Abstract
The host immune response plays a critical role not only in protection from human leishmaniasis, but also in promoting disease severity. Although candidate gene approaches in mouse models of leishmaniasis have been extremely informative, a global understanding of the immune pathways active in lesions from human patients is lacking. To address this issue, genome-wide transcriptional profiling of Leishmania braziliensis-infected cutaneous lesions and normal skin controls was carried out. A signature of the L. braziliensis skin lesion was defined that includes over 2,000 differentially regulated genes. Pathway-level analysis of this transcriptional response revealed key biological pathways present in cutaneous lesions, generating a testable ‘metapathway’ model of immunopathology, and providing new insights for treatment of human leishmaniasis.
Introduction
Leishmania braziliensis has a spectrum of clinical manifestations, all of which are associated with immunopathology (de Oliveira and Brodskyn, 2012). Patients develop small nodules at the site of infection that progress to chronic ulcerated lesions. We hypothesize that while parasite infection acts as an initial trigger for lesion development, it is the immunopathologic response that determines disease severity. Thus, defining the host inflammatory pathways within leishmania lesions is crucial for the development of new treatment modalities.
Many studies have examined the systemic immune response in L. braziliensis infected patients, and show that cells from patients release pro-inflammatory molecules in response to leishmania antigen (Bottrel et al., 2001; Follador et al., 2002; Vargas-Inchaustegui et al., 2010). These responses likely contribute to both the control of the parasites and the pathologic inflammatory response in the lesions (Bacellar et al., 2002; Bosque et al., 1998; de Oliveira and Brodskyn, 2012; Giudice et al., 2012). While important, these systemic responses may not reflect what is occurring at the site of infection. Indeed, recent studies of lesion biopsies from L. braziliensis patients have revealed an unexpected pathologic role for CD8 T cells during disease, which would not have been obvious from studies on systemic responses (Novais et al., 2013; Santos Cda et al., 2013).
Transcriptome analysis has helped to elucidate critical genes expressed during interactions between leishmania parasites and human macrophages (Ramirez et al., 2012). In addition, a genomic profiling has been reported for leishmania lesions from patients, in which the authors compared cutaneous leishmaniasis (CL) and mucosal leishmaniasis (Maretti-Mira et al., 2012). To our knowledge, however, ours is the first report to dissect the changes that occur in the skin after infection with leishmania when compared to normal skin. Using a genome-wide transcriptional analysis, we report on the pathways present in L. braziliensis lesions and propose a hypothetical ‘metapathway’ of immunopathology that drives disease.
Results
Comparative transcriptomics of L. braziliensis lesions and normal skin
We performed genome-wide transcriptional profiling on 25 biopsies from L. braziliensis patients (Table S1) and 10 normal skin biopsies obtained from non-endemic controls. Principal component analysis (PCA) of the entire dataset showed that principal component 1 (PC1) accounted for 54.3% of the variation in the data and resolved samples into two main groups, normal and lesion skin. PC2 accounted for a smaller amount of variation (12.4%) occurring within both these groups (Fig. 1a). The separation of lesion and control samples along a single principle component indicated that differentially expressed genes could be identified with high statistical confidence.
Figure 1. Defining the transcriptome of the human L. braziliensis skin lesion.
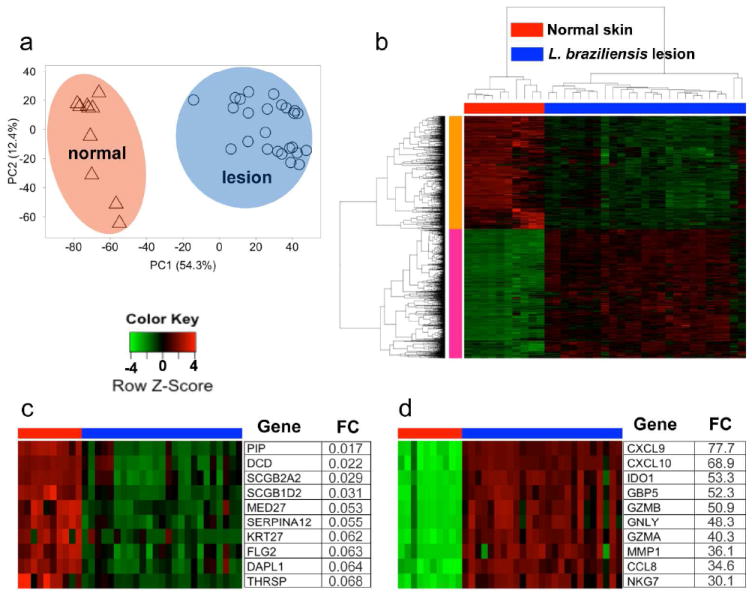
(a) Principal component analysis of entire normalized data set showing PC1 (x-axis) and PC2 (y-axis). (b) HC of all the differentially expressed genes between normal skin and L. braziliensis lesion (FC ≥ 2 and FDR ≤ 1%). (c and d) Top 10 ‘induced’ (c) and ‘repressed’ (d) genes. (b-d) Columns represent individual patients and rows individual genes, colored to indicate expression levels based on a Z-Score. Abbreviations: PC, principal component; FC, fold-change; HC, hierarchical clustering; FDR, false discovery rate.
