Abstract
Balancing mitophagy and mitochondrial biogenesis is essential for maintaining a healthy population of mitochondria and cellular homeostasis. Coordinated interplay between these two forces that govern mitochondrial turnover plays an important role as an adaptive response against various cellular stresses that can compromise cell survival. Failure to maintain the critical balance between mitophagy and mitochondrial biogenesis or homeostatic turnover of mitochondria results in a population of dysfunctional mitochondria that contribute to various disease processes. In this review we outline the mechanics and relationships between mitophagy and mitochondrial biogenesis, and discuss the implications of a disrupted balance between these two forces, with an emphasis on cardiac physiology.
Keywords: mitochondria, autophagy, mitophagy, mitochondrial biogenesis, cardiac, pathogenesis
Introduction
Mitochondria function as cellular power plants essential for meeting the energetic demands of eukaryotic cells. Their role extends to regulating fuel utilization, calcium stores, intracellular signaling and cell death. Because of the broad range of cellular functions they are involved in, mitochondria inherently occupy an important position as mediators of cellular homeostasis. Consequently, this crucial position associates the dysfunction of mitochondria to the development of various human diseases. Notably, studies to dissect the etiology of Parkinson Disease (PD) were among the first to highlight the physiological consequence of having poor mitochondrial quality control. Genetic models strongly implicate mitochondrial dysfunction as a common feature in development of this neurodegenerative disease that leads to the loss of dopaminergic neurons (reviewed in [1–5]). In support of this is the fact that aging increases the risk of developing PD, which correlates with higher incidence of mitochondrial DNA mutations in dopaminergic neurons [6]. Moreover, agents that induce mitochondrial toxicity have been shown to lead to PD-like symptoms in animal models [7].
The major chronic diseases we face today such as neurodegenerative diseases, cancer, aging, diabetes, and heart failure are accompanied by mitochondrial dysfunction, and in fact, many elements of these chronic diseases may be directly attributed to mitochondrial pathology [8]. Mitochondrial disorders may be inherited either through maternal transmission of an abnormal mitochondrial genome or through autosomal transmission of mutations in the nuclear-encoded mitochondrial genes. However, far more commonly, mitochondrial dysfunction is a consequence of derangements in the ordinarily robust systems that orchestrate and maintain the health and function of these vital organelles.
Mitochondrial quality control collectively describes the cellular systems used to maintain a population of optimally-functioning mitochondria. Mitochondria possess an internal protein quality control system to refold or eliminate misfolded proteins, comprising chaperones (Hps10, Hsp60 and others) and proteases (Lon and other AAA proteases). Import of nuclear-encoded proteins must be coordinated with expression of mitochondrial subunits for proper assembly of oxidative phosphorylation (OXPHOS) complexes. Homeostatic control of this is mediated through the mitochondrial unfolded protein response (UPRmt), which is activated by an imbalance of nuclear vs. mitochondrial OXPHOS subunits [9]. Mitochondrial turnover is another integral aspect of quality control in which dysfunctional mitochondria are selectively eliminated through autophagy (mitophagy) and replaced through expansion of preexisting mitochondria (biogenesis). Impaired mitochondrial quality control results in accumulation of damaged mitochondria that may generate more reactive oxygen species (ROS), produce ATP less efficiently, have a lower threshold for cytochrome c release (apoptosis) or mitochondrial permeability transition pore (MPTP) opening (necrosis), or may release mitochondrial components (mtHSP60, oxidized mitochondrial DNA) into cytosol where its recognition by receptors for damage-associated molecular patterns (DAMP) activates inflammation. In this way, impaired mitochondrial quality control gives rise to a myriad of disease states. Mitochondrial quality control is critically dependent on autophagy; factors that impair autophagy, such as advanced age or the metabolic syndrome (MetS), will impact mitochondrial quality control and accelerate the development of chronic disease phenotypes. In this review, we focus on the mechanics of mitophagy and mitochondrial biogenesis, and discuss the interplay between these two forces. We then discuss the pathophysiological consequences with an emphasis on the heart.
1. Mechanics of Mitophagy and Mitochondrial Biogenesis
1.1 Mechanics of mitophagy
Autophagy is a lysosome-dependent cellular degradation system in eukaryotic cells that allows for the bulk recycling of unwanted cytoplasmic aggregate proteins or dysfunctional organelles [10]. Along with the ubiquitin proteasome system (UPS), autophagy is important for maintaining proteostasis in the heart [11]. Mitophagy is the selective targeting and removal of mitochondria through autophagy. While some authors refer to the general process as mitochondrial autophagy and use the term mitophagy to mean Parkin-dependent elimination of mitochondria, in this review we will use ‘mitophagy’ to indicate autophagic removal of mitochondria, and where appropriate, will specify Parkin-dependent mitophagy. Mitophagy plays a critical role in protecting the heart during ischemia/reperfusion injury [12–14]. Depolarization of mitochondria is a prerequisite for Parkin-dependent mitophagy, but mitophagy mediated by Bnip3 and NIX may be triggered through other pathways including reactive oxygen species (ROS) [15], which promote dimerization of Bnip3 (and potentially NIX) on the mitochondrial outer membrane [16]. Nutrient stress (fasting) activates AMPK and general autophagy, which is associated with production of ROS from mitochondrial complex I [17]; however, fasting-induced mitophagy is impaired in cyclophilin D-deficient mice [18], which have hyperpolarized mitochondria. Thus there are hints that mitophagy initiated by nutrient stress may be initiated by mitochondrial depolarization and Parkin translocation, but a role for ROS and Bnip3 is not excluded.
Parkin-dependent (macro)mitophagy has been commonly studied using chemical uncouplers of mitochondria such as carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) or carbonyl cyanide m-chlorophenyl hydrazone (CCCP). Cellular stresses such as ischemia also trigger mitochondrial depolarization [13], resulting in stabilization of the serine/threonine kinase phosphatase and tensin homolog (PTEN)-induced kinase 1 (PINK1) on the outer mitochondrial membrane (OMM) and recruitment of the E3 ubiquitin ligase Parkin, key factors for mitophagy [19–22]. PINK1 and Parkin function as critical partners to mediate the clearance of dysfunctional mitochondria [23, 24]. Another Parkin-dependent mechanism for degrading mitochondrial components is through mitochondria-derived vesicles (MDV), which transit to multivesicular bodies and eventually the lysosome, or to the peroxisome [25].
Mitochondrial dynamics (fusion and fission) also play a critical role in mitochondrial quality control, and the process is closely linked to mitophagy, where fission is favored and fusion is suppressed, enabling engulfment by autophagosomes. Fission of reticulate mitochondria into smaller fragments is essential for mitophagy to occur [26, 27]. Key to this process is the dynamin-related protein 1 (Drp1), a GTPase in the dynamin super family of proteins, which is recruited to the mitochondria and facilitates the process of mitochondrial fragmentation [28]. Fission 1 (Fis1) is another key player in mitochondrial dynamics that interacts with Drp1 to facilitate mitochondria fragmentation [29]. Mfn1 and 2, which promote OMM fusion, are ubiquitinated and targeted for elimination by the UPS. Optic atrophy protein 1 (OPA1), important for fusion of the inner mitochondrial membrane, is degraded during mitophagy by the inner membrane zinc metalloprotease OMA1, which has overlapping activity with matrix AAA proteases [30–32].
PINK1 is constitutively made and continuously degraded by the mitochondria-specific proteases presenilin-associated rhomboid-like protein (PARL) and mitochondrial processing peptidase (MPP). Loss of membrane potential across the inner mitochondrial membrane inactivates PARL and MPP through an uncharacterized mechanism and permits the accumulation of PINK1 on the OMM. The kinase domain of PINK1 faces the cytosol and phosphorylates OMM proteins facilitating the recruitment of the E3-ubiquitin ligase Parkin [33–35]. PINK1 has been reported to phosphorylate a number of targets including Parkin itself [36, 37], mitofusin 2 (Mfn2) [15], and mitochondrial rho 1 (MIRO) [38], a component of the microtubule-associated motor complex that anchors kinesin to mitochondria. Mfn2, which functions in mitochondrial fusion events and links endoplasmic reticulum to mitochondria, functions as a Parkin receptor after phosphorylation by PINK1, thereby recruiting Parkin to the mitochondria, where it ubiquitinates a number of OMM targets. Voltage-dependent anion channel 1 (VDAC1) has been shown to be a Parkin target essential for mitophagy [19], although this finding has been contested [39]. Ubiquitination and proteasomal degradation of MIRO, Mfn2, and Mfn1 serve to immobilize the mitochondrion and prevent it from rejoining the mitochondrial network through fusion [15, 38, 40–42]. Ubiquitination of OMM proteins facilitates recruitment of autophagy adapter proteins such as neighbor of BRCA1 (NBR1) or sequestosome-1 (p62/SQSTM1). These bifunctional adaptor proteins have an ubiquitin binding domain (UBA) and microtubule-associated protein 1 light chain 3 (LC3) interacting region (LIR) to bring the developing autophagosomal membrane in proximity to the tagged mitochondrion in a zipper-like process [43, 44]. SMAD-specific E3 ubiquitin ligase 1 (SMURF1) has also been linked to Parkin-dependent mitophagy [45]. Surprisingly, its ability to facilitate mitophagy has been found to be independent of its E3 ubiquitin ligase function. Another Parkin-interacting autophagy promoter, activating molecule in Beclin 1-regulated autophagy (Ambra1) dissociates from mitochondrial Bcl-2 to bind Beclin1 to initiate autophagy [46, 47]. Ambra1 interacts with Parkin to promote mitophagy, but is not a substrate of Parkin [48].
Mitophagy that is independent of PINK1/Parkin/ubiquitin can be initiated through atypical members of the Bcl-2 homology domain 3 (BH3) family members such as BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3) and BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like protein (BNIP3L aka NIX). These proteins insert into the OMM and facilitate engulfment by the autophagosome through a LIR domain that can interact with LC3 isoforms including gamma-aminobutyric acid receptor-associated protein (GABARAP) and GABARAP-like 1 (GABARAPL1) [49, 50]. One study demonstrated in cardiomyocytes that Bnip3 recruited Parkin and Drp1 to the mitochondria to promote fission and mitophagy [51]. In hypoxic conditions mitophagy has been reported to be mediated by the OMM protein FUN14 domain containing 1 (FUNDC1) which contains a LIR [52].
