Abstract
Alternative splicing has emerged as a vital way to expand the functional repertoire of a set number of mammalian genes. For example, such changes can dramatically alter the function and cellular localization of transcription factors. With this in mind, we addressed whether Eklf/Klf1 mRNA, coding for a transcription factor that plays a critical role in erythropoietic gene regulation, is alternatively spliced. We find that Eklf mRNA undergoes exon skipping only in primary tissues and that this splice variant remains at a very low level in both embryonic and adult erythroid cells, and during terminal differentiation. The resultant protein is truncated and partially encodes a non-EKLF amino acid sequence. Its overexpression can alter full-length EKLF function at selected promoters. We discuss these results in the context of stress and with respect to recent global studies on the role of alternative splicing during terminal erythroid differentiation.
Keywords: erythropoiesis, alternative splicing, transcription factor, EKLF/KLF1
Introduction
Hematopoiesis is the process by which all terminally differentiated blood cells develop from a pool of uncommitted stem and progenitor cells. They comprise a small number of cells that have the ability to differentiate into larger numbers of lineage restricted progenitor cells, which in turn terminally differentiate into all the mature cell types belonging to the various myeloid and lymphoid lineages (reviewed in [1]). Transcription factors direct expression of lineage-restricted target genes that exert intracellular control of these commitment steps, resulting in the correct homeostatic balance of proliferation versus differentiation [2].
Among these lineage-restricted regulators is EKLF/KLF1 (erythroid Krüppel-like factor), an erythroid-enriched transcription factor with well-defined and critical roles in transcriptional control of genes relating to red cell differentiation, proliferation, and survival [3–5]. We and others have had a long-standing interest in dissecting the various post-translational modifications and protein-protein interactions that fine-tune EKLF’s function as a regulator of erythroid gene expression [6].
Despite the presence of a classical nuclear localization signal (NLS) in the EKLF transactivation region, and a zinc finger domain that itself functions as an NLS [7, 8], a large proportion of cellular EKLF localizes to the cytoplasm in primary erythroid cells and erythroid cell lines [9–12]. The differential localization of KLF6 splice variants [13] raises the question whether cytoplasmic EKLF might also arise from a differentially spliced isoform. In addition, Eklf isoforms arising from testis-specific promoters have recently been described [14]. To obtain a more complete understanding of EKLF function in erythropoiesis, we investigated whether there are alternative Eklf transcripts in erythroid cells.
Methods
K562 cells were maintained in RPMI (Gibco) supplemented with 10% FBS (Cellgro). 293T, MEL, and Cos7 cells were maintained in DMEM (Gibco) supplemented with 10% FBS. Cells were cultured at 37°C and 5% CO2. DMSO induction of MEL cells was performed using 0.6×10^6 cells/ml in 1.5% DMSO for five days. Fetal livers were cultured under extensively self-renewing erythroblast conditions and differentiated as described [15].
Full length Eklf expression vector (pSG5/FLAG-EKLF) has been described [16]. The EKLF SV version was constructed by PCR cloning of exons 1 and 3 and placement back into pSG5/Flag. The β-HS2/luciferase, Bklf(1b) promoter/luciferase, Ahsp promoter/luciferase, and p21 (site 3) promoter/luciferase reporter constructs have been described [17–20].
For analysis of stress effects, spleens were isolated from three phlebotomized female mice along with three untreated females as previously described [21].