Analysis of L. braziliensis lesions compared to normal skin identified 2,028 differentially expressed genes (≥ 2-fold, FDR ≤ 1%) (Fig. 1b). Hierarchical clustering (HC) based on Pearson correlation delineated two major clusters. Cluster 1 is comprised of 947 genes whose abundance is decreased in lesions, relative to normal skin. The 10 most ‘repressed’ genes from this cluster include genes associated with maintenance of skin barrier function, such as keratin-27 (KRT27), filaggrin-2 (FLG2), and dermcidin (DCD) (Fig. 1c). Cluster 2 is comprised of 1,081 transcripts that were more abundant in lesions compared to normal skin. The most strongly ‘induced’ members from this cluster included genes associated with inflammatory cell recruitment (CXCL9, CXCL10 and CCL8) and cytotoxicity (GZMA, GZMB and GLYN) (Fig. 1d).
Functional enrichment and pathway analysis of the L. braziliensis lesion
We next carried out a functional enrichment analysis using Gene Ontology (GO) terms (Ashburner et al., 2000). Genes upregulated in lesions were enriched in GO terms related to inflammation, host defense and chemotaxis (Fig 2a). In contrast, genes downregulated in lesions were associated primarily with fatty acid metabolism and epidermal development (Fig. 2a). This enrichment analysis suggests that lesion development is associated with a remodeling of the local skin environment, marked by induction of a potent pro-inflammatory signature and a concomitant loss of epidermal and fatty acid metabolic signatures. Although useful for identifying general functional categories, GO enrichment analysis is biased in that it requires a relatively arbitrary selection of differentially expressed genes as the input. Therefore, using GSEA analysis we leveraged manually curated pathway databases, including Reactome, Kyoto Encyclopedia of Genes and Genomes (KEGG), Biocarta and the Pathway Interaction Database (Kanehisa et al., 2014; Nishimura, 2001; Schaefer et al., 2009; Vastrik et al., 2007), to identify the key pathways enriched in lesions. Despite our finding that over 2,000 genes were differentially regulated in the L. braziliensis lesion, pathway analysis showed that much of this transcriptional response could be explained by a small number of pathways (Fig. 2b). GSEA results confirmed a potent repression of fatty acid metabolism in the L. braziliensis lesion. To further investigate this altered metabolic profile, we examined all genes known to be involved in either cholesterol or triglyceride and free fatty acid metabolism (Fig. S1). Interestingly, we identified a global repression of both cholesterol and free fatty acid biosynthesis (Fig. S1c-d), and a significant increase in expression of lipid exporters (Fig. S1 e-f), suggesting that L. braziliensis lesions are characterized by disregulated lipid biosynthesis. In contrast, lesions showed marked induction of at least five key pathways. As expected, cytotoxicity and pathways involved in the generation of reactive oxygen species (ROS) were strongly induced in the L. braziliensis lesion (Novais et al., 2013; Novais et al., 2014). In addition, this analysis identified at least three other pathways associated with the lesion transcriptome, including: 1) antigen processing and immunoproteasome activation; 2) nucleic acid sensing; and 3) inflammasome activation and apoptosis.
Figure 2. Functional enrichment analysis of L. braziliensis infected skin.
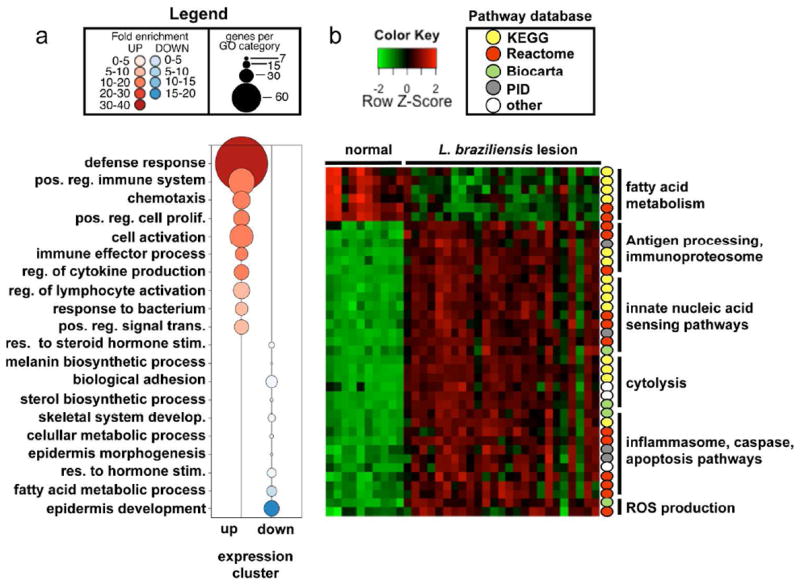
(a) GO enrichment analysis showing Biological Process terms enriched in induced genes from Fig. 1b (red) or repressed genes (blue) in lesions relative to normal skin. (b) GSEA showing enriched pathways from the MsigDB C2 canonical pathway collection. Color-coded circles to the right indicate pathway database provenance. Columns represent samples and rows individual pathways, colored to indicate expression levels based on a Z-Score. Abbreviations: GO, gene ontology; GSEA, gene set enrichment analysis; MSigDB, molecular signatures database.
