as TFUs were significantly elongated in both WT (57.69 to 61.65) and Terc+/− (42.65 to 46.94), and only slightly elongated in Terc−/− (25.64 to 27.05) ntESCs, compared with those of their donor TTF cells. Consistently, telomere signal-free ends, indicative of telomere loss, were only observed in Terc−/− (red arrows, Fig. 2D) donor and ntES cells, but not in WT and Terc+/− ntESCs.
Telomere lengths in cloned pups derived from telomerase haplo-insufficient ntESCs
We measured telomeres as RTLs of TTFs in cloned pups as well as their corresponding placentas (Fig. 3A). The clones were entirely derived from Terc+/− ntESCs of C57BL6 background with black coat color and confirmed by microsatellite genotyping, while the placentas were derived from WT tetraploid host embryo origin of ICR background.
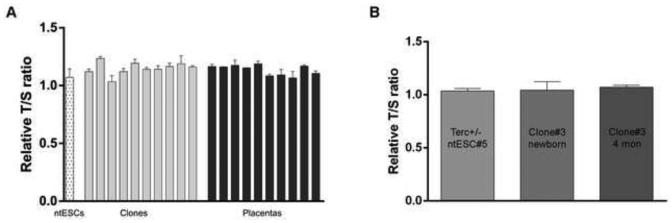
Fig. 3. Telomere lengths in all Terc+/− ntESC cloned pups.
(A) Relative T/S ratio (mean ±SEM) in Terc+/− ntESCs, in TTFs of clones and their corresponding placentas. ntESCs: Terc+/− ntESC line #5. (B) Relative T/S ratio (mean ±SEM) in ntESCs, and in the TTFs of Clone#3 at different ages.
RTLs of all clones measured (1.03-1.23) were similar to those of donor ntESCs (1.07±0.07). RTLs of all placentas fell in a comparable range (1.07-1.19, Fig. 3A). These data show that full-term clone pups from Terc+/− ntESC had telomere lengths like those of donor ntESCs.
Terc+/− clones are fertile and do not show signs of premature aging
Three clones produced from Terc+/− ntESCs have reached adulthood when we prepare this manuscript (Fig. 1I and J). They all appear normal and healthy and show no signs of premature aging. We compared RTLs of Terc+/− ntESCs, TTFs of Clone#3 (the oldest of all) at newborn and at the age of 4 month old, and found no differences in telomere lengths among them, indicating that telomere lengths are properly maintained after birth and at least till 4 months of age (Fig. 3B). Moreover, Clone#3 and #5 (male) were recently proven fertile. After breeding with WT females, they have fathered 8 and 6 apparently normal pups, respectively (pups fathered by Clone#3 shown in Fig. 1K).
Complete telomerase deficiency reduces proliferation of ntESCs
Considering the fundamental roles of telomeres and telomerase in proliferation, we assessed proliferation rates of ntESCs with different levels of telomerase activity (Fig. S1F). We seeded 100,000 cells on day 0 (D0) and counted the cell number on D2, D4 and D6. Number of cells on D6 was used as an indicator of cell proliferation rate. Terc−/− ntESCs, with shortest telomeres, turned out to proliferate at a much slower rate compared to Terc+/− and WT cells. Interestingly, Terc haplo-insufficient ntESCs proliferated at rates faster than WT ones. These data suggest that deficiency in self-renewal capacity may also contribute to the inability of Terc−/− ntESCs to support embryo generation by TEC assay, in addition to the compromised pluripotency.
Differential expression of genes involved in telomere regulation
To gain insights into molecular differences in telomere lengths and pluripotency, we performed microarray of ntESCs with three genotypes using Illumina Mouse Ref-8 chips. One hundred and nineteen genes out of over 25,697 (0.46%) genes were differentially expressed at 1.5 fold or higher between the WT and Terc−/− ntESCs.
Pairwise scatter plots (Fig. S3A) suggest that expression of genes routinely employed as markers of pluripotency, including Oct4, Nanog, Esrrb2, and Klf4, did not differ significantly among WT, Terc+/−, and Terc−/− ntESCs, consistent with data by immunostaining, RT-PCR (Fig. S1D and E), and qPCR (Fig. S3B). Our data support the notion that the conventional ES cell pluripotency markers cannot reliably distinguish ntESCs/ESCs with or without telomerase, nor can they indicate the naïve developmental potency (Huang et al., 2011).