RNA isolations were performed using TRI reagent (Sigma). For non-quantitative analyses, cDNA was synthesized using the Promega Reverse Transcription System with a combination of random primers and oligo dT(15). PCR amplification of Eklf transcripts was carried out with primers [22] for 30–35 PCR cycles. For quantitative analyses, RNA was subjected to DNAse digestion and cDNA synthesis using iScript cDNA synthesis kit (Bio Rad). qPCR was performed using Quantitect SYBR green reagent (Qiagen). Absolute quantification of splice variant (SV) Eklf and full length (FL) Eklf transcripts was done using standard curves generated with serial dilutions of SV Eklf or FL Eklf cDNA containing template plasmids and SV EKLF-specific (FP5′GGAGGACTTCCTCAAGGAGAG3′; RP5′CAGAGGTGACGCTTCATGTG3′) or FLEklf-specific (FP: 5′GAGGACTTCCTCAAGTGGTG3′; RP: 5′GGAACCTGGAAAGTTTGTAAGG3′) primers. Primer specificity and size of the qPCR products was confirmed on an agarose gel.
Transfections for transactivation studies were performed using DMRIE-C (Invitrogen) according to the manufacturer’s protocol as adjusted for K562 cells [23]. Luciferase reporter activity was assayed with the Dual-Luciferase Reporter Assay System (Promega).
Constructs coding for FLAG-EKLF and FLAG-SV proteins were transfected into COS7 cells using DMRIE-C. Protein extracts for analysis were derived from cells lysed with NP40 buffer [22]. Western blot analysis of the small EKLF SV protein was performed after slow electophoretic separation of proteins in a 16.5% Tris-Tricine gel (Biorad) followed by transfer to 0.2 uM PVDF (Hybond). FLAG-tagged Proteins were detected with anti-FLAG M2 antibody (Sigma) and normalized to GAPDH (Pierce).
Results
An alternatively spliced variant of Eklf is expressed in primary hematopoietic tissue
RT-PCR was performed on mRNA expressed in the murine erythroleukemia (MEL) cells using primers that spanned the full-length transcript. Only one PCR product (approximately 1.3 kb) corresponding to full length Eklf is observed from cDNA obtained from MEL cells (Fig. 1A). The absence of any isoforms other than full length in MEL cells that contain EKLF protein in both nucleus and cytoplasm (as monitored by western blot and immunofluorescence [10]) suggests that differential EKLF subcellular localization is not due to alternative splicing.
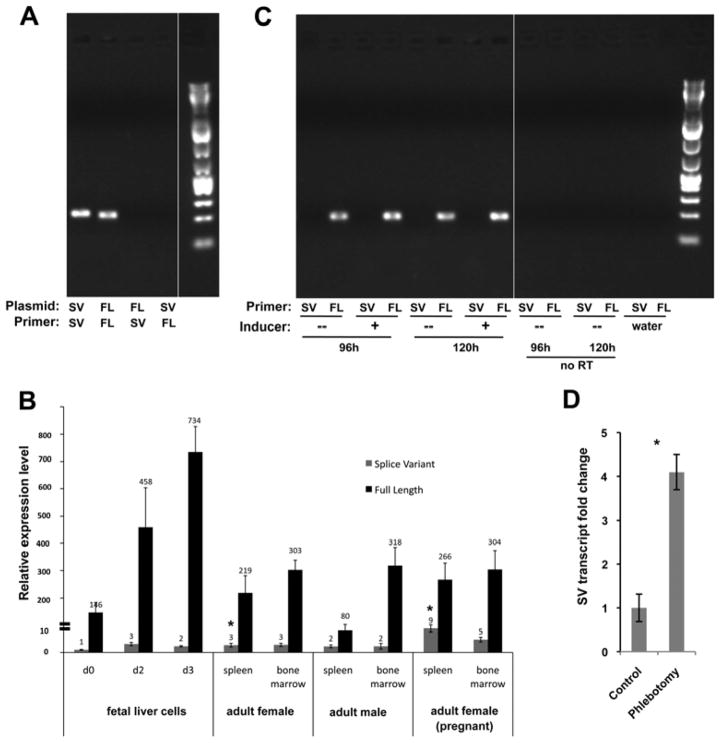
Figure 1.