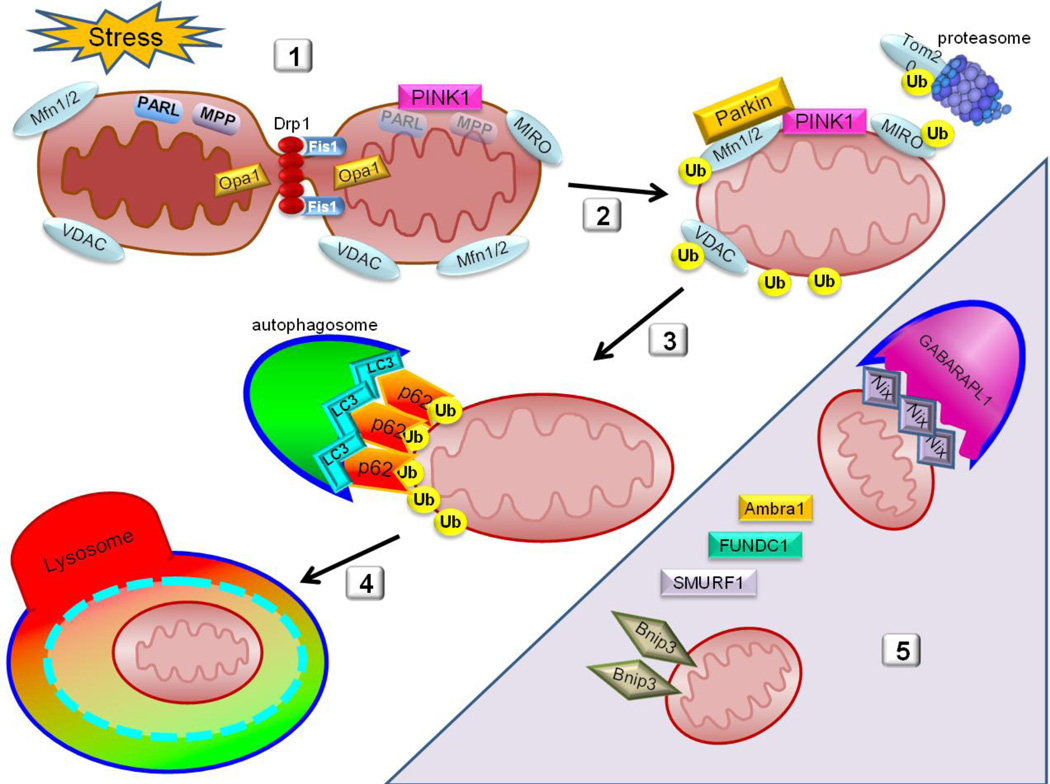
The process of mitophagy is rather complex and requires coordination of UPS-mediated degradation of outer membrane proteins with autophagic engulfment of the remainder of the organelle. The autophagosome, with its cargo, fuses with a lysosome, forming the acidic autophagolysosome. In cells in which the pH-sensitive fluorescent protein Keima is targeted to the mitochondria, one can monitor the delivery of mitochondria to the autophagolysosome by monitoring the pH-dependent shift in fluorescence of mitoKeima [53]. The cargo is degraded by lysosomal hydrolases, liberating amino acids and fatty acids which are exported to the cytosol via lysosomal permeases. A model representing our current knowledge of the process of mitophagy is shown in Figure 1. While mitophagy is responsible for bulk degradation of the organelle, turnover of individual components may proceed at asynchronous rates through redistribution of components via fusion events, selective degradation of proteins via mitochondrial proteases, and proteasomal elimination of some outer membrane proteins. Even in the case of Parkin-dependent mitophagy, some outer membrane proteins are “rescued” through transfer to the ER [54]. Proteomic studies using heavy isotope labeling [55] revealed that proteins of the IMM turn over with rates that are similar to mitochondrial turnover based on historic radiolabeling studies [56, 57], suggesting that IMM proteins (primarily OXPHOS constituents) may be primarily cleared via mitophagy. This also corresponds to studies which showed that matrix and OMM were readily redistributed across the mitochondrial network when fusion and fission were intact; however, IMM constituents redistributed much more slowly [27]. In contrast to the IMM proteins, OM proteins turned over faster in many cases [55], possibly because outer membrane proteins can be degraded by the UPS, by translocation to other sites (ER or peroxisomes), by the MDV pathway, or by mitophagy. Matrix and IMM proteins have fewer routes of degradation: while mitophagy predominates, matrix and IMM proteins can be found in MDVs, and are substrates for Lon and other AAA proteases in the matrix and intermembrane space. The rate of mitochondrial protein turnover in mice is much slower in heart (mean half-life 17d) than in liver (4d). A related study of mitochondrial protein synthesis in rat comparing heart and liver using isotopic labeling and mass spectrometry also showed a 6-fold slower rate of turnover in the heart. Turnover also varied according to subcellular location: protein turnover was ~15% faster in subsarcolemmal mitochondria than in interfibrillar mitochondria [58]. Mitochondrial protein import is inhomogeneous: the fluorescent MitoTimer protein revealed “hot spots” for synthesis and import of this mitochondrially-targeted protein [59].
Figure 1. Parkin-dependent Mitochondrial Autophagy (Mitophagy).
- Cellular stress, such as ischemia/reperfusion, triggers fragmentation of the mitochondria mediated by Drp1, segregating low-membrane potential mitochondria from the rest of the network. Ischemia/reperfusion injury also leads to the collapse of mitochondrial membrane potential which deactivates PARL and MPP, allowing for PINK1 stabilization on the OMM.
- Parkin is recruited to the OMM where it binds Mfn2 and ubiquitinates multiple OMM proteins, marking them for proteasomal degradation and targeted recognition of the ubiquitin-decorated mitochondrion.
- Autophagy adapter proteins such as p62 are then recruited to the mitochondria which in turn bind the ubiquitinated mitochondrion to the phagophore through interaction with LC3 or homologs.
- Once the autophagosome has fully engulfed the mitochondrion, it fuses with a lysosome to form the autophagolysosome where final degradation of bulk contents is completed.
- Shaded area indicates atypical players that participate in recognition and targeting of mitochondria for autophagic clearance. These include Nix and Bnip3 which bind LC3 or homologs including GABARAPL1.
The importance of mitophagy in the heart was highlighted in our previous work demonstrating the requirement for mitophagy in cardioprotection conferred by ischemic preconditioning [13] and acute statin administration [12]. These results suggest that mitophagy is part of the final common pathway for various cardioprotective interventions, and indeed, may be the ultimate effector. Beyond cardioprotection, Parkin-dependent mitophagy plays a role in ischemia tolerance [60], myocardial aging [61], and pathologic remodeling in response to pressure overload [62]. The cardiac effects of Parkin deficiency are phenocopied by PINK1 and Atg5 deletions [63, 64]. Similarly, deletion of Mfn2, considered an essential mitochondrial docking partner for Parkin, leads to mitochondrial dysfunction and heart failure [65]. These findings highlight the importance of autophagy and mitophagy in cardiac function.
1.2 Overview of mitochondrial biogenesis
Mitochondrial biogenesis, which acts in concert with mitophagy to maintain homeostasis in cells, depends on coordination of nuclear and mitochondrial-encoded gene expression. The nuclear-encoded genes are primarily controlled by the transcription cofactor peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) [66]. First identified as a binding partner of peroxisome proliferator-activated receptor γ (PPARγ) that increased its transcriptional activity during thermogenesis, PGC-1α is a member of the nuclear receptor superfamily of proteins that are responsible for assembling functional macrocomplexes of transcriptional machinery at specific DNA sequences [67, 68]. PGC-1α controls the expression of nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2), which in turn control the expression of mitochondrial transcription factor A (Tfam) [69]. Tfam plays a key role in mitochondrial biogenesis by regulating the expression of mitochondrial genes (tRNAs, rRNAs and 13 subunits of the respiratory chain) from the heavy and light strands of the mitochondrial genome; it is also essential for replication of mitochondrial DNA (mtDNA) [70].
Several studies have demonstrated the physiological importance of PGC-1α in mitochondrial biogenesis in response to cold and exercise. These external stimuli increase expression of PGC-1α leading to increased expression of mitochondrial enzymes such as ATP synthase (β-subunit), COX (cytochrome c oxidase) subunits (COX II and COX IV) and δ-aminolevulinate synthase (δ-ALAS) [71, 72]. Increased mitochondria content allows for more efficient thermogenesis in response to cold and enhanced capacity to generate ATP to sustain exercise bouts. An isoform of PGC-1α, PGC-1β has also been identified, and although overexpression of this protein increased mitochondrial biogenesis and basal oxygen consumption much like PGC-1α, it was not induced by cold or exercise, suggesting alternate pathways for induction of mitochondrial biogenesis [73].
Aside from influencing the transcription of key players in the respiratory chain, PGC-1α also interacts with and upregulates the expression of other transcription factors such as peroxisome proliferator-activated receptors (PPARs) [74], hormone receptors for estrogen and thyroxine, as well as estrogen-related receptors (ERRs) α and γ [75]. ERRs are a particularly interesting set of nuclear receptors known as orphan receptors due to a lack of an associated ligand [76]. These proteins are involved in PGC-1α-dependent regulation of mitochondrial biogenesis. For example, Schreiber et al have demonstrated that over-expression of PGC-1α results in upregulation of 151 genes that encode mitochondrial proteins involved in many metabolic functions of mitochondria such as fatty acid β-oxidation (FAO), tricarboxylic acid cycle and oxidative phosphorylation, as well as mitochondrial ribosomal machinery and mitochondrial membrane transport proteins. This effect is inhibited by siRNA targeted to ERRα, and conversely mimicked by overexpression of ERRα [77]. Endonuclease G is regulated by ERRα and PGC-1α, and its deletion results in cardiac hypertrophy and mitochondrial dysfunction [78]. Additional factors implicated in mitochondrial biogenesis include Lon protease and Hsp78 [79].