We also examined a number of genes related to telomere length regulation in the array data, followed by validation with qPCR. Probes for Terc were not in the array, unfortunately, but qPCR revealed that its expression was high in WT, moderate in Terc+/−, and undetectable in the Terc−/− cells, faithfully reflecting their corresponding genotypes (Fig. S3C). Both array and qPCR showed that expressions of Tert, along with several other genes (i.e. Pot1a, Terf1, Terf2, Rtel1, Tep1, and Pinx1) did not differ among these ntESCs (Fig. S3A and C). Notably, Zscan4, a key gene in telomerase-independent telomere elongation based on recombination (Zalzman et al., 2010), was found differentially expressed in these ntESCs (Fig. S3A). Quantitative PCR (Fig. S3C) and Western blot (Fig. S3D) confirmed that Zscan4 expression was up-regulated in the Terc−/− ntESCs, compared to WT and Terc+/− ntESCs.
In addition, we analyzed telomere sister chromatid exchange (T-SCE), indicative of telomere recombination, of WT, Terc+/− and Terc−/− TTFs and ntESCs using CO-FISH (Wang et al., 2012). Terc−/− ntESCs exhibited increased frequency of T-SCEs than did WT and Terc+/− cells (Fig. S2), a pattern similar to those of Terc−/− iPSCs (Wang et al., 2012), coinciding with Zscan4 expression levels of ntESCs of different Terc genotypes (Fig. S3C and D). To test whether Zscan4 is involved in maintaining telomeres of Terc−/− cells, we performed Zscan4 knockdown experiments in Terc−/− ntESCs using two independent shRNA sequences, based on the methods described (Dan et al., 2014). Indeed, Zscan4 knockdown resulted in telomere shortening in Terc−/− ntESCs (Fig. S3E, F and G). These data suggest that telomerase deficiency and short telomeres eventually induce telomere recombination events, likely through activation of Zscan4, and that Zscan4 plays important roles in maintaining telomeres in telomerase-deficient cells.
Discussion
We show that naïve pluripotent patient-specific stem cells can be established with robust telomere regeneration from telomerase haplo-insufficient cells via SCNT. This may have implications for cell based therapies for telomere syndrome patients. Human iPSCs derived from cells with haplo-insufficient telomerase genes have so far failed to fully restore telomeres (Batista et al., 2011; Winkler et al., 2013). Winkler et al. derived iPSCs from patients with aplastic anemia or hypocellular bone marrow carrying heterozygous mutations in TERT or TERC (Winkler et al., 2013). The telomeres in these telomerase haplo-insufficient iPSCs were elongated but at a lower extent comparing to the healthy iPSCs. Furthermore, these iPSCs showed defective hematopoietic differentiation in vitro (Winkler et al., 2013). Consistently, we show that iPSCs generated from telomerase haplo-insufficient isogenic mouse TTFs used in the present work also exhibit only limited telomere elongation and failure to produce complete iPSC pups (Wang et al., 2012).
Remarkably, telomeres of Terc+/− ntESCs are effectively elongated compared to the donor cells, and the Terc+/− ntESCs contribute to full-term development by TEC test, indicating naïve pluripotency. Moreover, Terc+/− ntESC clones are fertile and do not show signs of pre-mature aging, and their offspring appear normal. While reactivation of Terc may allow telomere elongation in telomerase defective iPSCs following prolonged passages (Agarwal et al., 2010), SCNT-mediated telomere reprogramming of Terc haplo-insufficient cells does not appear to engage over-activation of Terc (Fig. S3C). Instead, telomerase-independent mechanisms (Liu et al., 2007; Zalzman et al., 2010) may have contributed to the robust reprogramming of telomeres in Terc+/− ntESCs, which is supported by a recent report that telomeres of Terc−/− cloned blastocysts are longer than those of donor cells used for SCNT (Le et al., 2014), as well as the present study. Previously we and others have shown that Zscan4 plays important roles in telomerase-independent mechanisms for iPSC reprogramming (Amano et al., 2013; Falco et al., 2007; Hirata et al., 2012; Jiang et al., 2013; Zalzman et al., 2010). In the present work, we show that elevated Zscan4 levels are correlated with increased telomere recombination events in the Terc−/− ntESCs and that Zscan4 knockdown in these cells led to shortened telomeres, implicating the potential roles of Zscan4 in telomere maintenance of ntESCs.