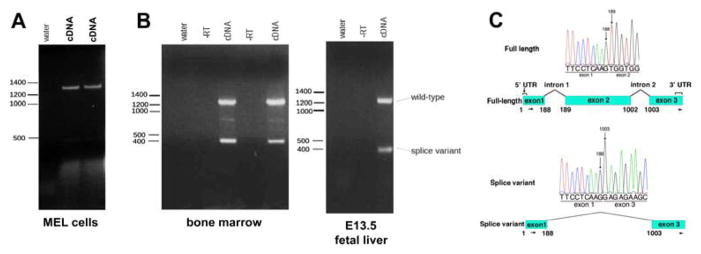
Eklf is alternatively spliced. Multiple samples of RNA from the MEL cell line (A) or from primary cells (B; bone marrow or fetal liver) were analyzed by RT-PCR using primers spanning the complete transcript. Controls (‘water’ or ‘–RT’) were included as indicated. (C) Sequence analysis of the gel-purified full length or splice variant is shown along with a schematic of the deduced exon-skipped product.
However, the same test using mRNA from primary murine E13.5 fetal liver and adult bone marrow cells unexpectedly reveal a second, much smaller transcript of 400 bp (Fig. 1B). We determined the sequence of the PCR product by direct sequencing. To confirm the sequence and to determine the existence of other lower abundance Eklf transcripts, we TA-cloned the PCR products and sequenced the resulting plasmids. In both cases, Sanger sequencing show that this transcript lacks exon 2, which is normally present in full-length Eklf (Fig. 1C); analysis of the TA-cloned PCR cloned products also show that full-length Eklf and the smaller splice variant presented here were the only Eklf-derived gene products that were amplified with our primers. These data uncover the presence of an additional Eklf splice variant (SV) that is expressed in primary murine hematopoietic tissues.
Eklf SV expression is not developmentally regulated and is expressed at a very low level in primary erythroid cells
To quantitatively determine the levels of SV expression, we designed isoform-specific PCR primers to monitor expression of the splice form by qRT-PCR (Fig. 2A). Using these primers, we find that the relative abundance of Eklf SV to full-length is consistently less than 2% in primary fetal liver, adult spleen, and bone marrow, demonstrating that SV expression remains quite low in embryonic and adult erythroid tissues (Fig. 2B).
Figure 2.
Quantitative analysis of splice variant levels. (A) Specificity of full-length (FL) or splice variant (SV) primer pairs was tested with plasmid DNA encoding the indicated isotype. (B) Quantitative analysis of FL (black) and SV (grey) levels in RNA derived from the indicated primary tissues is shown. Fetal liver cells underwent an erythroblast expansion phase (d0) followed by a terminal differentiation phase (d2, d3) [15]. All levels are normalized to the SV level seen in d0 fetal liver cells (=“1”). Note the discontinuity in the Y-axis as indicated. *p<10E-4 (based upon comparison of SV levels in spleens from pregnant compared to non-pregnant females). (C) RNA from MEL cells that had been induced for 96 or 120 hours with DMSO was analyzed for FL and SV expression by endpoint PCR along with uninduced and negative controls. (D) SV levels in spleens from phlebotomized or untreated adult females were determined and compared; SV level of untreated was given a value of “1”. Biological triplicates were each analyzed in triplicate. *p<0.002.
To determine if the presence of SV is regulated during erythroid differentiation, we cultured murine fetal liver cells under conditions that favor proliferation or terminal differentiation [15]. This protocol entails an expansion phase where robust generation of actively renewing erythroblasts are generated, followed by a differentiation phase that yields a high percentage of terminally maturing cells that include enucleated reticulocytes. While full-length transcripts increase during erythroid terminal differentiation consistent with increased expression of EKLF target genes, the levels of SV transcript remain constant (Fig. 2B). We also assayed SV expression by end-point PCR in MEL cells induced to differentiate with DMSO, and do not observe any new expression of SV (Fig. 2C). Collectively, these data indicate that SV expression in primary tissues is not regulated during development or differentiation, in contrast to full-length Eklf.