The expression of PGC-1α is controlled primarily through signaling cascades. Calcineurin A-dependent (CnA) and Ca2+/calmodulin-dependent protein kinase IV (CaMKIV) regulation of PGC-1α has been well characterized. CnA interacts with and activates myocyte enhancer factors 2C and 2D (MEF2C and MEF2D), which regulate the transcription of PGC-1α directly [80, 81]. Furthermore, activation of PGC-1α results in upregulation of MEF2C and 2D, creating a feed forward loop that allows PGC-1α to increase its own expression [82]. CaMKIV activates PGC-1α by phosphorylating the transcription factor cAMP response element (CRE)-binding protein (CREB). Phosphorylated CREB binds to promoter elements of the PGC-1α gene to drive its expression [80]. Other players controlling PGC-1α expression include p38 mitogen-activated protein kinase (p38 MAPK) and AMP-activated protein kinase (AMPK). p38 MAPK activity is increased following exercise, and this leads to the activation of MEF2 and activating transcription factor 2 (ATF2), both of which drive the expression of PGC-1α [83, 84]. The activation of AMPK in response to glucose depletion results in direct phosphorylation of PGC-1α on threonine-177 and serine-538, which is crucial for activation of PGC-1PGC-1α-dependent transcription from the PGC-1α promoter [85].
Post-translational modifications such as phosphorylation and acetylation regulate PGC-1α activity. Kinases that have been implicated in controlling of PGC-1α activity include: AMPK and Akt during caloric restriction and p38 MAPK after endurance exercise [85–87]. Likewise, p38 MAPK increases the activity of PGC-1α by directly phosphorylating threonine-262, serine-265, and threonine-268, which stabilizes the protein and disrupts the interaction between PGC-1α and its inhibitor p160MBP [86, 88]. Conversely, insulin inhibits the activity of PGC-1α through Akt, directly through phosphorylation of the serine-570 residue on PGC-1α, and indirectly through phosphorylation of the Clk2 kinase which in turn phosphorylates the C-terminal serine and threonine-rich regions of PGC-1α, thereby decreasing its co-transcriptional activity [89, 90]. In an even more indirect manner, glycogen synthase kinase 3b (GSK3β) has also been shown to inhibit PGC-1α activity in response to acute oxidative stress by increasing its proteasomal degradation and inhibiting the activity of Sirt1, an NAD-dependent deacetylase thought to activate PGC-1α [91]. This deacetylation event is crucial for the activation of PGC-1α, as the protein is very heavily acetylated by the acetyltransferase GCN5, inhibiting its activity and sequestering it in nuclear foci distant from promoter regions of its target genes [92]. Sirt1 activity is dependent upon the coenzyme nicotinamide adenine dinucleotide (NAD+), and it is therefore highly sensitive to the changes in the energetic state of the cell. Increased NAD+/NADH ratio–which may occur in response to fasting, exercise or redox stress—activates Sirt1, leading to PGC-1α deacetylation [93, 94]. The result of this deacetylation is an increase in transcription of PGC-1α targets, leading to mitochondrial biogenesis [95– 97]. Interestingly, AMPK may once again play a role in activating PGC-1α, not only by directly phosphorylating the protein, but also indirectly by increasing NAD+ levels in the cell by fatty acid oxidation, thereby increasing the activity of Sirt1 [97]. Other posttranslational modifications such as ubiquitination or methylation also play a role in modulating the activity of PGC-1α in response to energy demands and oxidative stress, states that require mitochondrial biogenesis [98].
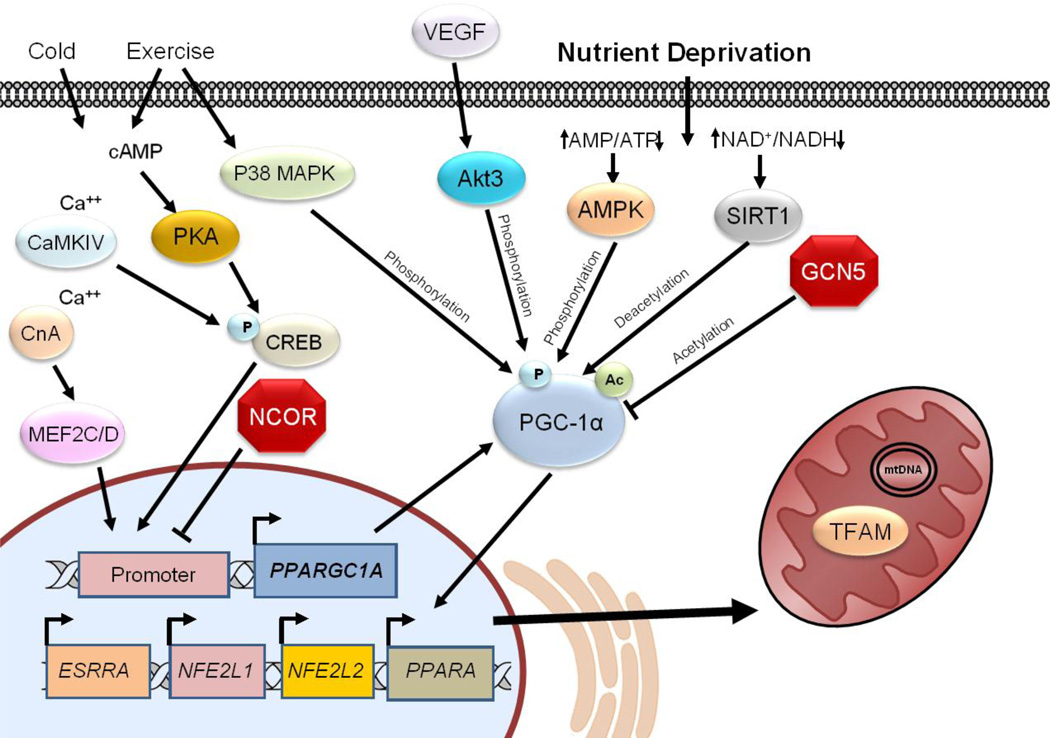
Other proteins that play a major part in mitochondrial biogenesis are vascular endothelial growth factor (VEGF) and hypoxia-inducible factor 1 (HIF-1). VEGF is a key regulator of angiogenesis which involves substantial cell proliferation and production and remodeling of extracellular matrix, processes which utilize large amounts of ATP. A study by Wright et al demonstrated that VEGF stimulates mitochondrial biogenesis by coordinated upregulation of OMM protein Tom70 and activation of PGC-1α [99]. Interestingly, PGC-1α can also activate VEGF, by coactivating ERR-α on conserved binding sites found in the promoter and in a cluster within the first intron of the VEGF gene, driving angiogenesis in response to oxygen deprivation independently of HIF-1 [100]. HIF-1 is a master regulator of the adaptive response to hypoxia, and as such is intimately linked to inducing mitochondrial biogenesis. Several studies have linked PGC-1α and PGC-1β with HIF-1 activity. O’Hagan et al reported that mitochondrial biogenesis driven by the expression of PGC-1α results in increased oxygen consumption and decreased intracellular oxygen tension, permitting stabilization of HIF-1 and activation of a gene expression program to increase oxygen delivery to the tissue [101]. Zhang et al demonstrated that HIF-1 represses mitochondrial biogenesis by controlling the transcription of PGC-1β through downregulation of c-MYC transcription factor [102]. Figure 2 highlights the central role of PGC-1α in regulating mitochondrial biogenesis.
Figure 2. PGC-1α Regulation of Mitochondrial Biogenesis.
PGC-1α is considered a master regulator of mitochondrial biogenesis. Transcriptional control of PGC-1α expression is closely linked to environmental cues of fuel availability, fuel type, and cellular energy requirements. PGC-1α transcription is governed by multiple transcription factors (trans) including PPAR/RXR, MEF2, C/EBP, FoxO, CREB/CRTC, ERRγ, and MyoD/E2A. These factors in turn are activated by specific signal pathways including free fatty acids, AMPK, calcineurin, p38 MAPK, CaMK IV, and PKA, and suppressed by other signals including GCN5, AKT and SHP. In addition to transcriptional control, PGC-1α activity is regulated by acetylation and phosphorylation by the factors illustrated here. Ultimately, PGC-1α increases mitochondrial biogenesis and the capacity to perform OXPHOS, in particular, fatty acid oxidation.
1.3 Interplay between mitophagy and mitochondrial biogenesis
Mitophagy and mitochondrial biogenesis are opposing forces that govern the rate of mitochondrial turnover. This dynamic tension allows for a readily adjustable population of mitochondria to match cellular demands. Here we discuss several players that participate in this regulatory cross-talk.
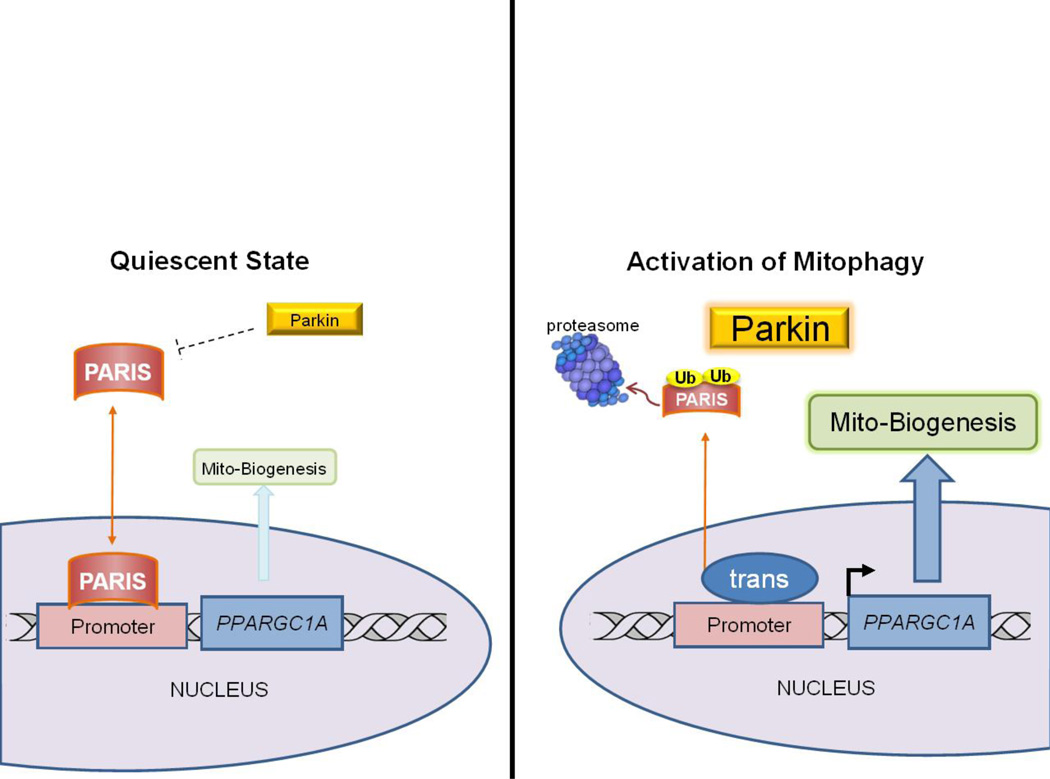
Sirt1, an NAD-dependent lysine deacetylase, stimulates autophagy directly by deacetylating various autophagy proteins including Atg7, Atg5, and Atg8 (LC3) [103]. Sirt1 also deacetylates and activates PGC-1α [104], thus positively regulating both mitophagy and biogenesis. Activation of PGC-1α is a key event in initiating mitochondrial biogenesis, but no less important are the repressors of the process. Parkin-interacting substrate (PARIS or ZNF746) has recently been identified as a direct transcriptional repressor of PGC-1α [105]. The accumulation of PARIS in the nucleus leads to direct inactivation of PGC-1α transcription and inhibition of expression of PGC-1α-dependent genes. Aside from its role in facilitating mitophagy, Parkin was shown to directly target PARIS for degradation through the UPS. Events leading to the upregulation of mitophagy also increase Parkin activity which then degrades PARIS, relieving repression of mitochondrial biogenesis. This relationship establishes an intricate system that links mitophagy with mitochondrial biogenesis. A model illustrating this mechanism linking mitophagy with mitochondrial biogenesis is shown in Figure 3. As the relationship between Parkin and PARIS has thus far been identified only in neuronal models, an important and exciting question remains as to whether this system exists in the heart. Moreover, what are its implications in the setting of ischemia/reperfusion injury?
