Abstract
Cancer cells rapidly evolve a multitude of defense mechanisms to evade the effects of the oncologist’s drug arsenal. Unfortunately, clinical strategies to overcome these lag far behind. This mismatch likely underlies our inability to implement new durable treatment strategies. Here, a new form of multi-drug resistance, inducible drug glucuronidation, is discussed. This form was discovered while developing means to target a specific oncogene, the eukaryotic translation initiation factor 4E (eIF4E), with its inhibitor ribavirin. In two clinical studies, ribavirin treatment led to substantial clinical responses, but all responding patients eventually relapsed. In most cases, this was due to the overexpression of the sonic hedgehog transcription factor Gli1 which elevated the UDP glucuronsyltransferase UGT1A enzymes. UGT1As add glucuronic acid to many drugs. Indeed, these cells are resistant to not only ribavirin, but also Ara-C, and likely other drugs. Inhibition of Gli1 reduced UGT1As, eliminated drug-glucuronides and renewed sensitivity to ribavirin and Ara-C. These studies highlight that cancer cells and their resistant counterparts metabolize drugs differently from each other as well as from normal cells. Likely these inducible modifications go beyond glucuronidation. Understanding the extent of inducible drug modifications and the pathways that drive expression of the corresponding enzymatic machinery will better position us to finally make resistance futile.
Keywords: drug modification, Gli1, glucuronidation, eIF4E, Ribavirin, Ara-C
Overview
Although cancer is many different diseases, nearly all share one inescapable feature: the remorseless onset of drug resistance. Resistance can emerge after drug exposure (acquired resistance) or prior to treatment (primary resistance). Unfortunately the majority of cancer patients will experience drug resistance, and associated clinical relapse, at some point during their treatment course(1). Thus understanding the molecular underpinnings of resistance is central to the development of durable treatment strategies.
Drug resistance most likely arises through a selection process with the emergence of dominant clones from pools of heterogeneous tumor cells(1). In acquired resistance, cells with features that permit survival and proliferation emerge upon the selective stress of drug treatment. Resistance in patients that have had other treatments can lead to cross-resistance i.e. cells that are resistant to more than one drug through some common underlying mechanism such as increased drug efflux. For frontline patients that do not respond to any type of therapy, there is likely some feature of the dominant population of cells, such as re-wiring of apoptotic pathways, that underpins this phenotype.
From the drug perspective, resistance studies typically explore uptake and pro-drug metabolism. In the first case, drug transporters such as MDR become elevated leading to increased drug efflux(2); or alternatively, transporters, such as the equilibrative nucleoside transporter ENT1, are mutated or downregulated impairing drug entry(3). Key enzymes in pro-drug metabolism can modulate the ability of the drug to stay in the cell, to bind its target or impair activity for other reasons. For instance, deoxycytosine kinase plays a critical role in the metabolism of cytarabine (Ara-C), the cornerstone of AML therapy. Deoxycytosine kinase can be mutated in resistant cells leading to reduced Ara-C efficacy(3). Unfortunately, these pathways have not been readily amenable to targeting. The majority of efforts to combat drug resistance have focused on MDR, with third generation inhibitors in clinical trial. However, addition of MDR inhibitors to at least some cancer regimens has not substantially enhanced clinical benefit(4, 5). Understanding the reasons for this is an active and important area of study.
Cells have many other ways to elude the effects of drugs. For example, protein targets for a given drug can be mutated or downregulated. Some forms of drug resistant CML arise due to mutations in the Bcr-Abl oncogene. The development of next generation drugs that bind to mutant Bcr-Abl allowed continued targeting of this oncogene(6-10). Aside from modulating the drug target, resistant cells use many other means to evade the effects of drugs. These have recently been reviewed in detail(11) and thus they are only briefly mentioned here. Such strategies include increased DNA repair, increased chromosome instability, re-wiring of apoptotic pathways and increased pro-survival signals(11). Pathway redundancy can play a major role in resistance as well. In this instance, drugs continue to inhibit their targets but alternative cellular pathways become activated in a process known as oncogenic bypass (11). The tumour microenvironment can also contribute to the development of drug resistance. For example, melanoma cells became resistant to the BRAF inhibitor PLX4720 when co-cultured with fibroblasts(12). The examples given so far are not a complete list, but simply a glimpse into the multitude of resistance pathways. Recently, we identified a new form of resistance which is described below.