It is intriguing that the second generation or even third generation Terc−/− mice in our breeding colony, similar to those reported by others previously (Herrera et al., 1999), are viable, while no viable pups generated from G2 Terc−/− ntESCs in the present work by TEC, and from conventional (i.e. derived from fertilized embryos) G1/3/4 Terc−/− ESCs in our earlier work (Huang et al., 2011). We speculate that at least two factors might have contributed to the failure of the development of Terc−/− ntESCs by TEC assays. First, telomere defects shown by loss of telomeres, rather than telomerase deficiency or absolute telomere length, could be the major factor for the failed development. Terc or telomerase deficiency itself unlikely immediately affects telomere function, since mouse telomeres are long, but the processes of ESC derivation of ESCs and high proliferation of ESCs can rapidly shorten telomeres with passage of ESCs, resulting in telomere loss and dysfunction. Although increased expression of Zscan4 can partly compensate for telomere shortening, thus maintains telomeres at relatively stable lengths without net shortening of average telomere lengths, telomere free-signals indicative of telomere loss and chromosome fusion were found in Terc−/− ntESCs, but not WT and Terc+/− ntESCs (Fig. 2D), like those of Terc−/−ESCs (Huang et al., 2011). These defects of telomeres likely contribute to failure in embryonic development of G2 Terc−/− ntESCs in the present study, and of G1, and G3/4 Terc−/− ESCs in earlier study (Huang et al., 2011). Second, reduced cell proliferation of Terc−/− ntESCs. We show that complete telomerase deficiency reduces proliferation of ntESCs (Fig. S1F), suggesting that deficiency in self-renewal capacity may also have contributed to the inability of Terc−/− ntESCs to support full term development by TEC assay.
Our work in chorus with several recent publications (Le et al., 2014; Li et al., 2014) arguably suggest that ntESCs appear to have certain advantages over their iPSC counterparts, and that at its minimum, researchers should aim to improve the iPSCs’ quality to the level of ntESCs. In particular, SCNT may provide a novel strategy to correct telomere deficiencies in telomere syndrome patients. Early elegant animal cloning study showed that telomere lengths of senescent somatic cells can be elongated by SCNT, even beyond those of newborns (Lanza et al., 2000). Understanding SCNT-mediated telomere reprogramming would likely provide insights to improve iPSCs by identifying novel factors in oocytes to optimally reset telomeres. These ntESC lines are also valuable in vitro models to study telomere biology, especially considering the fact that stem cell failure is a common pathophysiological mechanism underling many of the telomere syndromes.
Experiment Procedures
Full experimental procedures are available in the Supplementary Information accompany this paper.
Animals
Animal maintenance, care and use procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of National Taiwan University. Heterozygous telomerase-deficient (Terc+/−) mice C57B6.Cg-Terctm1Rdp/J were purchased from the Jackson Laboratory, housed and bred for generation of Terc−/− mice. The Terc deficient mice (Herrera et al., 1999; Huang et al., 2011) used in this work were at Generation 2 (G2).
SCNT, ntESC derivation, and tetraploid embryo complementation assay
SCNT was carried out using fusion method as we previously described (Sung et al., 2010). The spindle chromosome complex (SCC) was removed using a pipette with an inner diameter of 8-10 μm assisted by piezo-drill pulses. Nuclei from donor cells were transferred by electrofusion to produce cloned embryos. When embryos reached the blastocyst-stage after 4 days in culture, they were used to establish ES cell lines as we described previously (Sung et al., 2010). In TEC assay, ICR strain mice were used to generate tetraploid blastocysts. For ESC injections, 10 to 15 ES cells were injected into one blastocyst using a flat tip pipette with an inner diameter of 14–16 mm assisted by piezo-drill pulses. Tetraploid blastocysts containing injected ESCs were transferred to pseudopregnant recipient ICR females. The full-term pups were obtained from recipient mothers at day 18.5 by Caesarean section.