Figure 3. The Parkin-PARIS Axis Coordinates Mitophagy with Mitochondrial Biogenesis.
Basal state cellular homeostasis is characterized by balanced mitophagy and mitochondrial biogenesis (mitochondrial turnover). This maintains a network of healthy mitochondria. Mitophagy is linked to a transcriptional program for mitochondrial biogenesis. One pathway in this tightly coordinated process involves Parkin and PARIS. Triggers of mitophagy increase Parkin expression and activity, leading to proteasomal degradation of PARIS. Diminished PARIS levels relieve the transcriptional repression of PGC-1α, priming mitochondrial biogenesis.
Other repressors of mitochondrial biogenesis operate in less direct manner. Nuclear co-repressor 1 (Ncor1) acts as a repressor of PPARγ, PPARδ and ERR activity by interacting with histone deacetylases such as HDAC3 and SIRT1 to maintain tonic repression of MEF2, PPARδ, and ERR, thereby suppressing their participation in transcription programs involving mitochondrial oxidative metabolism [106]. mTOR is a serine/threonine kinase involved in numerous cell functions and can directly activate PGC-1α to control mitochondrial biogenesis. During fasting, mTOR is inhibited and autophagy/mitophagy is active. However, as lysosomal degradation releases amino acids, mTOR is reactivated, suppressing autophagy and supporting lysosomal and mitochondrial biogenesis [107]. Eukaryotic translation initiation factor 4E (eIF4E)-binding proteins (4E-BP) prevent translation of targets including nuclear encoded mitochondrial protein mRNAs including TFAM (transcription factor A, mitochondrial) and subunits of complex V and complex I. This inhibition is lifted by the action of mTORC1 which inhibits 4E-BP proteins from binding their targets [108, 109].
Autophagy and mitochondrial biogenesis are linked in both directions: PGC-1α induces the expression of transcription factor EB (TFEB) [110], the master regulator of lysosome biogenesis and the autophagy pathway [111]. TFEB and PGC-1α regulate one another’s expression, and the nutrient sensing regulator GCN5L1 suppresses both TFEB and PGC-1α [112]. Thus like the mythical ouroboros (serpent devouring its own tail), autophagy and mitochondrial biogenesis, mitophagy and lysosomal biogenesis, are elements of a system whose stability derives from its dynamic regulation.
2. Mitochondrial Turnover in the Context of the Organ and the Organism
2.1 Circadian Rhythms and Metabolic Status
The circadian rhythm is an important regulator of cellular and whole-body homeostasis. Disruption of this oscillatory system can cause and aggravate several health issues. The connection between autophagy and circadian rhythm was first demonstrated in the 1970’s using electron microscopic analysis of different rat tissues to show that the number of autophagic vacuoles changed over the course of the day [113, 114]. More recently an important work established a clear connection between circadian rhythm and the induction of autophagy [115]. Ma and collaborators showed that autophagy was upregulated during the dark cycle, preceded by increased mRNA expression of autophagy initiators including Ulk1, and Bnip3, but not factors involved in later phases of autophagy such as LC3B, Atg4, and Atg7. These events were regulated by the transcription factor CCAAT-enhancer-binding protein β (C/EBPβ). C/EBPβ is a liver clock protein that is tissue autonomous and regulates whole body bioenergetics. The induction of autophagy usually promotes mitophagy as well by upregulating mitochondria-targeting machinery such as the autophagy adapter p62/SQSTM1 and the E3 ubiquitin-ligase Parkin [116, 117]. Mitochondrial biogenesis is also directly regulated by the circadian clock, ensuring coordination of mitophagy with biogenesis [118–120]. Conditions that disrupt the circadian clock will also impact autophagy and mitochondrial biogenesis, with consequences for cell and organ function; conversely, disruption of autophagy may affect the clock [121].
The circadian rhythm is closely linked to eating behaviors (and diet types), another important regulator of mitophagy in animals. Mitophagy can be regulated by several different energy sensing stimuli: the availability of AAs in a cell regulates mTOR activity; the glucose level in blood regulates release of insulin and glucagon which have opposing effects on autophagy; and conditions such as metabolic syndrome and caloric restriction regulate autophagy and mitophagy. Insulin suppresses macroautophagy by activating the phosphatidyl-inositol triphosphate (PIP3) cascade leading to phosphorylation and activation of Akt and subsequently mTOR, which inhibits autophagy [122]. Usually high insulin is followed by an increase in AAs in the cytosol, which also activates mTOR, reinforcing the same pathway. Insulin also promotes mitochondrial fusion, whereas hypoglycemia and insulin resistance promote fission [123–125]; both processes are essential to mitochondrial quality control. The nutritional overload of a high fat diet suppresses autophagy and by extension, mitophagy [126–128]. More recently, impaired mitochondrial turnover in mice fed a high-fat diet was demonstrated using Timer protein targeted to mitochondria (MitoTimer). In this study, the authors electroporated a MitoTimer construct under a constitutive promoter into the skeletal muscle of mice which were then maintained on chow or a high-fat diet. They observed a shift in the MitoTimer ratio to red, indicating slower mitochondrial turnover [129]. Although they attributed the red shift to increased oxidative stress, previous work showed that MitoTimer maturation (color shift) was not sensitive to oxidants [130]. MitoTimer is a mitochondria-targeted fluorescent molecular clock that we developed as a tool to monitor mitochondrial turnover [59].
2.2 Cardiac Development, Cardioprotection and Cardiac Pathology
The development of stem cells into differentiated adult cells is a tightly regulated process. Autophagy is involved in the maturation of several different types of cells. Cardiomyocyte development is regulated by the fibroblast growth factor (FGF) signaling axis [131] and in skeletal muscle FoxO is the responsible factor [132]. In both cases, autophagy and mitophagy are tightly linked to differentiation and tissue plasticity [131, 133]. FGF suppresses autophagy and thereby prevents differentiation of cardiac progenitors [134], while FoxO signaling induces autophagy as part of the regeneration and growth of the muscle tissue. As discussed above, mitophagy and biogenesis are tightly linked. These examples highlight the importance of autophagy to tissue remodeling and repair beyond degradation.
Genetic deletions of mitofusin-2 and PINK1 illustrate the importance of mitophagy to heart development and homeostasis. Genetic deletion of mitofusin-2 is embryonic lethal [135] and is essential in the heart not only for mitochondrial dynamics [136], but also ER-mitochondrial calcium signaling [137], mitophagy [15], and autophagosome-lysosome fusion [138]. Absence of PINK1 has profound consequences for postnatal heart development [63] and exacerbates ischemia/reperfusion injury [139].
Among the most potent interventions to protect the heart from ischemia and reperfusion injury are ischemic pre- and post-conditioning [140, 141]. Pre and post conditioning require autophagy to deliver the protection [142–144], although this is controversial in the brain [145, 146]. Mitophagy is part of the autophagy response that is specifically required for protection [13]. Other interventions that protect the heart against ischemic injury, including chloramphenicol [147], caloric restriction [148] simvastatin [12], and SAHA [149] all act through the autophagy/mitophagy pathway, thus establishing autophagy/mitophagy as a hub for cardiac protection. There are few direct inducers of autophagy; rapamycin is an mTOR inhibitor widely used as a drug to induce autophagy. Rapamycin administration also decreased ischemia/reperfusion injury [149, 150] while upregulating autophagy. In chronic models of heart failure, rapamycin also helps to ameliorate the phenotype [151]. Taken together, these facts support the beneficial effects of inducing autophagy.