A new type of drug resistance: inducible drug modification
While developing new means to target the oncogene eIF4E in AML, we discovered a novel form of drug resistance arising from inducible drug modification(13)(Figure 1). First, some background information as to how we arrived at this point. eIF4E is dysregulated in an estimated 30% of cancers including specific subtypes of AML(14). eIF4E transforms these cells by driving nuclear export and translation of mRNAs which encode proteins involved in proliferation and survival(15). To associate with these transcripts, eIF4E binds the m7G cap on their 5′ end. We identified a direct cap competitor of eIF4E, ribavirin(16-18). Ribavirin impairs eIF4E’s biochemical activities in translation and mRNA export as well as its ability to transform cells in culture and represses tumor formation in mice(16-18). Our two clinical trials in AML revealed that ribavirin targeted eIF4E activity and that this correlated with clinical responses including remissions(19, 20). However, all responding patients ultimately developed ribavirin resistance, loss of eIF4E targeting and thus, relapsed. Furthermore, there was a population of patients that despite having highly elevated eIF4E, never responded to ribavirin. Thus we set out to understand the molecular bases for ribavirin resistance(13, 20).
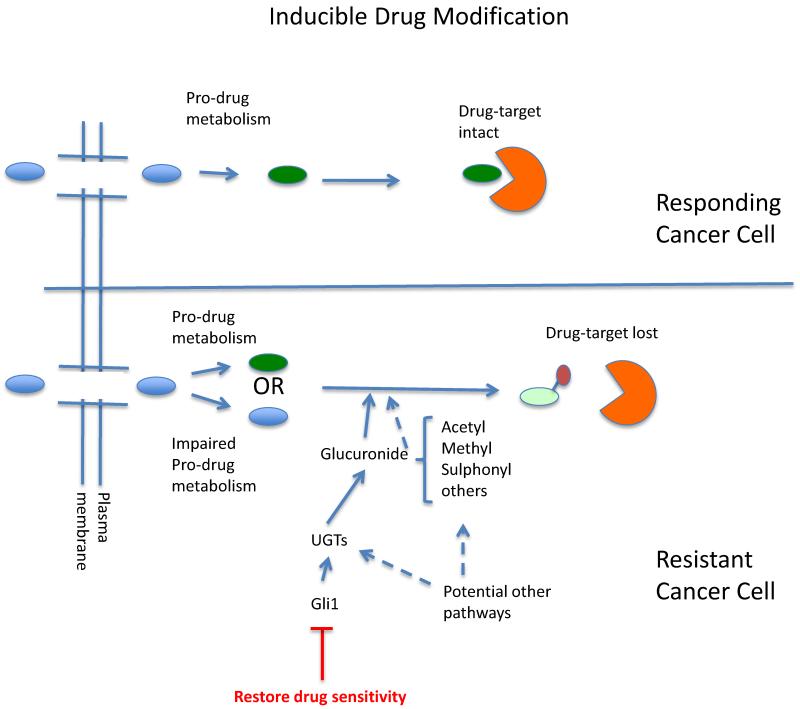
Figure 1. Schematic diagram depicting the role of inducible drug modification in resistance.
The protein target is in orange and the pro-drug is depicted as a light red oval, after pro-drug conversion in green and once modified in yellow with the covalent modification shown as a second smaller oval. Note that depending on the drug, it could be a target for modification in its pro-drug and/or active form. Drug sensitivity can be restored by Gli1 inhibition for the resistance mechanism we discuss, but may not be the case for other modifications or contexts. We speculate that other pathways could drive other enzymatic modifications as depicted by dashed arrows.
As a first step, we generated cell line models of ribavirin resistance as well as analyzed our clinical specimens before treatment, at response and at relapse. We identified two distinct forms of resistance. The first was due to impaired ribavirin uptake, as assessed by measuring 3H ribavirin. This occurred through two independent mechanisms: 1. The downregulation of the nucleoside transporter ENT1 which impaired cellular uptake and/or 2. The downregulation of adenosine kinase (ADK) an enzyme key to pro-drug metabolism of ribavirin. ADK is required for the first phosphorylation step of ribavirin to its active metabolite ribavirin triphosphate. Ribavirin, in its unphosphorylated form is readily exported from the cell using the ENT1 transporter. Thus, ribavirin net uptake is impaired when ADK is lost. In our patient group, only a few patients showed signs of reduced ADK or ENT1 suggesting that although this does occur clinically, it is not the predominant form of resistance in our patient cohorts.