Identification of unique and conserved pathways associated with skin lesion disease
Our data identified core pathways associated with L. braziliensis lesion, however it remained an open question as to whether they were a common feature of skin inflammation. To address this question, we compared our data to similar transcriptomic data generated from human psoriasis lesions (Fig. 3). With the use of data obtained from 334 paired microarrays from lesion and non-lesion sites on 167 patients (Tian et al., 2012), we quantitatively compared the enrichment of pathways in leishmania lesions with psoriasis (Fig. 3). As expected, only the L. braziliensis lesion was enriched for “JAK/STAT signaling”, “IFN-γ pathway” and the “Leishmaniasis” KEGG pathway, all of which include genes well known to be critical mediators of protection from this parasite. In addition, L. braziliensis lesions were uniquely enriched for “NK mediated cytotoxicity”, and “allograft rejection”, whereas our analysis showed that the “graft versus host disease” is enriched in both diseases. However, we found it to be much more strongly enriched in L. braziliensis lesions, suggesting that this pathologic response is a dominant feature of CL. Similarly, this analysis also identified inflammasome activation as a major pathway activated in L. braziliensis lesions, but not psoriasis. Several pathways were preferentially enriched in psoriasis and primarily included cell proliferation and nucleotide metabolism. Finally, several pathways were enriched in both diseases, including interferon-α/β signaling, NOD-like receptor signaling, cytosolic DNA sensing, defensins and regulation of apoptosis. Taken together, this comparison indicates that L. braziliensis induces a molecular signature of disease distinct from the psoriasis.
Figure 3. L. braziliensis and psoriasis skin lesions are associated with activation of distinct transcriptional modules.

Normalized enrichment scores (X axis) for select pathways (Y-axis) identified as being significantly enriched in psoriasis (blue squares) and/or L. braziliensis (red circles) skin lesions by gene set enrichment analysis. Open symbols indicate non-significant enrichment.
Early and late stage lesions are transcriptionally indistinguishable
Lesions from L. braziliensis patients could be classified into two categories based on the clinical stage of the disease, termed early and late (Fig. 4 and Table S1). Patients with early lesions had a small papule with no evident ulceration, a median lesion size of 38 mm2 (Fig. 4a), and illness duration of ≤ 30 days (Fig. 4b). In contrast, patients with late lesions had illness duration of ≥ 30 days, with ulcerated lesions with a median size of 250 mm2. Despite these marked differences, PCA of the entire transcriptome (data not shown) and a HC of the differentially expressed genes failed to resolve early and late stage lesions as transcriptionally distinct disease states (Fig. 4c). In addition, our analysis failed to find any significant differentially expressed genes between the two lesion stages, even when less stringent cutoffs were used (FC = 1.5 and p ≤ 0.05) (data not shown). The observation that L. braziliensis lesions at different clinical stages are indistinguishable by gene expression and pathway analysis (data not shown), reveals that the key pathways associated with L. braziliensis lesions are evident well before the development of ulcerated skin lesions, and therefore may be promoting cutaneous pathology, rather than simply arising as a consequence of disease.
Figure 4. Early and late lesions from L. braziliensis infected skin are transcriptionally indistinguishable.
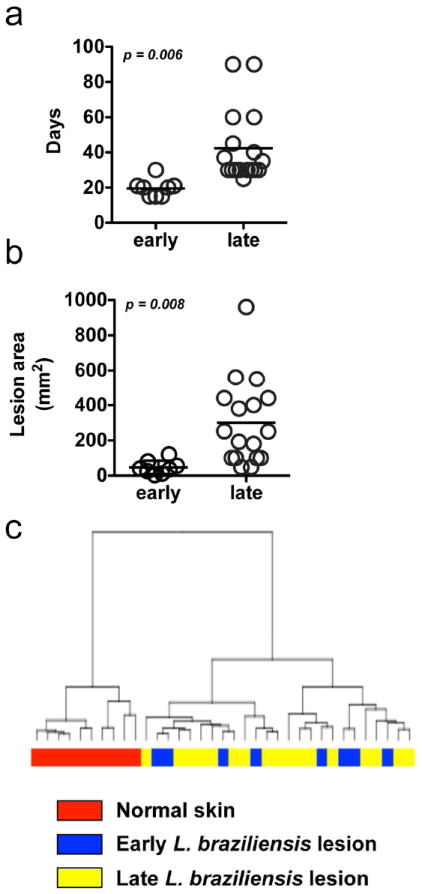
(a) Illness duration (days) and (b) lesion size from early and late stage L. braziliensis infected patients. (c) Hierarchical clustering of normal skin and early and late stage L. braziliensis infected patients based on differentially expressed genes.