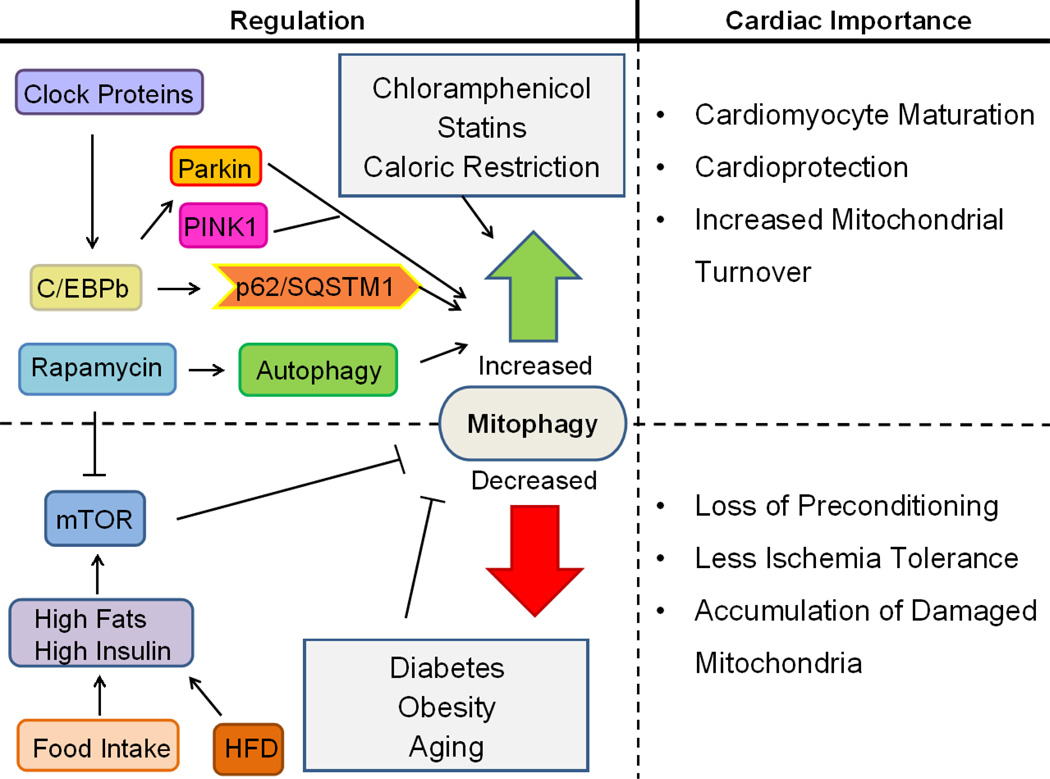
In conditions where autophagy/mitophagy is impaired the opposite is true: there is increased cardiac dysfunction and exacerbation of ischemia/reperfusion injury in the setting of Parkin and PINK1 deletion [60, 139], deletion of macrophage migration inhibitor factor, an inducer of homeostatic autophagy [152], and obesity [153]. Obesity [154] and diabetes [155] disrupt normal energy metabolism, changing basal activation of mTOR and other nutrient signaling cascades that regulate autophagy. High fat diets are known to increase ischemia/reperfusion damage in hearts [156, 157] and there is now a significant amount of work linking the high fat diet to impaired autophagy [154] and accumulation of dysfunctional mitochondria [158, 159], highlighting the importance of mitophagy to cardiac ischemia tolerance. The regulation of cardiac mitophagy and physiological importance of this process is illustrated in Figure 4.
Figure 4. Cardiac Mitophagy Regulation and Significance.
Mitophagy is essential during cardiomyocyte differentiation and for homeostatic mitochondrial turnover to maintain a healthy population of mitochondria. During cardiac stress such as ischemia/reperfusion, mitophagy functions to eliminate damaged mitochondria and reduce injury. Mitophagy is also critical for ischemic preconditioning. Circadian rhythm regulates basal levels of cardiac mitophagy. Nutrient overload, type 2 diabetes, obesity, and advanced age may compromise cardiac autophagy and mitophagy, disrupting this adaptive physiological response to stress.
Autophagy may not always be a good thing in the heart. Infection of juvenile mice with Coxsackievirus B virus can exacerbate stress-induced myocardial injury in adulthood [160]. Autophagy is needed for the spread and reproduction of the virus [161], and virus-induced autophagy triggers premature differentiation of cardiac-resident progenitor cells, contributing to heart failure later in life [160]. Excessive autophagy has been implicated in doxorubicin-mediated cardiac injury [162, 163], although other studies have reported a beneficial role for autophagy [164–167]. Interestingly, deletion of Nrf2, a transcriptional regulator of autophagy and mitochondrial biogenesis, exacerbated doxorubicin toxicity, but this was reversed by overexpression of Atg5 [168]. They did not examine whether restoring autophagy resulted in mitochondrial biogenesis independent of Nrf2. It seems likely that unless mitophagy is balanced by biogenesis, problems will ensue.
Mitochondrial biogenesis in the heart is tightly responsive to oxygen tension. This is manifest at the transition from the fetal hypoxic state to the postnatal aerobic environment, when HIF signaling is lost, thereby favoring mitochondrial fusion and mitochondrial biogenesis [169]. During cardiac hypertrophy in response to aortic banding, mitochondrial dysfunction and decreased biogenesis were noted [170]. Downregulation of PGC-1α is observed in animal models of heart failure, but attempts to restore mitochondrial biogenesis by overexpression of PGC-1α did not improve cardiac function despite a modest increase in mitochondrial content [171]. In fact, inducible overexpression of PGC-1α in the heart resulted in abnormal mitochondrial morphology and cardiomyopathy which was reversible upon normalization of PGC-1α levels [172]. These studies did not examine mitochondrial autophagy. However, in a porcine study of renovascular hypertension, hypertrophy was accompanied by upregulation of mTOR, increased abundance of markers of autophagy and mitophagy, and decreased mitochondrial protein content, all of which were reversed by the angiotensin II receptor blocker valsartan [173]. The authors concluded that hypertension increased autophagic clearance of mitochondria and valsartan suppressed autophagy and restored mitochondrial biogenesis. However, because they did not measure autophagic flux or p62 (a surrogate marker of flux), the data lend themselves to the opposite interpretation: that hypertension impaired autophagic flux, thereby limiting mitochondrial biogenesis possibly through the Parkin/Paris/PGC-1α network. Evidence in support of the latter interpretation is the finding that mTOR was strongly upregulated in the hypertensive hearts, which would suppress autophagy. The observed increase in Beclin 1 and LC3-II could reflect increased autophagy or impaired flux. Beclin 1 is known to interfere with autophagic flux [174, 175]. The perinuclear accumulation of Parkin-decorated mitochondria is also indicative of impaired lysosomal clearance of autophagosomes. This also illustrates the importance of determining autophagic flux and mitochondrial turnover before reaching a conclusion.
3. Prospects and Challenges for the Future
Mitochondrial quality control depends upon mitophagy, biogenesis, fusion, and fission, as well as selective protein quality control via AAA proteases and chaperones. To date, most studies have explored mitochondrial dynamics (fusion/fission) and mitophagy (Parkin-dependent and Parkin-independent mitochondrial autophagy). Mitophagy is increasingly recognized to play a significant role in the heart, yet in order to maintain homeostasis, biogenesis must keep pace. Therefore, approaches to monitoring mitochondrial turnover (the integrated outcome of these four processes) are needed. Recent advances include the analysis of the half-lives of mitochondrial proteins using mass spectrometry analysis and deuterium labeling [55], mito-Keima, which can report on mitochondria delivered to the lysosome [53], and MitoTimer, a fluorescent protein that can be used to monitor mitochondrial turnover [59, 129, 130]. What lies ahead is the application of these tools to study physiologic and pathologic processes in the heart.
A major challenge to overcome is imaging autophagy (or mitophagy) in humans. Relatively few studies have examined autophagy in the human heart, largely because of the challenges of accessing tissue, and none have examined mitophagy, although animal studies indicate that mitochondria are a frequent target of autophagy. There is a significant need to develop better tools for in vivo imaging of autophagy and mitophagy.
Still lacking is a thorough understanding of mitochondrial biogenesis: are all mitochondria equally capable of expanding and undergoing fission to give rise to daughter mitochondria enriched for newly-imported proteins and highly functional OXPHOS assemblies, or is there a subset of mitochondria that are specialized for mitochondrial regeneration? Studies of MitoTimer suggest that protein import preferentially takes place in mitochondria closest to the nucleus [59, 130]. This could be a trivial consequence of mRNA proximity, and import of MitoTimer may not necessarily reflect sites of biogenesis. It is exciting, however, to speculate that the subpopulation of mitochondria most actively engaged in importing newly-synthesized protein is indeed unique. Future studies may shed light on this.
Mitochondrial protein import is essential for biogenesis, but is also implicated in the regulation of mitophagy because PINK1 must transit through the intermembrane space in order to be degraded by PARL. Few studies have considered whether defective protein import is the red flag that signifies a mitochondrion due for autophagic elimination. It has not been demonstrated whether pre-amyloid oligomers might disrupt mitochondrial protein import, yet this might explain the deterioration of mitochondrial function (78) and impaired biogenesis (79) that often accompanies Alzheimer’s disease and potentially other protein folding disorders. We can expect that in the coming years investigators will integrate information exchange between mitochondria and cytosol/nucleus, for which the TOM/TIM complex and VDAC serve as important carriers.
A key to mitochondrial homeostasis is the ability to remove and replace components throughout the network: not only proteins, but also lipids and mtDNA copies. In the heart, where mitochondrial fusion and fission events seem to occur with a frequency approximately equal to the rate of turnover of the entire organelle, intra-mitochondrial degradation and protein import generalized across the network may play a larger role than regionally restricted biogenesis followed by redistribution via fusion events. The intriguing observation that proteins in subsarcolemmal mitochondria turn over faster than in interfibrillar mitochondria suggests that different turnover mechanisms may operate within the same cell. The thought-provoking discovery that individual mitochondrial proteins have widely differing half-lives raises questions about the mechanisms governing this process; regulation of these different mechanisms for degrading mitochondrial proteins may be quite complex, and their contribution to disease phenotypes will be equally so. The importance of mitochondrial protein import is emerging: recently it was reported that redirecting a mutant form of alanine:glyoxalate aminotransferase from mitochondria to peroxisomes corrects primary hyperoxaluria 1 (PH1), a lethal metabolic disease [176]. Enzymes that may traffic either to mitochondria or peroxisomes can have radically different consequences depending on their location; the potential significance of this process for heart disease is unknown at present.
Yet another emerging area is the role of miRNAs in regulating autophagy and mitochondrial biogenesis. miRNA-149 inhibits poly(ADP-ribose) polymerase-2 (PARP-2), thereby allowing an increase in cellular NAD+ and activation of sirtuin-1, leading to mitochondrial biogenesis [177]. miR-27a and miR-27b impair mitochondrial biogenesis [178]. miRNAs also regulate the Nrf2 pathway [179] and autophagy [180–184]. Elucidating the contribution of miRNAs to the dynamic regulation of mitophagy and biogenesis will require a systems biology approach.
Many open questions remain to be resolved, but technical advances continue to make new discoveries possible. The advent of novel gene therapy approaches, cell permeable proteins, and small molecule therapeutics targeting mitochondrial quality control mechanisms hold promise for treating a variety of diseases from the perspective of the underlying mitochondrial dysfunction. It is not too farfetched to envision mitochondrial medicine becoming a medical specialty as much as surgery, cardiology, or genetics.