Telomere measurements
Telomere lengths/contents were measured by quantitative real-time PCR (qPCR) (Callicott and Womack, 2006), telomere restriction fragment (TRF) analysis, and/or telomere QFISH. In qPCR, the telomere signal was normalized to the signal from the single-copy gene (36B4) to generate a T/S ratio indicative of relative telomere length of the given sample. The TRF analysis was performed using a commercial kit (TeloTAGGG Telomere Length Assay, Catalog# 12209136001, Roche Diagnostics Corporation, Indianapolis, IN, USA). Telomere QFISH was used to estimate telomere length and function (telomere integrity and chromosome stability) as described previously (Huang et al., 2011; Wang et al., 2012).
Statistical analysis
Percentages were transformed using arcsin transformation. Percentage transformed data and other numbers were analyzed by ANOVA and means compared by Fisher’s LSD using Graphpad Prism (v6.02, Graphpad Software, Inc., La Jolla, CA). Significant differences were defined as P<0.05 (*), 0.01 (**), 0.001 (***).
Supplementary Material
Acknowledgments
This study was supported by grants from the National Science Council, Taiwan, R.O.C. (Grant # 100-2313-B-002-045-MY3) and National Taiwan University (Grant# 103R7885) to L.Y.S, China MOST National Major Basic Research Program (Grant# 2012CB911202, 2011CBA01002) and China Ministry of Education (PCSIRT #IRT13023) to L.L., and National Institutes of Health (Grant# 1R03HD053066) to J.X. We thank the Bioinformatics & Biostatistics Core Lab, Center of Genomic Medicine at National Taiwan University for analysis of microarray data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal S, Loh YH, McLoughlin EM, Huang J, Park IH, Miller JD, Huo H, Okuka M, Dos Reis RM, Loewer S, et al. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292–296. doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T, Hirata T, Falco G, Monti M, Sharova LV, Amano M, Sheer S, Hoang HG, Piao Y, Stagg CA, et al. Zscan4 restores the developmental potency of embryonic stem cells. Nature Communications. 2013;4:1966. doi: 10.1038/ncomms2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M, Blackburn EH. The telomere syndromes. Nature Reviews Genetics. 2012;13:693–704. doi: 10.1038/nrg3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista LF, Pech MF, Zhong FL, Nguyen HN, Xie KT, Zaug AJ, Crary SM, Choi J, Sebastiano V, Cherry A, et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature. 2011;474:399–402. doi: 10.1038/nature10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Brambrink T, Hochedlinger K, Bell G, Jaenisch R. ES cells derived from cloned and fertilized blastocysts are transcriptionally and functionally indistinguishable. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:933–938. doi: 10.1073/pnas.0510485103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott RJ, Womack JE. Real-time PCR assay for measurement of mouse telomeres. Comparative Medicine. 2006;56:17–22. [PubMed] [Google Scholar]
- Chung YG, Eum JH, Lee JE, Shim SH, Sepilian V, Hong SW, Lee Y, Treff NR, Choi YH, Kimbrel EA, et al. >Human Somatic Cell Nuclear Transfer Using Adult Cells. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.03.015. [DOI] [PubMed] [Google Scholar]
- Dan J, Liu Y, Liu N, Chiourea M, Okuka M, Wu T, Ye X, Mou C, Wang L, Wang L, et al. Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Developmental Cell. 2014;29:7–19. doi: 10.1016/j.devcel.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Developmental Biology. 2007;307:539–550. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunes C, Rudolph KL. The role of telomeres in stem cells and cancer. Cell. 2013;152:390–393. doi: 10.1016/j.cell.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Herrera E, Samper E, Martin-Caballero J, Flores JM, Lee HW, Blasco MA. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. The EMBO Journal. 