Identification of genes associated with a molecular signature of skin pathology
We next sought to identify gene signatures that contributed to patient-to-patient variability in the lesion transcriptome. A PCA was carried out using only the 2,028 differentially expressed genes from lesion samples. PC1 accounted for 35.2% of the variability within the group of lesion samples, followed by 11.1% in PC2 (Fig. 5a), and PC1 and PC2 together explained almost half of variation between patients’ samples. This analysis showed that patients varied in their induction of this transcriptional program, but this variation was independent of age, sex, drug sensitivity (data not shown) and lesion stage (Fig. 5a). To determine which genes had the strongest influence on these two principal components, and therefore contributed the most to variability in the lesion transcriptome between patients, we plotted the PCA ‘scores’ from all differentially expressed genes for PC1 and PC2 (Fig. S2). This analysis identified a subset of immune and skin barrier function genes, whose expression is variable across the patients. A subset of immune genes and skin barrier genes (Fig. S2) from our PCA scores plot was selected for correlation analysis (Fig. 5b). As expected, there is a strong positive correlation between functionally related genes (Fig. 5b), such as components of the cytolytic granule (GZMB, GNLY and PRF1) (Fig. 5b), meaning that patients with high levels of granzyme B transcript in the lesion often had high levels of granulysin and perforin. In contrast, the cytolytic genes and components of the inflammasome, as well as IL8 and LAG3 showed a strong negative correlation with skin barrier genes, such as filaggrin-1 and -2, and loricrin (FLG, FLG2 and LOR) (Fig. 5b). Indeed, the more GLNY (Fig. 5c) or IL1B (Fig. 5d) expressed in a lesion, the less FLG or LOR were expressed. We also observed a negative correlation between the expression AIM2 and LOR (data not shown). This inverse correlation between immune genes and skin barrier genes was not a general relationship seen with just any strongly induced immune effector gene, as there was no significant correlation between expression of NCF1 and skin barrier genes (Fig. 5e). Taken together, these results show that patients exhibit variability in induction of specific genes, and that a subset of these genes is associated with a more severe molecular signature of skin pathology.
Figure 5. Cytotoxicity and inflammasome related genes inversely correlate with expression of skin barrier function genes.
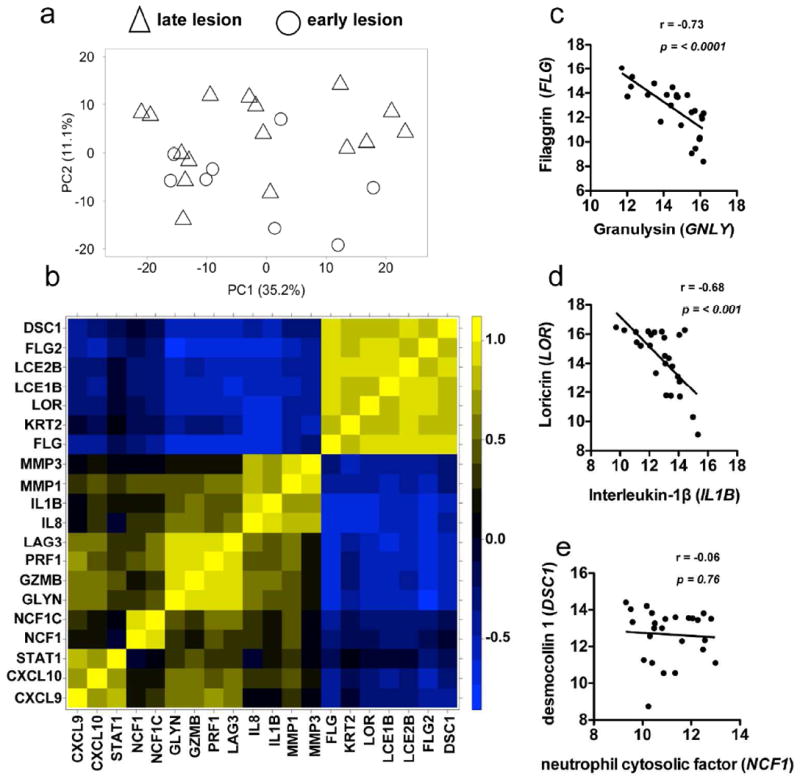
(a) Principal component analysis of differentially expressed genes from L. braziliensis infected patients. (b) Correlation heatmap showing selected modules of genes. Columns and rows represent individual genes, colored to indicate the correlation coefficient (r). (c-e) Log2 expression of (c) filaggrin and granulysin, (d) loricrin and interleukin-1β, and (e) desmocolin-1 and neutrophil cytosolic factor-1 in L. braziliensis infected patients.