By comparing SV levels across conditions, there is little variation except for a small but significant (~3-fold) increase in the relative SV level in the spleen from an adult pregnant female (Fig. 2B), raising the possibility of a stress-induced effect. To more directly address this, we determined the SV levels in spleens from phlebotomized female mice and compared them to those from untreated animals. We find they are also increased to a similar relative extent (~4-fold) as that seen in the pregnant mouse (Fig. 2D).
Expression of predicted protein from the SV transcript
Eklf SV contains exon 1 in common with full-length and thus encodes the expected first 29 amino acids (aa). Skipping exon 2 predicts a frame-shift that causes truncation of an open reading frame after an additional 21 aa, followed by a long 3′UTR (Fig. 3A). Due to the absence of exon 2, the 400 bp Eklf SV is predicted to yield a small protein of 50 aa that does not contain any EKLF functional domains (as described in [6]).
Figure 3.
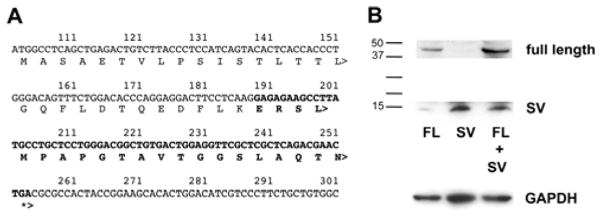
Expression testing of SV protein. (A) Predicted protein following exon skipping; bold indicates amino acids not encoded by full-length EKLF. (B) Plasmids expressing either FLAG-tagged FL or SV Eklf were transfected as indicated into COS7 cells. Inclusion of MG132 to inhibit proteasome degradation of the small SV product had no effect (not shown). Molecular weight markers are on left; anti-GAPDH was monitored as a loading control (bottom). Exposure of the bottom half of the gel was ~30x greater than the top half.
To test whether this small region can be translated in cells, we transfected FLAG-tagged full-length Eklf and/or SV expression constructs into cells and probed for their presence by western blot analysis. We find that the SV protein can be expressed (Fig. 3B), although its levels are quite low compared to full-length.
Alternative splicing occurs in erythroid cells as a mechanism to control gene expression via nonsense-mediated mRNA decay of splice variants [24, 25]. However, qRT-PCR of Eklf +/− fetal liver shows similar levels of splice variant expression when compared to wild-type fetal livers. Together with our data demonstrating that splice variant production is not altered during erythroid terminal differentiation, we conclude that alternative splicing of Eklf is not a mechanism by which to control full-length EKLF protein synthesis during differentiation.
Overexpression of Eklf SV downregulates EKLF function at specific target promoters
KLF6 splice variant proteins antagonize the transcription activity of full length KLF6 [13]. To determine if this was also the case for EKLF, we co-transfected EKLF target promoters fused to luciferase reporters into K562 cells along with equimolar amounts of EKLF full-length and/or SV expression constructs (Fig. 4). We expected that the EKLF SV would enter the nucleus, as molecules smaller than 40kDa in size can passively diffuse through the nuclear pore complex [26]. In the presence of the splice variant, EKLF activity is reduced at the p21 and AHSP promoters but remains unaffected at the β-globin and Bklf promoters. This demonstrates that SV antagonizes EKLF at specific loci, suggesting that the splice variant may be one factor that contributes to context-specific regulation of EKLF transactivation ([27] and discussed in [6]) when expressed at significant levels.
Figure 4.