Given our observation that impaired drug uptake did not appear to be a major cause of drug resistance in our patients, we further studied a second form of drug resistance we had identified in the lab. Here we found that resistant cells had normal 3H-ribavirin uptake but by using an immunoprecipitation strategy, noted that eIF4E no longer interacted with 3H-ribavirin. Analysis of eIF4E levels, sequence and activity suggested that eIF4E was fully functional. Indeed, RNAi knockdown of eIF4E in resistant cells inhibited their growth indicating these cells still required eIF4E.
In order to understand the molecular basis for this second form of resistance, we carried out RNA sequencing and analyzed differences in transcript levels between resistant and sensitive cells. We found that the transcription factor Glioma associated protein 1 (Gli1), was highly elevated in these samples. Gli1 is a major transcription factor downstream of sonic hedgehog signaling (Figure 1). Strikingly, Gli1 overexpression alone was sufficient to drive ribavirin resistance and importantly did not modulate eIF4E protein levels. Here, we observed that Gli1 overexpression also underlied resistance to cytarabine (Ara-C) indicating that Gli1 could instigate resistance to multiple drugs. Gli1 knockdown led to drug re-sensitization to ribavirin and Ara-C. Analysis of our patient specimens indicated that Gli1 became highly elevated at relapse relative to response. Gli1 was also elevated at relapse relative to diagnosis in patients that received standard of care for AML, Ara-C plus an anthracycline. We note that Gli1 levels alone could vary substantially and that it was the ratio of Gli1 levels at relapse to diagnosis (or best response) that were the most telling. In summary, Gli1 was driving resistance to two distinct drugs, but how?
We found that Gli1 increased levels of the family of glucuronidation enzymes known as the UDP glucuronsyltransferases UGT1As (Figure 1). These enzymes chemically modify a wide variety of metabolites and xenobiotics through the covalent addition of glucuronic acid. Indeed we observed resistant cells had highly elevated UGT1A enzyme levels, as did AML patients at relapse from either ribavirin or standard of care therapies. Gli1 elevation alone was sufficient to drive UGT1A protein expression. Conversely, Gli1 knockdown lowered UGT1A protein levels. It is important to note that UGT1A is not a direct transcription target of Gli1. Indeed UGT1A RNAs are actually lower in resistant cells while its protein levels are highly elevated. Experiments with the proteasome inhibitor MG132 suggest that Gli1 increases the protein stability of UGT1As. Polysomal loading analysis of resistant and parental cells indicated that there was no difference in translation efficiency of UGT1A transcripts between these cell lines suggesting that Gli1 does not modulate translation of UGT1A transcripts or eIF4E levels. We propose that the disconnect between UGT1A protein and RNA levels is likely due to an attempted feedback loop by the cell to lower UGT1A RNA levels in the face of increased UGT1A protein expression. It is also possible that microRNAs play a key role in this process. These possibilities need to be experimentally tested. Irrespective of the particular mechanism, these studies clearly highlight the difficulties of predicting drug resistance mechanisms based only on gene expression data. In summary, there is a clear correlation between Gli1 elevation, overexpression of UGT1A enzymes and resistance in our patients. Indeed, Gli1 and UGT1A elevation were more prevalent in our patient cohorts than impaired drug uptake or pro-drug metabolism.
Given the role of these enzymes in drug modification, we postulated that drug resistance in both our lab models and in these AML patients arose due to drug glucuronidation. Using mass spectrometry techniques, we directly observed the ribavirin-glucuronides and ara-C-glucuronides in resistant cells. We also observed that ribavirin-glucuronides could no longer directly interact with eIF4E thereby providing a molecular basis for drug resistance. Consistent with the strong link between Gli1 and UGT1A protein levels, we found that Gli1 knockdown led to the loss of the drug-glucuronides correlating with drug re-sensitization.