1999;18:2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T, Amano T, Nakatake Y, Amano M, Piao Y, Hoang HG, Ko MS. Zscan4 transiently reactivates early embryonic genes during the generation of induced pluripotent stem cells. Scientific Reports. 2012;2:208. doi: 10.1038/srep00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang F, Okuka M, Liu N, Ji G, Ye X, Zuo B, Li M, Liang P, Ge WW, et al. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Research. 2011;21:779–792. doi: 10.1038/cr.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Lv W, Ye X, Wang L, Zhang M, Yang H, Okuka M, Zhou C, Zhang X, Liu L, et al. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Research. 2013;23:92–106. doi: 10.1038/cr.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanza RP, Cibelli JB, Blackwell C, Cristofalo VJ, Francis MK, Baerlocher GM, Mak J, Schertzer M, Chavez EA, Sawyer N, et al. Extension of cell life-span and telomere length in animals cloned from senescent somatic cells. Science. 2000;288:665–669. doi: 10.1126/science.288.5466.665. [DOI] [PubMed] [Google Scholar]
- Le R, Kou Z, Jiang Y, Li M, Huang B, Liu W, Li H, Kou X, He W, Rudolph KL, et al. Enhanced telomere rejuvenation in pluripotent cells reprogrammed via nuclear transfer relative to induced pluripotent stem cells. Cell Stem Cell. 2014;14:27–39. doi: 10.1016/j.stem.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Li Z, Lu H, Yang W, Yong J, Zhang Z, Zhang K, Deng H, Xu Y. Mouse SCNT ESCs Have Lower Somatic Mutation Load Than Syngeneic iPSCs. Stem Cell Reports. 2014:399–405. doi: 10.1016/j.stemcr.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, et al. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- Sung LY, Chang CC, Amano T, Lin CJ, Amano M, Treaster SB, Xu J, Chang WF, Nagy ZP, Yang X, et al. Efficient derivation of embryonic stem cells from nuclear transfer and parthenogenetic embryos derived from cryopreserved oocytes. Cellular Reprogramming. 2010;12:203–211. doi: 10.1089/cell.2009.0072. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee HS, Sritanaudomchai H, et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama S, Kohda T, Obokata H, Tokoro M, Li C, Terashita Y, Mizutani E, Nguyen VT, Kishigami S, Ishino F, et al. Successful serial recloning in the mouse over multiple generations. Cell Stem Cell. 2013;12:293–297. doi: 10.1016/j.stem.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Wakayama T, Tabar V, Rodriguez I, Perry AC, Studer L, Mombaerts P. Differentiation of embryonic stem cell lines generated from adult somatic cells by nuclear transfer. Science. 2001;292:740–743. doi: 10.1126/science.1059399. [DOI] [PubMed] [Google Scholar]
- Wang F, Yin Y, Ye X, Liu K, Zhu H, Wang L, Chiourea M, Okuka M, Ji G, Dan J, et al. Molecular insights into the heterogeneity of telomere reprogramming in induced pluripotent stem cells. Cell Research. 2012;22:757–768. doi: 10.1038/cr.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler T, Hong SG, Decker JE, Morgan MJ, Wu C, Hughes W.M.t., Yang Y, Wangsa D, Padilla-Nash HM, Ried T, et al. Defective telomere elongation and hematopoiesis from telomerase-mutant aplastic anemia iPSCs. The Journal of Clinical Investigation. 2013;123:1952–1963. doi: 10.1172/JCI67146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Johannesson B, Sagi I, Burnett LC, Kort DH, Prosser RW, Paull D, Nestor MW, Freeby M, Greenberg E, et al. Human oocytes reprogram adult somatic nuclei of a type 1 diabetic to diploid pluripotent stem cells. Nature. 2014 doi: 10.1038/nature13287. [DOI] [PubMed] [Google Scholar]
- Yan L, Wu S, Zhang S, Ji G, Gu A. Genetic variants in telomerase reverse transcriptase (TERT) and telomerase-associated protein 1 (TEP1) and the risk of male infertility. Gene. 2014;534:139–143. doi: 10.1016/j.gene.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Young NS, Calado RT, Scheinberg P. Current concepts in the pathophysiology and treatment of aplastic anemia. Blood. 2006;108:2509–2519. doi: 10.1182/blood-2006-03-010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.