Abstract
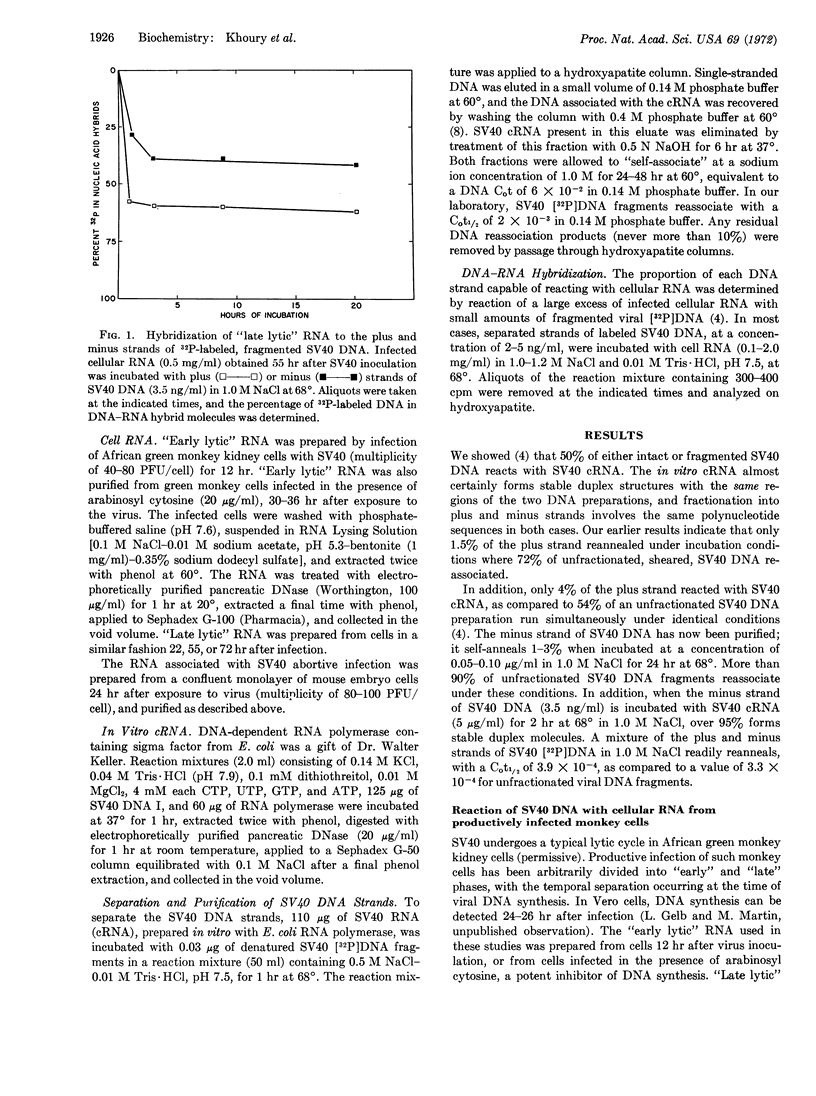
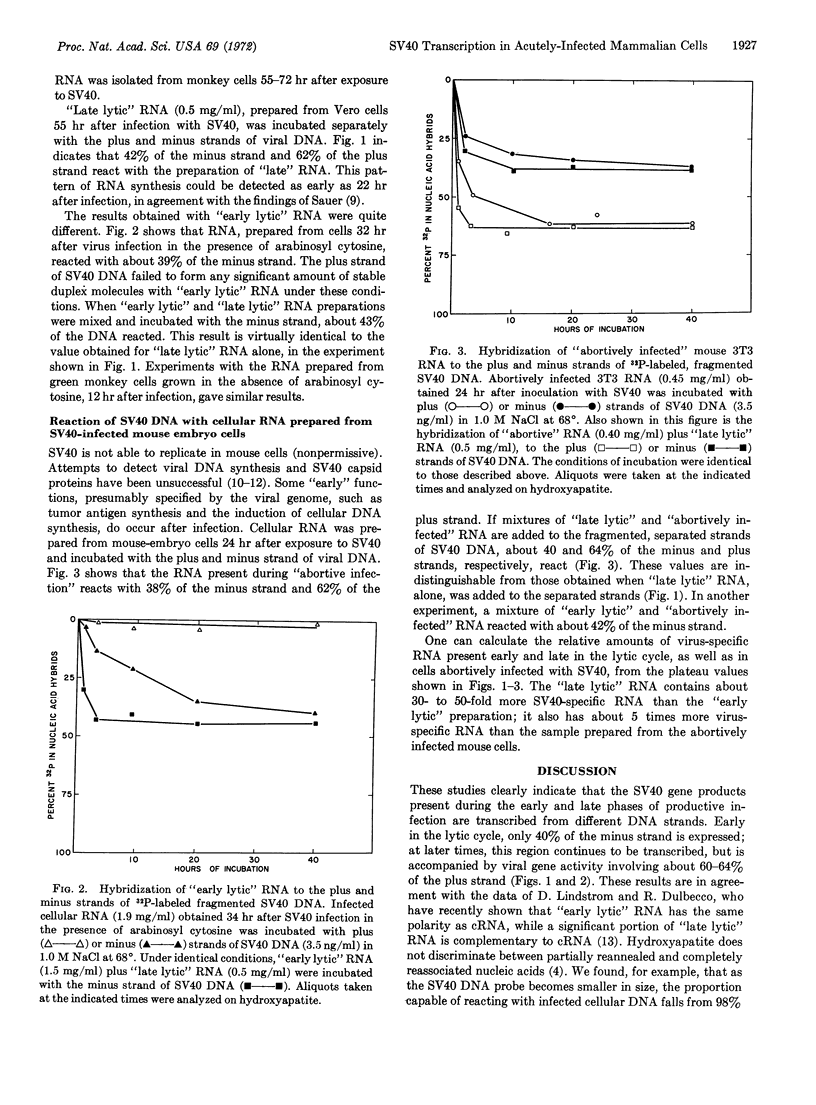
Small amounts of fractionated, denatured, 32P-labeled DNA from SV40 virus were incubated with a large excess of the complementary RNA of SV40 prepared in vitro with Escherichia coli RNA polymerase; the viral DNA strands were separated on hydroxyapatite columns. The RNA present in green monkey cells late in the lytic cycle reacted with 40-42% of the strand complementary to the in vitro complementary RNA (minus strand), and 60-64% of the opposite (plus) strand. “Early lytic” RNA failed to significantly interact with the plus strand, but formed stable duplex molecules with 35-39% of the minus strand. The RNA prepared from mouse embryo cells 24 hr after infection with SV40 combined with 35-38% of the minus strand and 60-62% of the plus strand. In all cases, the same regions of either the plus or minus strand appear to be transcribed in permissive and nonpermissive infections.
Keywords: SV40, tissue culture, monkey, mouse, plus and minus strands, RNA synthesis
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aloni Y., Winocour E., Sachs L. Characterization of the simian virus 40-specific RNA in virus-yielding and transformed cells. J Mol Biol. 1968 Feb 14;31(3):415–429. doi: 10.1016/0022-2836(68)90418-x. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., McGrath J., Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970 Oct 17;228(5268):227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Earley E., Peralta P. H., Johnson K. M. A plaque neutralization method for arboviruses. Proc Soc Exp Biol Med. 1967 Jul;125(3):741–747. doi: 10.3181/00379727-125-32194. [DOI] [PubMed] [Google Scholar]
- Gelb L. D., Kohne D. E., Martin M. A. Quantitation of Simian virus 40 sequences in African green monkey, mouse and virus-transformed cell genomes. J Mol Biol. 1971 Apr 14;57(1):129–145. doi: 10.1016/0022-2836(71)90123-9. [DOI] [PubMed] [Google Scholar]