Our goal was to develop a pharmacological strategy to overcome Gli1 inducible drug resistance (Figure 1). We used two Gli1 inhibitors, the FDA approved Vismodegib which targets the extracellular receptor for this pathway and also GANT-61, a direct Gli1 inhibitor. In both cases, Gli1 inhibition resulted in the loss of ribavirin-glucuronides, re-association of eIF4E with ribavirin, and re-sensitization of cells to drug. Gli1 inhibition similarly reverted ara-C glucuronidation and resistance. These studies serve as the starting point for a clinical trial using Gli1 inhibitors to revert drug resistance (ClinicalTrials.gov NCT02073838). This trial is scheduled to open in Fall 2014. Importantly, Gli1 elevation alone was sufficient to drive drug glucuronidation not only in AML cells, but also in head and neck cell lines suggesting that this form of resistance will be applicable to at least some solid tumours.
In this way, our studies revealed a novel mode of drug resistance we denote inducible drug modification (Figure 1). These modifications do not occur in corresponding normal cells and are not prevalent in the bulk population of cancer cells prior to treatment. In this model, the cellular machinery covalently modifies multiple drugs in order to impair activity (Figure 1). This is a distinct mechanism from inactivation through impairing uptake or pro-drug metabolism. In our case, the activated machinery could potentially target a wide variety of chemically distinct drugs. This is because glucuronidation enzymes target nitrogens, sulphurs or oxygens available for nucleophilic attack(21, 22). To better predict the repertoire of drugs that are clients for this resistance mechanism, HTS screening of resistance cells will be carried out. It is difficult to determine drugs that will be clients a priori because although specific UGT1As target different chemical moieties (but with substantial overlap), there are no antibodies available to each specific UGT1A family member. Our enzymatic studies implicate UGT1A4, UGT1A6 and UGT1A9 in ribavirin glucuronidation, strongly suggesting that N-glucuronidation will be elevated here. However, it is not possible, or wise, to rule out other moieties at this point. Consistent with our initial studies on N-glucurondiation of ribavirin and Ara-C, more recently we showed that azacytidine is also a client of this mechanism (Zahreddine and Borden, unpublished result). Importantly, neither ribavirin nor Ara-C glucuronides are observed in normal tissues, and thus this modification is an adaptive responsive in resistant cancer cells. In this way, inducible drug glucuronidation could play wide-ranging roles in drug resistance. Further, future compounds could be designed to reduce the potential modification in resistant cells.
Glucuronidation and other drug modifications
Glucuronidation itself was first described in the early 1950s(22). There are two main families of UGT enzymes, the aforementioned UGT1As as well as UGT2Bs. There are nine UGT1A family members which arise due to alternative exon sharing and all contain a common C-terminal domain which specifically binds to glucuronic acid(23). Different enzymes target specific subsets of chemical groups. For instance, UGTA4, 1A6 and 1A9 are known to target nitrogens, consistent with our results that these likely contribute to glucuronidation of the carboxyamide of ribavirin(21, 23, 24). Other enzymes in the family target other chemical groups such as sulphurs or oxygens. Most enzymes have a broad target base with significant overlap between enzymes and targets(21). Originally these enzymes were thought to be restricted to the liver, but it is now known that glucuronidation occurs in a broad range of tissues(21).
Glucuronidation is typically considered a detoxification mechanism increasing drug clearance(22). However, the effects of this modification are not always predictable, i.e. do not always increase efflux and can even increase toxicity(22). Indeed, in many cases glucuronidation modulates the binding partners of metabolites and drugs. For instance, testosterone glucuronides are better substrates for cytoplasmic β-reductase than testosterone and worse substrates for Δ-5α-reductase (22). These activities are unrelated to efflux. It is entirely possible that the effects of inducible glucuronidation will be drug dependent, depending on the chemical structure.