Mitochondrial Mysteries.
Roberta A. Gottlieb
We know so much yet understand so little
About mitochondrial ox-phos and fusion and fission
Mitochondrial autophagy and biogenesis
MitoTimer and lenses have given us celluvision
Though heart cells live years it’s quite different within
Mitochondrial life is counted in weeks
Outer and inner membrane proteins vary yet more
In their lifespans revealed by mass spectrum peaks.
Protein import must match what’s inside
Lest proteins unfold and fall prey to Lon
The peptides escape to the cytosol
To trigger transcription of chaperones.
Try we must to describe and define
The complex nature of the proteome
As mitochondria expand and divide
Fragment and fall into autophagosomes
Yet for all we know and all we learn
The mysteries grow and questions expand
Like Mandelbrot sets of fractal images
We see the work of divinity’s hand
HIGHLIGHTS.
The role of mitophagy and biogenesis in the heart are discussed
Mitochondrial quality control depends on balanced mitophagy and biogenesis
Factors regulating these processes are summarized
Mitochondrial turnover is discussed in the context of heart disease
Major open questions are enumerated
ACKNOWLEDGMENTS
RAG is a consultant for Takeda Pharmaceuticals and is a cofounder of TissueNetix, Inc. RAG holds the Dorothy and E. Phillip Lyon Chair in Molecular Cardiology in honor of Clarence M. Agress, MD. This work was funded in part by NIH P01 HL112730 (RAG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
The other authors have no potential conflicts of interest to disclose.
REFERENCES
- 1.Bogaerts V, Theuns J, van Broeckhoven C. Genetic findings in Parkinson's disease and translation into treatment: a leading role for mitochondria? Genes, brain, and behavior. 2008;7:129–151. doi: 10.1111/j.1601-183X.2007.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bueler H. Impaired mitochondrial dynamics and function in the pathogenesis of Parkinson's disease. Exp Neurol. 2009;218:235–246. doi: 10.1016/j.expneurol.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Park J, Kim Y, Chung J. Mitochondrial dysfunction and Parkinson's disease genes: insights from Drosophila. Disease models & mechanisms. 2009;2:336–340. doi: 10.1242/dmm.003178. [DOI] [PubMed] [Google Scholar]
- 4.Schapira AH. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet neurology. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- 5.Trancikova A, Tsika E, Moore DJ. Mitochondrial dysfunction in genetic animal models of Parkinson's disease. Antioxid Redox Signal. 2012;16:896–919. doi: 10.1089/ars.2011.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biskup S, Moore DJ. Detrimental deletions: mitochondria, aging and Parkinson's disease. Bioessays. 2006;28:963–967. doi: 10.1002/bies.20471. [DOI] [PubMed] [Google Scholar]
- 7.Kotake Y, Ohta S. MPP+ analogs acting on mitochondria and inducing neuro-degeneration. Curr Med Chem. 2003;10:2507–2516. doi: 10.2174/0929867033456558. [DOI] [PubMed] [Google Scholar]
- 8.Duchen MR, Szabadkai G. Roles of mitochondria in human disease. Essays in biochemistry. 2010;47:115–137. doi: 10.1042/bse0470115. [DOI] [PubMed] [Google Scholar]
- 9.Pellegrino MW, Nargund AM, Haynes CM. Signaling the mitochondrial unfolded protein response. Biochim Biophys Acta. 2013;1833:410–416. doi: 10.1016/j.bbamcr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Wang X. The interplay between autophagy and the ubiquitin-proteasome system in cardiac proteotoxicity. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbadis.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andres AM, Hernandez G, Lee P, Huang C, Ratliff EP, Sin J, et al. Mitophagy is Required for Acute Cardioprotection by Simvastatin. Antioxid Redox Signal. 2013 doi: 10.1089/ars.2013.5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, et al. Parkin Protein Deficiency Exacerbates Cardiac Injury and Reduces Survival following Myocardial Infarction. Journal of Biological Chemistry. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubli DA, Quinsay MN, Huang C, Lee Y, Gustafsson AB. Bnip3 functions as a mitochondrial sensor of oxidative stress during myocardial ischemia and reperfusion. Am J Physiol Heart Circ Physiol. 2008;295:H2025–H2031. doi: 10.1152/ajpheart.00552.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. Embo J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carreira RS, Lee Y, Ghochani M, Gustafsson AB, Gottlieb RA. Cyclophilin D is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 2010;6:462–472. doi: 10.4161/auto.6.4.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 20.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 24.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 25.Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014 doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacVicar TD, Lane JD. Impaired OMA1-dependent cleavage of OPA1 and reduced DRP1 fission activity combine to prevent mitophagy in cells that are dependent on oxidative phosphorylation. Journal of cell Science. 2014;127:2313–2325. doi: 10.1242/jcs.144337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaser M, Kambacheld M, Kisters-Woike B, Langer T. Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J Biol Chem. 2003;278:46414–46423. doi: 10.1074/jbc.M305584200. [DOI] [PubMed] [Google Scholar]
- 32.McBride H, Soubannier V. Mitochondrial function: OMA1 and OPA1, the grandmasters of mitochondrial health. Current biology : CB. 2010;20:R274–R276. doi: 10.1016/j.cub.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 33.Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, et al. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012;13:378–385. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, et al. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iguchi M, Kujuro Y, Okatsu K, Koyano F, Kosako H, Kimura M, et al. Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J Biol Chem. 2013;288:20027–22032. doi: 10.1074/jbc.M113.467530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Developmental cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glauser L, Sonnay S, Stafa K, Moore DJ. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J Neurochem. 2011;118:636–645. doi: 10.1111/j.1471-4159.2011.07318.x. [DOI] [PubMed] [Google Scholar]
- 42.Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 44.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 45.Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, Tang Y, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 47.Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, et al. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30:1195–1208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Humbeeck C, Cornelissen T, Hofkens H, Mandemakers W, Gevaert K, De Strooper B, et al. Parkin interacts with Ambra1 to induce mitophagy. J Neurosci. 2011;31:10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakrama FZ, Seguin-Py S, Le Grand JN, Fraichard A, Delage-Mourroux R, Despouy G, et al. GABARAPL1 (GEC1) associates with autophagic vesicles. Autophagy. 2010;6:495–505. doi: 10.4161/auto.6.4.11819. [DOI] [PubMed] [Google Scholar]
- 50.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee Y, Lee HY, Hanna RA, Gustafsson AB. Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. Am J Physiol Heart Circ Physiol. 2011;301:H1924–H1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 53.Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chemistry & biology. 2011;18:1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 54.Saita S, Shirane M, Nakayama KI. Selective escape of proteins from the mitochondria during mitophagy. Nature communications. 2013;4:1410. doi: 10.1038/ncomms2400. [DOI] [PubMed] [Google Scholar]
- 55.Kim TY, Wang D, Kim AK, Lau E, Lin AJ, Liem DA, et al. Metabolic Labeling Reveals Proteome Dynamics of Mouse Mitochondria. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.M112.021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aschenbrenner B, Druyan R, Albin R, Rabinowitz M. Haem a, cytochrome c and total protein turnover in mitochondria from rat heart and liver. Biochem J. 1970;119:157–160. doi: 10.1042/bj1190157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lipsky NG, Pedersen P. Mitochondrial turnover in animal cells. Half-lives of mitochondria and mitochondrial subfractions of rat liver based on [14C]bicarbonate incorporation. J Biol Chem. 1981;256:8652–8657. [PubMed] [Google Scholar]
- 58.Kasumov T, Dabkowski ER, Shekar KC, Li L, Ribeiro RF, Jr, Walsh K, et al. Assessment of cardiac proteome dynamics with heavy water: slower protein synthesis rates in interfibrillar than subsarcolemmal mitochondria. Am J Physiol Heart Circ Physiol. 2013;304:H1201–H1214. doi: 10.1152/ajpheart.00933.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hernandez G, Thornton C, Stotland A, Lui D, Sin J, Ramil J, et al. MitoTimer: A novel tool for monitoring mitochondrial turnover. Autophagy. 2013;9:1852–1861. doi: 10.4161/auto.26501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kubli DA, Zhang X, Lee Y, Hanna RA, Quinsay MN, Nguyen CK, et al. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kubli DA, Quinsay MN, Gustafsson AB. Parkin deficiency results in accumulation of abnormal mitochondria in aging myocytes. Communicative & integrative biology. 2013;6:e24511. doi: 10.4161/cib.24511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci U S A. 2011;108:9572–9577. doi: 10.1073/pnas.1106291108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 65.Chen Y, Liu Y, Dorn GW., 2nd Mitochondrial fusion is essential for organelle function and cardiac homeostasis. Circ Res. 2011;109:1327–1331. doi: 10.1161/CIRCRESAHA.111.258723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu Z, Puigserver P, Spiegelman BM. Transcriptional activation of adipogenesis. Curr Opin Cell Biol. 1999;11:689–6894. doi: 10.1016/s0955-0674(99)00037-x. [DOI] [PubMed] [Google Scholar]
- 67.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 68.Xiao X, Wang P, Chou KC. Recent progresses in identifying nuclear receptors and their families. Current topics in medicinal chemistry. 2013;13:1192–1200. doi: 10.2174/15680266113139990006. [DOI] [PubMed] [Google Scholar]
- 69.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci U S A. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fisher RP, Clayton DA. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J Biol Chem. 1985;260:11330–11338. [PubMed] [Google Scholar]