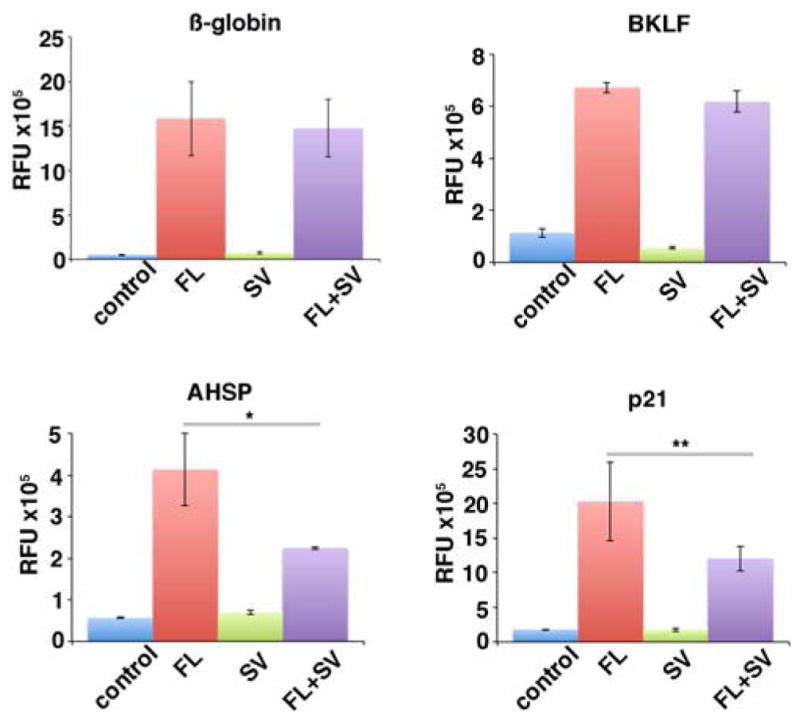
Effect of SV expression on transactivation of EKLF targets. Cotransfections of the indicated luciferase reporters (β-globin, Bklf, Ahsp, p21) were performed with combinations of EKLF FL or SV as indicated. *p<0.05; **p<0.1. All values were normalized to cotransfected Renilla plasmid.
Discussion
EKLF/KLF1 plays a global role in erythropoiesis as a master regulator of transcription [4, 6]; however, a significant level of protein resides in the cytoplasm [9–12]. Our initial impetus for these studies had been the precedent of KLF6 alternative splicing, which not only leads to altered subcellular localization of the variant protein, but also affects the function of full-length KLF6 [13]. In the present studies we find that an alternatively spliced Eklf product results from exon skipping. However, the expression level of Eklf SV is quite low, and the resultant out of frame protein product is only 50 amino acids. This variant is not only missing the carboxyl-located NLS and zinc finger sequences, but also the amino terminal-located PEST sequences [23], TFIIH interaction region [28], and ubiquitin binding domain [29], making it unlikely that EKLF SV functions as a transcription factor or that it has contributed to EKLF visualization in the cytoplasm.
However, its small size suggests that it can freely diffuse into the nucleus [26]. The exon 1-encoded portion potentially allows it to act as a competitor for EKLF binding partners or serve as a structural component in protein complexes, as this small amino-terminus encodes the CKII modification site [30] and partially encompasses the region that comprises the EKLF minimal activation domain [31]. Our studies suggest that that the SV could have such an effect at selected EKLF target promoters when expressed at appropriate levels. Although these levels are not altered in primary tissues, the elevated SV amounts in the spleen of a pregnant mouse and during phlebotomy-induced anemia imply that SV levels change during stress erythropoiesis.
Recent studies have shown that alternative splicing plays a major role in altering gene expression patterns during terminal erythroid differentiation [24, 25, 32], leading to a significant degree of splicing switches by both exon inclusion and skipping, possibly via RBM38 [33] or MBNL1 [32]. We suggest that post-transcriptional, splicing-mediated regulatory mechanisms postulated to occur during terminal differentiation [24] may also be operant in erythropoietic tissues in response to stress.
EKLF utilizes alternate modes of activation at different target promoters [6, 27]. We have shown that co-expression of the splice variant with full-length EKLF down-regulates its transactivation activity at the Ahsp and p21 promoters, but not the β-globin and Bklf promoter. These data suggest that the splice variant may regulate EKLF interactions with protein partners at target promoters that are selectively regulated in a context specific manner. Of note, downregulation of p21 by the splice variant may function to facilitate proliferation of erythroid progenitors during situations of physiological stress, e.g., during pregnancy or during times of extreme anemia.