Interestingly, glucuronidation enzyme levels are reduced in some cancer cells relative to normal tissue(24, 25). This observation is attributed to the loss of the ability to detoxify certain environmental carcinogens such as hydroxy-benzo(a)pyrenes in cigarette smoke, leading to DNA damage and carcinogenesis(24). Some genetic disorders such as Gilbert’s and Crigler-Najjar’s syndromes are characterized by a reduction in UGT enzyme activity due to mutations which lead to impaired bilirubin glucuronidation which must be appropriately managed(24, 26). Other familial polymorphisms have been identified leading to impaired glucuronidation of specific drugs(26). Finally, in rare cases, patients who have polymorphisms in UGT1A7, have reduced enzyme activity and increased risk of tobacco related lung cancer(26). In all, these genetic disorders are related to a loss of UGT1 activity. By way of contrast, we observe elevated UGT1As in patients at relapse. Thus, it is clear that there is a “Goldilocks” zone for UGT1A expression making it important to elucidate the factors, besides Gli1, that control expression of this family of enzymes. Further, Gli1 may also be driving resistance through both glucuronidation and other pathways simultaneously.
Phase II drug metabolism involves conjugation of drug via multiple pathways including (but not limited to) glutathione addition, sulphation, acetylation, methylation as well as glucuronidation. We propose that similar to glucuronidation, these other modifications could also become induced upon drug resistance (Figure 1). Consistent with this idea, elevated glutathione levels correlate with inactivation of platinum drugs, but whether this a direct modification or a secondary effect remains to be established (27). In all, phase II drug modifications are usually considered only in the initial stages of drug development in normal tissues and not as part of an adaptive response specific to resistant cancer cells. Our findings suggest that drug modifications and the phase II machinery should be characterized in cancer patients, particularly at resistance. Further, a better understanding of how signaling pathways such as Gli1 can impact on phase II metabolism in normal and cancer contexts could reveal new means to target these pathways at resistance.
How can we make resistance futile?
Drug resistance may be more common than we think and is likely significantly impacting failure of many drugs to move from the lab to the clinic. For instance, many early phase trials targeting important cellular pathways are clinical failures. For example in AML, many trials end without any patients achieving even a modest response much less a remission(28). Generally, these trials test drugs on patient cohorts that have failed other therapies i.e. have developed some form(s) of drug resistance. Clearly, many drugs may fail here, not because targeting a given pathway is not important nor because the drug did not target the pathway, but rather, that cross-resistance mechanisms are already in play and these impair the experimental agent’s efficacy. Further, we do not take into account the fact that onset of drug resistance in patients could be rapid, occurring even within weeks i.e. during the early phase of experimental treatments. Clearly, we must understand the pathways of drug resistance by more in depth molecular analysis of patient material from clinical trials that fail, as well as those that succeed. Such evaluations should include (but not be limited to): PK analysis to ensure all drugs are absorbed as expected, examination of drug uptake into the cell, pro-drug metabolism, assessment of whether drug modifications emerge, determination of molecular status of relevant pathways and whether the drug target interaction is intact or lost. Patient therapies should be tailored based on their resistance marker profile (MDR, transport loss etc), in conjunction with other relevant markers. In other words, drug resistance should be targeted, just as relevant oncogenes are.
We propose that inducible drug glucuronidation is the first in an emerging class of drug modifications that occur in resistant cells but are absent from normal counterparts and even, in some cases, in the initial bulk cancer cell population. A mechanistic understanding of the enzymatic pathways that underlie these modifications will enable one to develop therapies which include impairing, overcoming or evading this form of resistance clinically. For our case, we will endeavor to prevent, or at least slow down, resistance by combining ribavirin with Gli1 inhibitors. However, it seems certain that other pathways and modifications will be implicated and thus, a more global understanding of these modifications is warranted.