Discussion
Here, we identified key immunological pathways induced following infection and developed a putative model explaining immunopathology in L. braziliensis lesions (Fig. 6). Activation of CD8 T cells requires recognition of antigens presented via MHC class I, and the immunoproteasome plays an important role in this process (Groettrup et al., 2010). We hypothesize that immunoproteasome activation drives cytolytic CD8 T cells in the skin. Since cell death leads to the release of danger associated molecular patterns (DAMPs), we propose that DAMPs act as a positive feedback that potentiates CD8 T cell (Bonilla et al., 2012; Kim et al., 2014) and inflammasome (Latz et al., 2013) activation. The inflammasome has been implicated in detrimental responses to several inflammatory diseases (Davis et al., 2011), and its definitive role in human leishmaniasis is still unclear. Finally, the most highly induced genes, CXCL10 and CXCL9 are chemokines that recruit T cells, and we propose that excessive expression of these chemokines brings more CD8 T cells to the skin thereby exacerbating immunopathology.
Figure 6. A putative ‘metapathway’ driving immunopathology in cutaneous leishmaniasis.
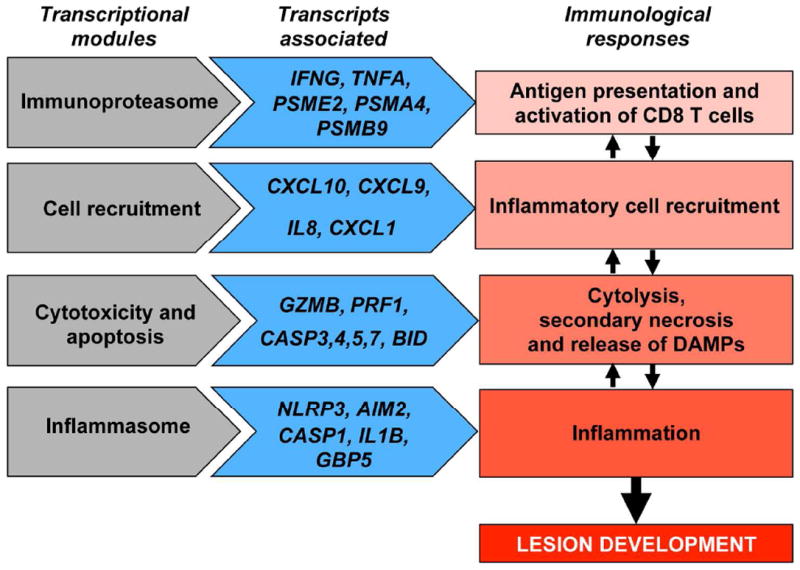
Transcriptional modules (pathways) induced during leishmaniasis (shown on the left), with examples of genes that are differentially expressed in lesions (shown on the center), suggest a testable model of immune responses (shown on the right) that lead to immunopathology within leishmanial lesions, in which cytotoxicity plays a central role.
Genes associated with the development and function of T helper type 1 (Th1) responses were highly expressed in L. braziliensis lesions while genes associated with a Th2 (Novais et al., 2014) or Th17 responses (data not shown) were not induced. This contrasts with the observations that Th2 and Th17 responses are induced in mucosal disease (Boaventura et al., 2010; Maretti-Mira et al., 2012). Our results are consistent with the strong Th1 response observed systemically in CL patients (Carvalho et al., 2012). Several genes downstream of IFN-γ were upregulated and may contribute to pathology. These data show an increased expression of immunoproteasome genes in CL, which helps generating MHC class I epitopes from the parasite, and ultimately increase CD8 T cell activation. Also, studies indicate that the immunoproteasome contributes to inflammation (Muchamuel et al., 2009) and CD8 T cell survival (Moebius et al., 2010). In addition to immunoproteasome related genes, IFN-γ also induces expression of CXCL10 and CXCL9, both of which recruit activated T cells and NK cells (Dufour et al., 2002). Therefore, we propose that in addition to its well-known function in parasite control (Kaye and Scott, 2011), IFN-γ participates indirectly in immunopathological responses in L. braziliensis infection by inducing the recruitment of CD8 T cells and NK cells to the skin and triggering cytotoxicity by stimulating the immunoproteasome activation and antigen presentation to CD8 T cells.
We found that the cytotoxicity is one of the main signatures of disease induced by L. braziliensis, a finding consistent with a previous study performed with a smaller number of samples (Novais et al., 2013). Although we find Th1 responses induced in lesions, the dominance of the cytolytic pathway is evident when one compares the fold change in IFNG and GZMB expression between normal skin and leishmanial lesions. As expected IFNG is increased in expression (8.8 FC) (Novais et al., 2014), but GZMB has a significantly higher fold change (50.9) (Novais et al., 2013). In L. braziliensis patients’ lesions CD4, but not CD8 T cells, make IFN-γ and thus the main function of CD8 T cells in the lesions of patients appears to be cytotoxicity (Santos Cda et al., 2013). We found that cytotoxic CD8 T cells mediated immunopathology in mice, but the mechanism by which cytotoxicity enhanced disease was unclear (Novais et al., 2013). In light of our transcriptome analysis, we now hypothesize that the increased pathology mediated by CD8 T cells is due to activation of the inflammasome by release of DAMPs.