Highlights.
An alternatively spliced Eklf variant is expressed in primary erythropoietic tissue
Eklf SV is not developmentally regulated and is expressed at a very low level
The predicted protein is truncated and partially encodes a non-EKLF sequence
Overexpression of Eklf SV downregulates EKLF function at specific target promoters
Acknowledgments
We thank Li Xue for isolation of mouse tissues and Dr. Barry Paw for technical advice. We thank Drs. Scott Friedman and Xiajun Li for productive scientific discussions. This work was supported by PHS grants R01 DK046865 to JJB, T32 HL007574 and F32 DK098866 to YYY, and R01 HL102449 and R01 DK090554 to SR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novershtern N, Subramanian A, Lawton LN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borg J, Patrinos GP, Felice AE, Philipsen S. Erythroid phenotypes associated with KLF1 mutations. Haematologica. 2011;96:635–638. doi: 10.3324/haematol.2011.043265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siatecka M, Bieker JJ. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood. 2011;118:2044–2054. doi: 10.1182/blood-2011-03-331371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tallack MR, Perkins AC. KLF1 directly coordinates almost all aspects of terminal erythroid differentiation. IUBMB Life. 2010;62:886–890. doi: 10.1002/iub.404. [DOI] [PubMed] [Google Scholar]
- 6.Yien YY, Bieker JJ. EKLF/KLF1, a tissue-restricted integrator of transcriptional control, chromatin remodeling, and lineage determination. Mol Cell Biol. 2013;33:4–13. doi: 10.1128/MCB.01058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandya K, Townes TM. Basic residues within the Kruppel zinc finger DNA binding domains are the critical nuclear localization determinants of EKLF/KLF-1. J Biol Chem. 2002;277:16304–16312. doi: 10.1074/jbc.M200866200. [DOI] [PubMed] [Google Scholar]
- 8.Quadrini KJ, Bieker JJ. Kruppel-like zinc fingers bind to nuclear import proteins and are required for efficient nuclear localization of EKLF. J Biol Chem. 2002;18:18. doi: 10.1074/jbc.M205677200. [DOI] [PubMed] [Google Scholar]
- 9.Pilon AM, Ajay SS, Kumar SA, et al. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood. 2011;118:e139–148. doi: 10.1182/blood-2011-05-355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quadrini KJ, Gruzglin E, Bieker JJ. Non-random subcellular distribution of variant EKLF in erythroid cells. Experimental Cell Research. 2008;314:1595–1604. doi: 10.1016/j.yexcr.2008.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoenfelder S, Sexton T, Chakalova L, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shyu YC, Lee TL, Wen SC, et al. Subcellular transport of EKLF and switch-on of murine adult beta maj globin gene transcription. Mol Cell Biol. 2007;27:2309–2323. doi: 10.1128/MCB.01875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narla G, Difeo A, Reeves HL, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65:1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- 14.Southwood CM, Lipovich L, Gow A. Tissue-restricted transcription from a conserved intragenic CpG island in the Klf1 gene in mice. Biol Reprod. 2012;87:108. doi: 10.1095/biolreprod.112.099879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.England SJ, McGrath KE, Frame JM, Palis J. Immature erythroblasts with extensive ex vivo self-renewal capacity emerge from the early mammalian fetus. Blood. 2011;117:2708–2717. doi: 10.1182/blood-2010-07-299743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caterina JJ, Donze D, Sun CW, Ciavatta DJ, Townes TM. Cloning and functional characterization of LCR-F1: a bZIP transcription factor that activates erythroid-specific, human globin gene expression. Nucleic Acids Res. 1994;22:2383–2391. doi: 10.1093/nar/22.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funnell AP, Maloney CA, Thompson LJ, et al. Erythroid Kruppel-like factor directly activates the basic Kruppel-like factor gene in erythroid cells. Mol Cell Biol. 2007;27:2777–2790. doi: 10.1128/MCB.