- Gershon D., Sachs L., Winocour E. The induction of cellular DNA synthesis by simian virus 40 in contact-inhibited and in x-irradiated cells. Proc Natl Acad Sci U S A. 1966 Sep;56(3):918–925. doi: 10.1073/pnas.56.3.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry P., Black P. H., Oxman M. N., Weissman S. M. Stimulation of DNA synthesis in mouse cell line 3T3 by Simian virus 40. Proc Natl Acad Sci U S A. 1966 Oct;56(4):1170–1176. doi: 10.1073/pnas.56.4.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai K., Defendi V. Integration of simian virus 40 deoxyribonucleic acid into the deoxyribonucleic acid of permissive monkey kidney cells. J Virol. 1972 Apr;9(4):705–707. doi: 10.1128/jvi.9.4.705-707.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson J., Goldstein D., Weil R. A study on the transcription of the polyoma viral genome. Proc Natl Acad Sci U S A. 1970 Jan;65(1):226–233. doi: 10.1073/pnas.65.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom D. M., Dulbecco R. Strand orientation of simian virus 40 transcription in productively infected cells. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1517–1520. doi: 10.1073/pnas.69.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. A., Axelrod D. SV40 gene activity during lytic infection and in a series of SV40 transformed mouse cells. Proc Natl Acad Sci U S A. 1969 Dec;64(4):1203–1210. doi: 10.1073/pnas.64.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Westphal H., Srinivasan P. R., Dulbecco R. The integrated state of viral DNA in SV40-transformed cells. Proc Natl Acad Sci U S A. 1968 Aug;60(4):1288–1295. doi: 10.1073/pnas.60.4.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer G. Apparent differences in transcriptional control in cells productively infected and transformed by SV40. Nat New Biol. 1971 Jun 2;231(22):135–138. doi: 10.1038/newbio231135a0. [DOI] [PubMed] [Google Scholar]
- Summers W. C., Siegel R. B. Transcription of late phage RNA by T7 RNA polymerase. Nature. 1970 Dec 19;228(5277):1160–1162. doi: 10.1038/2281160a0. [DOI] [PubMed] [Google Scholar]
- Todaro G. J., Green H. Cell growth and the initiation of transformation by SV40. Proc Natl Acad Sci U S A. 1966 Feb;55(2):302–308. doi: 10.1073/pnas.55.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal H. SV40 DNA strand selection by Escherichia coli RNA polymerase. J Mol Biol. 1970 Jun 14;50(2):407–420. doi: 10.1016/0022-2836(70)90201-9. [DOI] [PubMed] [Google Scholar]