Activation of the inflammasome generates mature IL-1β, which promotes increased inflammation by stimulating the production of chemokines, such as IL-8, and also matrix metalloproteinases, which degrade the extracellular matrix leading to more damage to the skin. Our study indicates that genes associated with the inflammasome pathway (such as IL1B, AIM2, NLRP3, CASP1, CASP5) are highly expressed in L. braziliensis lesions, suggesting that there is inflammasome activation and secretion of IL-1β during disease. In fact, ex vivo cultured human L. braziliensis lesions release IL-1β protein into culture supernatants (Carvalho et al., unpublished data). However, the role that the inflammasome and subsequent IL-1β play in human disease is still unclear. IL-1β mRNA was previously found in lesions from L. braziliensis patients (Pirmez et al., 1993), and in individuals infected with L. mexicana IL-1β production has been linked to disease severity (Fernandez-Figueroa et al., 2012). Here we expand those results by demonstrating that genes associated with two inflammasome pathways, AIM2 and NLRP3, are upregulated in lesions, and thus may play a previously unappreciated role in L. braziliensis human disease.
Skin diseases can share some characteristics. For example, dysbiosis of the skin has recently been considered as a distinctive feature of both CL and psoriasis (Cho and Blaser, 2012; Naik et al., 2012). In addition, IFN-γ has been associated with immunopathology in both diseases, although by different mechanisms. In L. braziliensis infection, IFN-γ is thought to induce immunopathology by activating innate cells. In psoriasis, IFN-γ synergizes with other pro-inflammatory cytokines, notably IL-17, and induces activation of keratinocytes. Although Th17 responses have been implicated in L. braziliensis infection in mucosal leishmaniasis (Boaventura et al., 2010; Maretti-Mira et al., 2012), we could not detect differences in IL-17 transcripts in L. braziliensis patients, suggesting that unlike psoriasis, L. braziliensis CL is not associated with a Th17 response. Our comparison of pathways enriched in these two diseases revealed additional differences. For example, although cytotoxicity has been implicated in both leishmaniasis (Novais et al., 2013) and psoriasis (Prpic Massari et al., 2007; Yawalkar et al., 2001), our data show that cytotoxicity is a more pronounced signature in L. braziliensis infection.
A surprising finding of our study was that the transcriptional profile of non-ulcerated lesions was similar to those of patients with ulcerated lesion. This result suggests that early after infection, inflammatory pathways are activated in the skin, which may explain why lesions often develop despite early detection and treatment (Machado et al., 2002). Although our data is based on a fraction of the total lesion, as biopsies were collected from the border of the ulcer, we believe the results appropriately reflect the ongoing immune response as the ulcer is mainly composed of dead cells. Since disease signatures are present before the ulcer develops, our data position cytotoxicity, immunoproteasome and inflammasome as potential causes of lesion development, rather than simply arising as consequence of disease.
Therapies that target the inflammatory response, without affecting mechanisms that kill the parasites, would be an ideal adjunct to drug treatment in leishmaniasis. Here, we have identified a hypothetical metapathway that leads from CD8 T cell activation and cytolysis to IL-1β production. Since cytotoxicity does not control L. braziliensis parasites (Novais et al., 2013; Santos Cda et al., 2013), nor does IL-1β appear to be protective in humans (Fernandez-Figueroa et al., 2012), blocking the major components of this metapathway should limit pathology without affecting parasite control.
Materials and Methods
Ethics statement
This study was conducted according to the principles specified in the Declaration of Helsinki and under local ethical guidelines and this study was approved by the Ethical Committee of the Federal University of Bahia (Salvador,Bahia, Brazil)(010/10) and the University of Pennsylvania IRB (Philadelphia, Pa)(813390). All patients provided written informed consent for the collection of samples and subsequent analysis.
Patients and biopsies
All CL patients were seen at the health post in Corte de Pedra, Bahia, Brazil, which is a well-known area of L. braziliensis transmission. The criteria for diagnosis were a clinical picture characteristic of CL in conjunction with parasite detection or a positive delayed-type hypersensitivity response to leishmania antigen. Prior to therapy, biopsies were collected at the border of the lesions using a 4 mm punch before therapy. Normal skin samples were taken from volunteers that were living in a non-endemic area without a history of leishmaniasis.
Transcriptional profiling and functional enrichment analysis
Microarrays and data analyses were carried out as previously described (Beiting et al., 2014). Briefly, Illumina HumanHT-12 version-4 beadchips were hybridized with biotin-labeled cRNA generated from 10 normal skin, 8 early and 17 late lesion samples. Data analyses were carried out using the statistical computing environment, R (v3.0.2), the Bioconductor suite of packages for R, and RStudio (v0.97). Probes sets that were differentially regulated ≥ 2fold (FDR ≤ 1%); after controlling for multiple testing using the Bonferroni-Hochberg method (Reiner et al., 2003)) were used for HC and heatmap generation. Data have been deposited on the GEO database for public access (GSE# GSE55664). GSEA (Mootha et al., 2003; Subramanian et al., 2005) was carried out using the MSigDB (v4.0) and either the GSVA bioconductor package (Fig. 2) (Hanzelmann et al., 2013) or the GSEA preranked tool (Fig. 3) to query the “C2: Canonical Pathways” collection in the MSigDB, which consists of 1,310 gene sets, or ‘signatures’, representing annotated pathways.