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siatecka M, Lohmann F, Bao S, Bieker JJ. EKLF directly activates the p21WAF1/CIP1 gene by proximal promoter and novel intronic regulatory regions during erythroid differentiation. Mol Cell Biol. 2010;30:2811–2822. doi: 10.1128/MCB.01016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pilon A, Nilson D, Zhou D, et al. Alterations in Expression and Chromatin Configuration of the Alpha Hemoglobin-Stabilizing Protein Gene in Erythroid Kruppel-Like Factor-Deficient Mice. Mol Cell Biol. 2006;26:4368–4377. doi: 10.1128/MCB.02216-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos P, Casu C, Gardenghi S, et al. Macrophages support pathological erythropoiesis in polycythemia vera and beta-thalassemia. Nat Med. 2013;19:437–445. doi: 10.1038/nm.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yien YY, Bieker JJ. Functional interactions between erythroid Kruppel-like factor (EKLF/KLF1) and protein phosphatase PPM1B/PP2Cbeta. J Biol Chem. 2012;287:15193–15204. doi: 10.1074/jbc.M112.350496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quadrini KJ, Bieker JJ. EKLF/KLF1 is ubiquitinated in vivo and its stability is regulated by activation domain sequences through the 26S proteasome. FEBS Lett. 2006;580:2285–2293. doi: 10.1016/j.febslet.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 24.Pimentel H, Parra M, Gee S, et al. A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2014 doi: 10.1093/nar/gkt1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto ML, Clark TA, Gee SL, et al. Alternative pre-mRNA splicing switches modulate gene expression in late erythropoiesis. Blood. 2009;113:3363–3370. doi: 10.1182/blood-2008-05-160325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marfori M, Mynott A, Ellis JJ, et al. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta. 2011;1813:1562–1577. doi: 10.1016/j.bbamcr.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Sengupta T, Cohet N, Morle F, Bieker JJ. Distinct modes of gene regulation by a cell-specific transcriptional activator. Proc Natl Acad Sci U S A. 2009;106:4213–4218. doi: 10.1073/pnas.0808347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mas C, Lussier-Price M, Soni S, et al. Structural and functional characterization of an atypical activation domain in erythroid Kruppel-like factor (EKLF) Proc Natl Acad Sci U S A. 2011;108:10484–10489. doi: 10.1073/pnas.1017029108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raiola L, Lussier-Price M, Gagnon D, et al. Structural characterization of a noncovalent complex between ubiquitin and the transactivation domain of the erythroid-specific factor EKLF. Structure. 2013;21:2014–2024. doi: 10.1016/j.str.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 30.Ouyang L, Chen X, Bieker JJ. Regulation of erythroid Kruppel-like factor (EKLF) transcriptional activity by phosphorylation of a protein kinase casein kinase II site within its interaction domain. J Biol Chem. 1998;273:23019–23025. doi: 10.1074/jbc.273.36.23019. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Bieker JJ. Erythroid Krüppel-like Factor (EKLF) contains a multifunctional transcriptional activation domain important for inter- and intramolecular interactions. EMBO Journal. 1996;15:5888–5896. [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng AW, Shi J, Wong P, et al. Muscleblind-like 1 (Mbnl1) regulates pre-mRNA alternative splicing during terminal erythropoiesis. Blood. 2014 doi: 10.1182/blood-2013-12-542209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinicke LA, Nabet B, Shen S, et al. The RNA binding protein RBM38 (RNPC1) regulates splicing during late erythroid differentiation. PLoS One. 2013;8:e78031. doi: 10.1371/journal.pone.0078031. [DOI] [PMC free article] [PubMed] [Google Scholar]