Differences between phase II drug metabolism in humans and rodents underpin the necessity for development of more human-like model systems when studying processes such as glucuronidation. For instance, rodents lack UGT1A4 and thus most forms of N-glucuronidation of primary, secondary and tertiary amines of xenobiotics (23, 29). Glucuronidation of some drugs containing hydroxyl groups for instance is also different between species(29). Indeed, inter-species variation in drug metabolism is likely a larger issue in the development of experimental therapeutics. For instance, pre-clinical studies with Fialuridine for Hepatitis B treatment were very promising and led to a clinical trial(30). Unfortunately, the Fialuridine, despite promising responses in the first weeks, led to the death of 5/15 trial patients and two others required emergency liver transplants. Such toxicity was not predicted in the dog, primates or rodent models used prior to the human trial. However, the use of mice with humanized livers showed similar toxicity to the patients and suggested that the presence of the ENT1 transporter on mitochondria as well as plasma membranes in humans could underlie the tragic responses seen in the patients(30). Similar approaches have been used to study glucuronidation, where mice with the full complement of human UGT1As in the liver were developed(29). Here, the humanized mice could glucuronidate amine containing xenobiotics. However, a complete humanization was not observed. This could arise due to many factors including this model did not include the UGT2B family of enzymes, and further, that other aspects of drug absorption, uptake and metabolism could be very different between species. Additionally, there are the UGT1As in the gastro-intestinal track (e.g. UGT1A8 and UGT1A10) and elsewhere that are clearly not taken into account in such a model. Thus, these humanized mice are valuable systems to study pharmacology, but still may not fully recapitulate many key aspects of human drug metabolism. Ultimately, these human mice differences could impact predictions of drug efficacy, toxicity and mechanism of drug resistance.
In summary, we demonstrate a new form of drug resistance, inducible drug modification. Moreover, our studies demonstrate that cancer cells can metabolize drugs differently than their normal counterparts, and that this should be taken into account not only during initial drug development, but also upon resistance. If “an ounce of prevention is worth a pound of cure (B. Franklin),” it is clear that onset of drug resistance should be planned for at the outset of treatment.
Acknowledgments
Financial Support: NIH RO1 98571 and 80728, LLS Translational Research Program grants (6160 and 1670) as well as Canada Research Chair in Molecular Biology of the Cell Nucleus.
Footnotes
Conflicts of interest. None.
References
- 1.Zahreddine H, Borden KL. Mechanisms and insights into drug resistance in cancer. Front Pharmacol. 2013;4:28. doi: 10.3389/fphar.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nature reviews Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 3.Cai J, Damaraju VL, Groulx N, Mowles D, Peng Y, Robins MJ, et al. Two distinct molecular mechanisms underlying cytarabine resistance in human leukemic cells. Cancer research. 2008;68:2349–57. doi: 10.1158/0008-5472.CAN-07-5528. [DOI] [PubMed] [Google Scholar]
- 4.Ruff P, Vorobiof DA, Jordaan JP, Demetriou GS, Moodley SD, Nosworthy AL, et al. A randomized, placebo-controlled, double-blind phase 2 study of docetaxel compared to docetaxel plus zosuquidar (LY335979) in women with metastatic or locally recurrent breast cancer who have received one prior chemotherapy regimen. Cancer chemotherapy and pharmacology. 2009;64:763–8. doi: 10.1007/s00280-009-0925-9. [DOI] [PubMed] [Google Scholar]
- 5.Pusztai L, Wagner P, Ibrahim N, Rivera E, Theriault R, Booser D, et al. Phase II study of tariquidar, a selective P-glycoprotein inhibitor, in patients with chemotherapy-resistant, advanced breast carcinoma. Cancer. 2005;104:682–91. doi: 10.1002/cncr.21227. [DOI] [PubMed] [Google Scholar]
- 6.Zhou T, Commodore L, Huang WS, Wang Y, Thomas M, Keats J, et al. Structural mechanism of the Pan-BCR-ABL inhibitor ponatinib (AP24534): lessons for overcoming kinase inhibitor resistance. Chemical biology & drug design. 2011;77:1–11. doi: 10.1111/j.1747-0285.2010.01054.x. [DOI] [PubMed] [Google Scholar]
- 7.Golas JM, Arndt K, Etienne C, Lucas J, Nardin D, Gibbons J, et al. SKI-606, a 4-anilino-3-quinolinecarbonitrile dual inhibitor of Src and Abl kinases, is a potent antiproliferative agent against chronic myelogenous leukemia cells in culture and causes regression of K562 xenografts in nude mice. Cancer research. 2003;63:375–81. [PubMed] [Google Scholar]