Comparison of L. braziliensis and psoriasis lesion transcriptomes
L. braziliensis data were compared to the ‘MAD-3’ human psoriasis data set (Tian et al., 2012), a meta-analysis of three independent psoriasis gene expression studies including 334 paired samples (lesion and non-lesion biopsies) from 167 patients (Gudjonsson et al., 2009; Suarez-Farinas et al., 2012; Yao et al., 2008). The ‘MAD-3’ data set was first filtered to remove non-specific Affymetrix probesets (probeset identifiers ending in ‘_x_at’). Genes with multiple probesets were used to calculate a mean FC relative to non-lesion controls. 17,061 genes in common between the psoriasis ‘MAD-3’ dataset and our L. braziliensis lesion data were used for carrying out a competitive GSEA analysis. Both data sets were rank ordered by Log2 FC in expression between lesion and control, and used as input for the GSEA preranked algorithm (Mootha et al., 2003; Subramanian et al., 2005). GSEA results were explored using the network analysis software, Cytoscape (Cline et al., 2007; Smoot et al., 2011) and the Enrichment Map plugin (Merico et al., 2010), in order to identify common and unique pathways.
Supplementary Material
Acknowledgments
The authors thank Dr. Mayte Suarez-Farinas (The Rockefeller University) for providing the ‘MAD3’ psoriasis data set, Ednaldo Lago, and Drs. Adriano Queiroz and Luiz Guimarães for assistance.
Funding: These studies were funded by the National Institutes of Health International Centers for Infectious Disease Research (U01-AUI 088650).
Abbreviations
- PCA
principal component analysis
- GO
gene ontology
- GSEA
gene set enrichment analysis
- MSigDB
molecular signatures database
- HC
hierarchical clustering
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacellar O, Lessa H, Schriefer A, et al. Up-regulation of Th1-type responses in mucosal leishmaniasis patients. Infection and immunity. 2002;70:6734–40. doi: 10.1128/IAI.70.12.6734-6740.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiting DP, Peixoto L, Akopyants NS, et al. Differential Induction of TLR3-Dependent Innate Immune Signaling by Closely Related Parasite Species. PLoS One. 2014;9:e88398. doi: 10.1371/journal.pone.0088398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boaventura VS, Santos CS, Cardoso CR, et al. Human mucosal leishmaniasis: neutrophils infiltrate areas of tissue damage that express high levels of Th17-related cytokines. European journal of immunology. 2010;40:2830–6. doi: 10.1002/eji.200940115. [DOI] [PubMed] [Google Scholar]
- Bonilla WV, Frohlich A, Senn K, et al. The alarmin interleukin-33 drives protective antiviral CD8(+) T cell responses. Science. 2012;335:984–9. doi: 10.1126/science.1215418. [DOI] [PubMed] [Google Scholar]
- Bosque F, Milon G, Valderrama L, et al. Permissiveness of human monocytes and monocyte-derived macrophages to infection by promastigotes of Leishmania (Viannia) panamensis. The Journal of parasitology. 1998;84:1250–6. [PubMed] [Google Scholar]
- Bottrel R, Dutra W, Martins F, et al. Flow cytometric determination of cellular sources and frequencies of key cytokine-producing lymphocytes directed against recombinant LACK and soluble Leishmania antigen in human cutaneous leishmaniasis. Infection and immunity. 2001;69:3232–9. doi: 10.1128/IAI.69.5.3232-3239.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho LP, Passos S, Schriefer A, et al. Protective and pathologic immune responses in human tegumentary leishmaniasis. Frontiers in immunology. 2012;3:301. doi: 10.3389/fimmu.2012.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature reviews Genetics. 2012;13:260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using Cytoscape. Nature protocols. 2007;2:2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annual review of immunology. 2011;29:707–35. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira CI, Brodskyn CI. The immunobiology of Leishmania braziliensis infection. Frontiers in immunology. 2012;3:145. doi: 10.3389/fimmu.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour JH, Dziejman M, Liu MT, et al. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. Journal of immunology. 2002;168:3195–204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- Fernandez-Figueroa EA, Rangel-Escareno C, Espinosa-Mateos V, et al. Disease severity in patients infected with Leishmania mexicana relates to IL-1beta. PLoS neglected tropical diseases. 2012;6:e1533. doi: 10.1371/journal.pntd.0001533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follador I, Araújo C, Bacellar O, et al. Epidemiologic and immunologic findings for the subclinical form of Leishmania braziliensis infection. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2002;34:8. doi: 10.1086/340261. [DOI] [PubMed] [Google Scholar]
- Giudice A, Vendrame C, Bezerra C, et al. Macrophages participate in host protection and the disease pathology associated with Leishmania braziliensis infection. BMC infectious diseases. 2012;12:75. doi: 10.1186/1471-2334-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groettrup M, Kirk CJ, Basler M. Proteasomes in immune cells: more than peptide producers? Nature reviews Immunology. 2010;10:73–8. doi: 10.1038/nri2687. [DOI] [PubMed] [Google Scholar]