- 71.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 72.Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, et al. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 73.Meirhaeghe A, Crowley V, Lenaghan C, Lelliott C, Green K, Stewart A, et al. Characterization of the human, mouse and rat PGC1 beta (peroxisome-proliferator-activated receptor-gamma co-activator 1 beta) gene in vitro and in vivo. Biochem J. 2003;373:155–165. doi: 10.1042/BJ20030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Madrazo JA, Kelly DP. The PPAR trio: regulators of myocardial energy metabolism in health and disease. J Mol Cell Cardiol. 2008;44:968–975. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 75.Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 76.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocrine reviews. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 77.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McDermott-Roe C, Ye J, Ahmed R, Sun XM, Serafin A, Ware J, et al. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;478:114–118. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marton O, Koltai E, Takeda M, Koch LG, Britton SL, Davies KJ, et al. Mitochondrial biogenesis-associated factors underlie the magnitude of response to aerobic endurance training in rats. Pflugers Arch. 2014 doi: 10.1007/s00424-014-1554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 81.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci U S A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 83.Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, et al. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280:19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 84.Zhao M, New L, Kravchenko VV, Kato Y, Gram H, di Padova F, et al. Regulation of the MEF2 family of transcription factors by p38. Mol Cell Biol. 1999;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 87.Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, et al. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci U S A. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sano M, Tokudome S, Shimizu N, Yoshikawa N, Ogawa C, Shirakawa K, et al. Intramolecular control of protein stability, subnuclear compartmentalization, and coactivator function of peroxisome proliferator-activated receptor gamma coactivator 1alpha. J Biol Chem. 2007;282:25970–25980. doi: 10.1074/jbc.M703634200. [DOI] [PubMed] [Google Scholar]
- 89.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 90.Rodgers JT, Haas W, Gygi SP, Puigserver P. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell Metab. 2010;11:23–34. doi: 10.1016/j.cmet.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anderson RM, Barger JL, Edwards MG, Braun KH, O'Connor CE, Prolla TA, et al. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008;7:101–111. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 93.Hayashida S, Arimoto A, Kuramoto Y, Kozako T, Honda S, Shimeno H, et al. Fasting promotes the expression of SIRT1, an NAD+ -dependent protein deacetylase, via activation of PPARalpha in mice. Mol Cell Biochem. 2010;339:285–292. doi: 10.1007/s11010-010-0391-z. [DOI] [PubMed] [Google Scholar]
- 94.White AT, Schenk S. NAD(+)/NADH and skeletal muscle mitochondrial adaptations to exercise. American journal of physiology Endocrinology and metabolism. 2012;303:E308–E321. doi: 10.1152/ajpendo.00054.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocrine reviews. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. The American journal of clinical nutrition. 2011;93:884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wright GL, Maroulakou IG, Eldridge J, Liby TL, Sridharan V, Tsichlis PN, et al. VEGF stimulation of mitochondrial biogenesis: requirement of AKT3 kinase. FASEB J. 2008;22:3264–3275. doi: 10.1096/fj.08-106468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 101.O'Hagan KA, Cocchiglia S, Zhdanov AV, Tambuwala MM, Cummins EP, Monfared M, et al. PGC-1alpha is coupled to HIF-1alpha-dependent gene expression by increasing mitochondrial oxygen consumption in skeletal muscle cells. Proc Natl Acad Sci U S A. 2009;106:2188–2193. doi: 10.1073/pnas.0808801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 103.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 105.Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, et al. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yamamoto H, Williams EG, Mouchiroud L, Canto C, Fan W, Downes M, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147:827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 109.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 110.Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, et al. PGC-1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Science translational medicine. 2012;4:142ra97. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Scott I, Webster BR, Chan CK, Okonkwo JU, Han K, Sack MN. GCN5-like protein 1 (GCN5L1) controls mitochondrial content through coordinated regulation of mitochondrial biogenesis and mitophagy. J Biol Chem. 2014;289:2864–2872. doi: 10.1074/jbc.M113.521641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pfeifer U, Scheller H. A morphometric study of cellular autophagy including diurnal variations in kidney tubules of normal rats. J Cell Biol. 1975;64:608–621. doi: 10.1083/jcb.64.3.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reme CE, Sulser M. Diurnal variation of autophagy in rod visual cells in the rat. Albrecht von Graefes Archiv fur klinische und experimentelle Ophthalmologie Albrecht von Graefe's archive for clinical and experimental ophthalmology. 1977;203:261–270. doi: 10.1007/BF00409832. [DOI] [PubMed] [Google Scholar]
- 115.Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J. 2011;30:4642–4651. doi: 10.1038/emboj.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cali T, Ottolini D, Negro A, Brini M. Enhanced parkin levels favor ER-mitochondria crosstalk and guarantee Ca(2+) transfer to sustain cell bioenergetics. Biochim Biophys Acta. 2013;1832:495–508. doi: 10.1016/j.bbadis.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 117.Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc Natl Acad Sci U S A. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bray MS, Young ME. Diurnal variations in myocardial metabolism. Cardiovasc Res. 2008;79:228–237. doi: 10.1093/cvr/cvn054. [DOI] [PubMed] [Google Scholar]
- 119.Bray MS, Shaw CA, Moore MW, Garcia RA, Zanquetta MM, Durgan DJ, et al. Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol Heart Circ Physiol. 2008;294:H1036–H1047. doi: 10.1152/ajpheart.01291.2007. [DOI] [PubMed] [Google Scholar]
- 120.Young ME, Razeghi P, Cedars AM, Guthrie PH, Taegtmeyer H. Intrinsic diurnal variations in cardiac metabolism and contractile function. Circ Res. 2001;89:1199–1208. doi: 10.1161/hh2401.100741. [DOI] [PubMed] [Google Scholar]
- 121.Ma D, Li S, Molusky MM, Lin JD. Circadian autophagy rhythm: a link between clock and metabolism? Trends in endocrinology and metabolism: TEM. 2012;23:319–325. doi: 10.1016/j.tem.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Troncoso R, Diaz-Elizondo J, Espinoza SP, Navarro-Marquez MF, Oyarzun AP, Riquelme JA, et al. Regulation of cardiac autophagy by insulin-like growth factor 1. IUBMB Life. 2013;65:593–601. doi: 10.1002/iub.1172. [DOI] [PubMed] [Google Scholar]
- 123.Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jhun BS, Lee H, Jin ZG, Yoon Y. Glucose stimulation induces dynamic change of mitochondrial morphology to promote insulin secretion in the insulinoma cell line INS-1E. PLoS One. 2013;8:e60810. doi: 10.1371/journal.pone.0060810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Holmstrom MH, Iglesias-Gutierrez E, Zierath JR, Garcia-Roves PM. Tissue-specific control of mitochondrial respiration in obesity-related insulin resistance and diabetes. American journal of physiology Endocrinology and metabolism. 2012;302:E731–E739. doi: 10.1152/ajpendo.00159.2011. [DOI] [PubMed] [Google Scholar]
- 126.Giricz Z, Mentzer RM, Jr, Gottlieb RA. Autophagy, myocardial protection, and the metabolic syndrome. J Cardiovasc Pharmacol. 2012;60:125–132. doi: 10.1097/FJC.0b013e318256ce10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu X, Ren J. Unmasking the janus faces of autophagy in obesity-associated insulin resistance and cardiac dysfunction. Clinical and experimental pharmacology & physiology. 2012;39:200–208. doi: 10.1111/j.1440-1681.2011.05638.x. [DOI] [PubMed] [Google Scholar]
- 128.Sciarretta S, Zhai P, Shao D, Maejima Y, Robbins J, Volpe M, et al. Rheb is a critical regulator of autophagy during myocardial ischemia: pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012;125:1134–146. doi: 10.1161/CIRCULATIONAHA.111.078212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Laker RC, Xu P, Ryall KA, Sujkowski A, Kenwood BM, Chain KH, et al. A novel MitoTimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. J Biol Chem. 2014;289:12005–12015. doi: 10.1074/jbc.M113.530527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ferree AW, Trudeau K, Zik E, Benador IY, Twig G, Gottlieb RA, et al. MitoTimer probe reveals the impact of autophagy, fusion, and motility on subcellular distribution of young and old mitochondrial protein and on relative mitochondrial protein age. Autophagy. 2013;9:1887–1896. doi: 10.4161/auto.26503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang J, Liu J, Liu L, McKeehan WL, Wang F. The fibroblast growth factor signaling axis controls cardiac stem cell differentiation through regulating autophagy. Autophagy. 2012;8:690–691. doi: 10.4161/auto.19290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sanchez AM, Candau RB, Bernardi H. FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell Mol Life Sci. 2014;71:1657–1671. doi: 10.1007/s00018-013-1513-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sanchez AM, Bernardi H, Py G, Candau R. Autophagy is essential to support skeletal muscle plasticity in response to endurance exercise. Am J Physiol Regul Integr Comp Physiol. 2014 doi: 10.1152/ajpregu.00187.2014. [DOI] [PubMed] [Google Scholar]
- 134.Zhang J, Liu J, Huang Y, Chang JY, Liu L, McKeehan WL, et al. FRS2alpha-mediated FGF signals suppress premature differentiation of cardiac stem cells through regulating autophagy activity. Circ Res. 2012;110:e29–e39. doi: 10.1161/CIRCRESAHA.111.255950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Eura Y, Ishihara N, Yokota S, Mihara K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J Biochem. 2003;134:333–344. doi: 10.1093/jb/mvg150. [DOI] [PubMed] [Google Scholar]
- 137.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 138.Zhao T, Huang X, Han L, Wang X, Cheng H, Zhao Y, et al. Central role of mitofusin 2 in autophagosome-lysosome fusion in cardiomyocytes. J Biol Chem. 2012;287:23615–23625. doi: 10.1074/jbc.M112.379164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Siddall HK, Yellon DM, Ong SB, Mukherjee UA, Burke N, Hall AR, et al. Loss of PINK1 increases the heart's vulnerability to ischemia-reperfusion injury. PLoS One. 2013;8:e62400. doi: 10.1371/journal.pone.0062400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Murry CE, Jennings RB, Reimer KA. Preconditioning with ischemia: a delay of lethal cell injury in ischemic myocardium. Circulation. 1986;74:1124–1136. doi: 10.1161/01.cir.74.5.1124. [DOI] [PubMed] [Google Scholar]