- 8.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 9.Weisberg E, Manley PW, Breitenstein W, Bruggen J, Cowan-Jacob SW, Ray A, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer cell. 2005;7:129–41. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 11.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nature reviews Cancer. 2013;13:714–26. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 12.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, Gendron P, Romeo AA, Morris SJ, et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 2014;511:90–3. doi: 10.1038/nature13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borden KL, Culjkovic-Kraljacic B. Ribavirin as an anti-cancer therapy: Acute Myeloid Leukemia and beyond? Leukemia and Lymphoma. 2010 doi: 10.3109/10428194.2010.496506. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Culjkovic-Kraljacic B, Borden KL. Aiding and abetting cancer: mRNA export and the nuclear pore. Trends Cell Biol. 2013 doi: 10.1016/j.tcb.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004 doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kentsis A, Volpon L, Topisirovic I, Soll CE, Culjkovic B, Shao L, et al. Further evidence that ribavirin interacts with eIF4E. Rna. 2005;11:1762–6. doi: 10.1261/rna.2238705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volpon L, Osborne MJ, Zahreddine H, Romeo AA, Borden KL. Conformational changes induced in the eukaryotic translation initiation factor eIF4E by a clinically relevant inhibitor, ribavirin triphosphate. Biochem Biophys Res Commun. 2013 doi: 10.1016/j.bbrc.2013.03.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood. 2009;114:257–60. doi: 10.1182/blood-2009-02-205153. [DOI] [PubMed] [Google Scholar]
- 20.Assouline S, Culjkovic-Kraljacic B, Bergeron J, Caplan S, Cocolakis E, Lambert C, et al. A Phase I trial of ribavirin and low-dose cytarabine for the treatment of relapsed and refractory acute myeloid leukemia FAB subtypes M4 and M5 or high eIF4E. Haematologica. doi: 10.3324/haematol.2014.111245. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- 22.Dutton G. Glucuronidation of Drugs and Other Compounds. CRC Press; Boca Rotan, Florida: 1980. [Google Scholar]
- 23.Rowland A, Miners JO, Mackenzie PI. The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. The international journal of biochemistry & cell biology. 2013;45:1121–32. doi: 10.1016/j.biocel.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 24.Strassburg CP, Lankisch TO, Manns MP, Ehmer U. Family 1 uridine-5′-diphosphate glucuronosyltransferases (UGT1A): from Gilbert’s syndrome to genetic organization and variability. Archives of toxicology. 2008;82:415–33. doi: 10.1007/s00204-008-0314-x. [DOI] [PubMed] [Google Scholar]
- 25.Strassburg CP, Strassburg A, Nguyen N, Li Q, Manns MP, Tukey RH. Regulation and function of family 1 and family 2 UDP-glucuronosyltransferase genes (UGT1A, UGT2B) in human oesophagus. The Biochemical journal. 1999;338(Pt 2):489–98. [PMC free article] [PubMed] [Google Scholar]
- 26.Burchell B. Genetic variation of human UDP-glucuronosyltransferase: implications in disease and drug glucuronidation. American journal of pharmacogenomics: genomics-related research in drug development and clinical practice. 2003;3:37–52. doi: 10.2165/00129785-200303010-00006. [DOI] [PubMed] [Google Scholar]
- 27.Meijer C, Mulder NH, Timmer-Bosscha H, Sluiter WJ, Meersma GJ, de Vries EG. Relationship of cellular glutathione to the cytotoxicity and resistance of seven platinum compounds. Cancer research. 1992;52:6885–9. [PubMed] [Google Scholar]
- 28.Assouline S, Cocolakis E, Borden KL. The Development of Novel Therapies for the Treatment of Acute Myeloid Leukemia (AML) Cancers (Basel) 2012;4:1161–79. doi: 10.3390/cancers4041161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kutsuno Y, Sumida K, Itoh T, Tukey RH, Fujiwara R. Glucuronidation of drugs in humanized UDP-glucuronosyltransferase 1 mice: Similarity with glucuronidation in human liver microsomes. Pharmacology Research & Perspectives. 2013;1 doi: 10.1002/prp2.2. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu D, Nishimura T, Nishimura S, Zhang H, Zheng M, Guo YY, et al. Fialuridine induces acute liver failure in chimeric TK-NOG mice: a model for detecting hepatic drug toxicity prior to human testing. PLoS medicine. 2014;11:e1001628. doi: 10.1371/journal.pmed.1001628. [DOI] [PMC free article] [PubMed] [Google Scholar]