- Gudjonsson JE, Ding J, Li X, et al. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. The Journal of investigative dermatology. 2009;129:2795–804. doi: 10.1038/jid.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic acids research. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye P, Scott P. Leishmaniasis: complexity at the host-pathogen interface. Nature reviews Microbiology. 2011;9:604–15. doi: 10.1038/nrmicro2608. [DOI] [PubMed] [Google Scholar]
- Kim TS, Gorski SA, Hahn S, et al. Distinct Dendritic Cell Subsets Dictate the Fate Decision between Effector and Memory CD8(+) T Cell Differentiation by a CD24-Dependent Mechanism. Immunity. 2014;40:400–13. doi: 10.1016/j.immuni.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nature reviews Immunology. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado P, Araujo C, Da Silva AT, et al. Failure of early treatment of cutaneous leishmaniasis in preventing the development of an ulcer. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2002;34:E69–73. doi: 10.1086/340526. [DOI] [PubMed] [Google Scholar]
- Maretti-Mira AC, Bittner J, Oliveira-Neto MP, et al. Transcriptome patterns from primary cutaneous Leishmania braziliensis infections associate with eventual development of mucosal disease in humans. PLoS neglected tropical diseases. 2012;6:e1816. doi: 10.1371/journal.pntd.0001816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, et al. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS one. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moebius J, van den Broek M, Groettrup M, et al. Immunoproteasomes are essential for survival and expansion of T cells in virus-infected mice. European journal of immunology. 2010;40:3439–49. doi: 10.1002/eji.201040620. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Muchamuel T, Basler M, Aujay MA, et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nature medicine. 2009;15:781–7. doi: 10.1038/nm.1978. [DOI] [PubMed] [Google Scholar]
- Naik S, Bouladoux N, Wilhelm C, et al. Compartmentalized control of skin immunity by resident commensals. Science(New York, NY) 2012;337:1115–9. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura D. BioCarta. Biotech Software & Internet Report. 2001;2:117–20. [Google Scholar]
- Novais FO, Carvalho LP, Graff JW, et al. Cytotoxic T cells mediate pathology and metastasis in cutaneous leishmaniasis. PLoS pathogens. 2013;9:e1003504. doi: 10.1371/journal.ppat.1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais FO, Nguyen BT, Beiting DP, et al. Human classical monocytes control the intracellular stage of Leishmania braziliensis by reactive oxygen species. The Journal of infectious diseases. 2014 doi: 10.1093/infdis/jiu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirmez C, Yamamura M, Uyemura K, et al. Cytokine patterns in the pathogenesis of human leishmaniasis. The Journal of clinical investigation. 1993;91:1390–5. doi: 10.1172/JCI116341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prpic Massari L, Kastelan M, Laskarin G, et al. Analysis of perforin expression in peripheral blood and lesions in severe and mild psoriasis. Journal of dermatological science. 2007;47:29–36. doi: 10.1016/j.jdermsci.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Ramirez C, Diaz-Toro Y, Tellez J, et al. Human macrophage response to L. (Viannia) panamensis: microarray evidence for an early inflammatory response. PLoS neglected tropical diseases. 2012;6:e1866. doi: 10.1371/journal.pntd.0001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–75. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Santos Cda S, Boaventura V, Ribeiro Cardoso C, et al. CD8(+) granzyme B(+)-mediated tissue injury vs. CD4(+)IFNgamma(+)-mediated parasite killing in human cutaneous leishmaniasis. The Journal of investigative dermatology. 2013;133:1533–40. doi: 10.1038/jid.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CF, Anthony K, Krupa S, et al. PID: the Pathway Interaction Database. Nucleic acids research. 2009;37:D674–9. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, et al. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Farinas M, Li K, Fuentes-Duculan J, et al. Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate-to-severe psoriasis. The Journal of investigative dermatology. 2012;132:2552–64. doi: 10.1038/jid.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Krueger JG, Li K, et al. Meta-analysis derived (MAD) transcriptome of psoriasis defines the “core” pathogenesis of disease. PloS one. 2012;7:e44274. doi: 10.1371/journal.pone.0044274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Inchaustegui D, Hogg A, Tulliano G, et al. CXCL10 production by human monocytes in response to Leishmania braziliensis infection. Infection and immunity. 2010;78:301–8. doi: 10.1128/IAI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastrik I, D’Eustachio P, Schmidt E, et al. Reactome: a knowledge base of biologic pathways and processes. Genome biology. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Richman L, Morehouse C, et al. Type I interferon: potential therapeutic target for psoriasis? PloS one. 2008;3:e2737. doi: 10.1371/journal.pone.0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawalkar N, Schmid S, Braathen LR, et al. Perforin and granzyme B may contribute to skin inflammation in atopic dermatitis and psoriasis. The British journal of dermatology. 2001;144:1133–9. doi: 10.1046/j.1365-2133.2001.04222.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