- 141.Zhao ZQ, Corvera JS, Halkos ME, Kerendi F, Wang NP, Guyton RA, et al. Inhibition of myocardial injury by ischemic postconditioning during reperfusion: comparison with ischemic preconditioning. American journal of physiology Heart and circulatory physiology. 2003;285:H579–H588. doi: 10.1152/ajpheart.01064.2002. [DOI] [PubMed] [Google Scholar]
- 142.Han Z, Cao J, Song D, Tian L, Chen K, Wang Y, et al. Autophagy is involved in the cardioprotection effect of remote limb ischemic postconditioning on myocardial ischemia/reperfusion injury in normal mice, but not diabetic mice. PLoS One. 2014;9:e86838. doi: 10.1371/journal.pone.0086838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huang C, Yitzhaki S, Perry CN, Liu W, Giricz Z, Mentzer RM, Jr, et al. Autophagy induced by ischemic preconditioning is essential for cardioprotection. J Cardiovasc Transl Res. 2010;3:365–373. doi: 10.1007/s12265-010-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wei C, Li H, Han L, Zhang L, Yang X. Activation of autophagy in ischemic postconditioning contributes to cardioprotective effects against ischemia/reperfusion injury in rat hearts. J Cardiovasc Pharmacol. 2013;61:416–422. doi: 10.1097/FJC.0b013e318287d501. [DOI] [PubMed] [Google Scholar]
- 145.Gao L, Jiang T, Guo J, Liu Y, Cui G, Gu L, et al. Inhibition of autophagy contributes to ischemic postconditioning-induced neuroprotection against focal cerebral ischemia in rats. PLoS One. 2012;7:e46092. doi: 10.1371/journal.pone.0046092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Qi ZF, Luo YM, Liu XR, Wang RL, Zhao HP, Yan F, et al. AKT/GSK3beta-Dependent Autophagy Contributes to the Neuroprotection of Limb Remote Ischemic Postconditioning in the Transient Cerebral Ischemic Rat Model. CNS neuroscience & therapeutics. 2012:965–973. doi: 10.1111/cns.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sala-Mercado JA, Wider J, Undyala VV, Jahania S, Yoo W, Mentzer RM, Jr, et al. Profound cardioprotection with chloramphenicol succinate in the swine model of myocardial ischemia-reperfusion injury. Circulation. 2010;122:S179–S184. doi: 10.1161/CIRCULATIONAHA.109.928242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shinmura K, Tamaki K, Sano M, Murata M, Yamakawa H, Ishida H, et al. Impact of long-term caloric restriction on cardiac senescence: caloric restriction ameliorates cardiac diastolic dysfunction associated with aging. J Mol Cell Cardiol. 2011;50:117–127. doi: 10.1016/j.yjmcc.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 149.Xie M, Kong Y, Tan W, May H, Battiprolu PK, Pedrozo Z, et al. Histone deacetylase inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation. 2014;129:1139–1151. doi: 10.1161/CIRCULATIONAHA.113.002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dutta D, Xu J, Kim JS, Dunn WA, Jr, Leeuwenburgh C. Upregulated autophagy protects cardiomyocytes from oxidative stress-induced toxicity. Autophagy. 2013;9:328–344. doi: 10.4161/auto.22971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sciarretta S, Volpe M, Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res. 2014;114:549–564. doi: 10.1161/CIRCRESAHA.114.302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu X, Hua Y, Nair S, Bucala R, Ren J. Macrophage migration inhibitory factor deletion exacerbates pressure overload-induced cardiac hypertrophy through mitigating autophagy. Hypertension. 2014;63:490–499. doi: 10.1161/HYPERTENSIONAHA.113.02219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Giricz Z, Mentzer RM, Jr, Gottlieb RA. Autophagy, Myocardial Protection and the Metabolic Syndrome. J Cardiovasc Pharmacol. 2012 doi: 10.1097/FJC.0b013e318256ce10. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ren SY, Xu X. Role of autophagy in metabolic syndrome-associated heart disease. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbadis.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kobayashi S, Liang Q. Autophagy and mitophagy in diabetic cardiomyopathy. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbadis.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 156.Chess DJ, Stanley WC. Role of diet and fuel overabundance in the development and progression of heart failure. Cardiovasc Res. 2008;79:269–278. doi: 10.1093/cvr/cvn074. [DOI] [PubMed] [Google Scholar]
- 157.Mozaffari MS, Schaffer SW. Myocardial ischemic-reperfusion injury in a rat model of metabolic syndrome. Obesity (Silver Spring, Md) 2008;16:2253–2258. doi: 10.1038/oby.2008.356. [DOI] [PubMed] [Google Scholar]
- 158.Essop MF, Anna Chan WY, Valle A, Garcia-Palmer FJ, Du Toit EF. Impaired contractile function and mitochondrial respiratory capacity in response to oxygen deprivation in a rat model of pre-diabetes. Acta physiologica. 2009;197:289–296. doi: 10.1111/j.1748-1716.2009.02024.x. [DOI] [PubMed] [Google Scholar]
- 159.Yu L, Fink BD, Herlein JA, Oltman CL, Lamping KG, Sivitz WI. Dietary fat, fatty acid saturation and mitochondrial bioenergetics. J Bioenerg Biomembr. 2014;46:33–44. doi: 10.1007/s10863-013-9530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sin J, Puccini JM, Huang C, Konstandin MH, Gilbert PE, Sussman MA, et al. The impact of juvenile coxsackievirus infection on cardiac progenitor cells and postnatal heart development. PLoS pathogens. 2014;10:e1004249. doi: 10.1371/journal.ppat.1004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Robinson SM, Tsueng G, Sin J, Mangale V, Rahawi S, McIntyre LL, et al. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS pathogens. 2014;10:e1004045. doi: 10.1371/journal.ppat.1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dirks-Naylor AJ. The role of autophagy in doxorubicin-induced cardiotoxicity. Life sciences. 2013;93:913–916. [PubMed] [Google Scholar]
- 163.Wang X, Wang XL, Chen HL, Wu D, Chen JX, Wang XX, et al. Ghrelin inhibits doxorubicin cardiotoxicity by inhibiting excessive autophagy through AMPK and p38-MAPK. Biochem Pharmacol. 2014;88:334–350. doi: 10.1016/j.bcp.2014.01.040. [DOI] [PubMed] [Google Scholar]
- 164.Kawaguchi T, Takemura G, Kanamori H, Takeyama T, Watanabe T, Morishita K, et al. Prior starvation mitigates acute doxorubicin cardiotoxicity through restoration of autophagy in affected cardiomyocytes. Cardiovasc Res. 2012;96:456–465. doi: 10.1093/cvr/cvs282. [DOI] [PubMed] [Google Scholar]
- 165.Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, et al. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nature communications. 2013;4:2308. doi: 10.1038/ncomms3308. [DOI] [PubMed] [Google Scholar]
- 166.Sishi BJ, Loos B, van Rooyen J, Engelbrecht AM. Autophagy upregulation promotes survival and attenuates doxorubicin-induced cardiotoxicity. Biochem Pharmacol. 2013;85:124–134. doi: 10.1016/j.bcp.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 167.Xu X, Bucala R, Ren J. Macrophage migration inhibitory factor deficiency augments doxorubicin-induced cardiomyopathy. Journal of the American Heart Association. 2013;2:e000439. doi: 10.1161/JAHA.113.000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li S, Wang W, Niu T, Wang H, Li B, Shao L, et al. Nrf2 deficiency exaggerates doxorubicin-induced cardiotoxicity and cardiac dysfunction. Oxidative medicine and cellular longevity. 2014;2014:748524. doi: 10.1155/2014/748524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Neary MT, Ng KE, Ludtmann MH, Hall AR, Piotrowska I, Ong SB, et al. Hypoxia signaling controls postnatal changes in cardiac mitochondrial morphology and function. J Mol Cell Cardiol. 2014;74:340–352. doi: 10.1016/j.yjmcc.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mei Z, Wang X, Liu W, Gong J, Gao X, Zhang T, et al. Mitochondrial adaptations during myocardial hypertrophy induced by abdominal aortic constriction. Cardiovasc Pathol. 2014;23:283–288. doi: 10.1016/j.carpath.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 171.Karamanlidis G, Garcia Menendez L, Kolwicz SC, Jr, Lee CF, Tian R. Promoting PGC1alpha-driven Mitochondrial Biogenesis Is Detrimental In Pressure Overloaded Mouse Hearts. Am J Physiol Heart Circ Physiol. 2014 doi: 10.1152/ajpheart.00280.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, et al. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- 173.Zhang X, Li ZL, Crane JA, Jordan KL, Pawar AS, Textor SC, et al. Valsartan regulates myocardial autophagy and mitochondrial turnover in experimental hypertension. Hypertension. 2014;64:87–93. doi: 10.1161/HYPERTENSIONAHA.113.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ma X, Liu H, Foyil SR, Godar RJ, Weinheimer CJ, Hill JA, et al. Impaired Autophagosome Clearance Contributes to Cardiomyocyte Death in Ischemia-Reperfusion Injury. Circulation. 2012 doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Miyata N, Steffen J, Johnson ME, Fargue S, Danpure CJ, Koehler CM. Pharmacologic rescue of an enzyme-trafficking defect in primary hyperoxaluria 1. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1408401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mohamed JS, Hajira A, Pardo PS, Boriek AM. MicroRNA-149 inhibits PARP-2 and promotes mitochondrial biogenesis via SIRT-1/PGC-1alpha network in skeletal muscle. Diabetes. 2014;63:1546–1559. doi: 10.2337/db13-1364. [DOI] [PubMed] [Google Scholar]
- 178.Kang T, Lu W, Xu W, Anderson L, Bacanamwo M, Thompson W, et al. MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. J Biol Chem. 2013;288:34394–34402. doi: 10.1074/jbc.M113.514372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Cheng X, Ku CH, Siow RC. Regulation of the Nrf2 antioxidant pathway by microRNAs: New players in micromanaging redox homeostasis. Free Radic Biol Med. 2013;64:4–11. doi: 10.1016/j.freeradbiomed.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 180.Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie T, et al. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget. 2014;5:7013–7026. doi: 10.18632/oncotarget.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Song L, Su M, Wang S, Zou Y, Wang X, Wang Y, et al. MiR-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. J Cell Mol Med. 2014 doi: 10.1111/jcmm.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Seca H, Lima RT, Lopes-Rodrigues V, Guimaraes JE, Almeida GM, Vasconcelos MH. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Current drug targets. 2013;14:1135–1143. doi: 10.2174/13894501113149990185. [DOI] [PubMed] [Google Scholar]
- 183.Pan W, Zhong Y, Cheng C, Liu B, Wang L, Li A, et al. MiR-30-regulated autophagy mediates angiotensin II-induced myocardial hypertrophy. PLoS One. 2013;8:e53950. doi: 10.1371/journal.pone.0053950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yu Y, Yang L, Zhao M, Zhu S, Kang R, Vernon P, et al. Targeting microRNA-30a-mediated autophagy enhances imatinib activity against human chronic myeloid leukemia cells. Leukemia. 2012;26:1752–1760. doi: 10.1038/leu.2012.65. [DOI] [PubMed] [Google Scholar]