Abstract
Rotavirus, a cause of pediatric gastroenteritis, has a genome consisting of 11 segments of double-stranded (ds)RNA surrounded by a triple-layered protein capsid. The rotavirus RNA-dependent RNA polymerase, VP1, synthesizes both dsRNA and plus-strand RNA (+RNA) within subviral particles. Structural analyses of the rotavirus capsid and polymerase, combined with functional studies of purified capsid proteins, indicate that the inner capsid protein controls the initiation of RNA synthesis by VP1. Whether VP1 directs dsRNA versus +RNA synthesis may be regulated by the impact of the viral RNA capping enzyme on the position of the polymerase plug, a flexible element that inserts into one of the polymerase’s RNA exit tunnels. This review discusses recent findings and ideas into the mechanisms used by rotavirus capsid proteins to control the activities of its viral polymerase and to coordinate RNA synthesis with the assembly of virus particles.
Keywords: RNA-dependent RNA polymerase, rotavirus, capsid protein, dsRNA virus, RNA chaperone
Introduction
In the life cycle of RNA viruses, the activity of the viral RNA-dependent RNA polymerase (RdRP) must be carefully regulated to ensure that RNA products are made at levels and in appropriate sites to support genome replication and protein translation. Such regulation is particularly intriguing in the case of rotavirus, as its RdRP (VP1) has a dual role, sometimes acting as a replicase and, at other times, as a transcriptase. The observation that rotavirus dsRNA genome segments are made at equimolar levels, while the viral transcripts are not [1, 2], makes the question of how RdRP activity is regulated even more interesting. A critical discovery made by analysis of intracellular subviral particles [3, 4] and by in vitro assays with recombinant proteins [5••] is that rotavirus dsRNAs and +RNAs are only made by particle-associated VP1 [6••, 7]. These findings have established that rotavirus inner capsid proteins are essential for VP1 polymerase activity and determine whether VP1 acts as a replicase or transcriptase. Regulation of VP1 activity by capsid proteins allows the coordination of genome replication with the packaging of newly made dsRNAs into progeny particles and likely prevents the induction of host antiviral responses to exposed dsRNAs. Here we review insights gained from structural and functional studies into the mechanism by which VP1 polymerase activity is regulated by the virion capsid protein.
Virion architecture
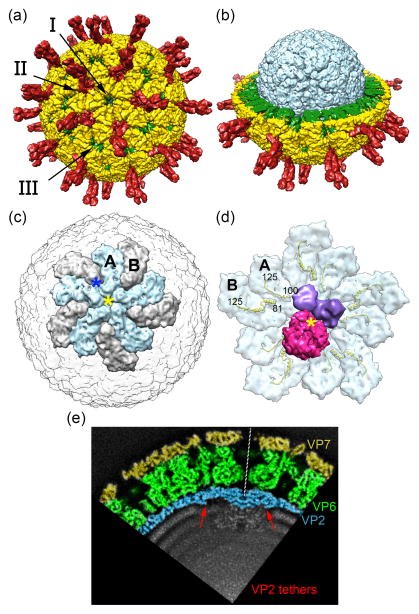
Rotavirus, a member of the Reoviridae family, is a non-enveloped icosahedral triple-layered particle (TLP) with a genome consisting of 11 segments of dsRNA (Figure 1). The outer and intermediate protein layers are formed by trimers of VP7 and VP6, respectively, each organized with T=13 symmetry [8, 9]. Anchored into the VP6 layer and projecting out of the VP7 layer is the viral attachment protein VP4. The inner layer defines the core shell and is composed of 60 VP2 dimers, organized with T=1 symmetry [9, 10]. Twelve VP2 decamers interact to form the core, with the VP2 components of each decamer present in either of two conformers, VP2-A or VP2-B. Five VP2-A conformers meet at the center of each decamer (five-fold axis), while five VP2-B conformers are pulled back, interdigitating between the A-forms [10]. Three types of channels (I, II, and III) pass through the core shell and serve as conduits for the translocation of substrates and products in and out of the core [11]. The Type I channel is positioned at the center of each decamer.
Figure 1. Architecture of the rotavirus virion.
(a) The mature rotavirus TLP is shown with the VP7 glycoprotein (yellow) and VP4 attachment spike protein (red) coating the surface. The virion contains 132 channels that enable small molecule exchange: 12 Type I channels, 60 Type II channels, and 60 Type III channels. (b) Removing the outermost VP7/VP4 protein layer and the intermediate VP6 (green) from half of the particle reveals the innermost VP2 layer (light blue) (a–b, PDB: 3IYU and 3N09). (c) The inner core shell is formed by 12 decamers of VP2. Each decamer assembles from two VP2 conformers: A (light blue) and B (gray). The Type I channel sits at the center of the five VP2-A conformers (yellow asterisk), while the Type II channel sits at the inner tip of the five VP2-B conformers (dark blue asterisk) (PDB: 4F5X). (d) Packaged within the core, just offset from the fivefold axis, is the RdRP, VP1 (pink), and the RNA capping enzyme, VP3 (purple) (VP2-VP1, PDB: 4F5X; VP3, modeled). The VP2 tethers (yellow ribbons) are 80–100 residues in length and extend into the core. Residues 1–80, or 1–100, of VP2-B and VP2-A, respectively, are too flexible to be resolved. The tethers are required to encapsidate VP1 and VP3 [16••], but their role in RNA synthesis, if any, is unknown. (e) A slice of a cryoelectron microscopy image of the TLP, colored to identify the three protein layers (yellow, VP7; green, VP6; blue, VP2). The dsRNA viral genome is organized in concentric rings within the TLP. Part of the VP2 tethers are observed (red arrows), oriented towards the five-fold axes (dashed line). The extra density beneath the five-fold axes belongs to VP1 and VP3, after five-fold averaging.
Inside of the core, copies of VP1 and the RNA capping enzyme (VP3) are bound near the fivefold axes formed by the VP2 decamers (Figure 1d) [12••]. Each genome segment is believed to interact with one specific VP1-VP3 complex. Since the rotavirus genome consists of 11 segments of dsRNA, but 12 decamers are present in the core, one decamer may lack a VP1-VP3 complex. Estrozi and colleagues revealed the position and orientation of VP1 in the core [13••]. Their analysis indicates that VP1 is anchored to the VP2 decamer at a position slightly offset from the exact five-fold axis, barely covering the Type I channel. The VP1 footprint on the VP2 decamer is quite large, covering portions of at least four VP2 conformers, of both A and B forms [13••]. Modeling suggests that VP3 has a structure similar to the RNA capping enzyme of bluetongue virus, another member of Reoviridae [14]. Although the orientation and location of VP3 within the core is unknown, it is believed to reside adjacent to, and possibly bound to, VP1.
The VP2 molecule (880 amino acids, aa) consists of two domains: the C-terminal scaffold domain that forms the core shell (126–880 aa) and the N-terminal tether domain (1–125 aa) [12••]. The tethers extend across the interior face of the VP2 decamers, generally running towards the five-fold axes, with the tethers of the VP2-B conformers crossing over onto the face of the VP2-A conformers (Figure 1d). In the crystal structure solved for the rotavirus particle, the first 99 residues of the VP2-A tethers and the first 80 residues of the VP2-B tethers were not resolved [12••], but these missing regions are predicted to extend around VP1 and VP3, forming a cradle that helps stabilize the enzymes in position. This concept is supported by studies showing that deletion of the tethers prevents encapsidation of VP1 and VP3 into virus-like particles [15, 16••, 17]. The tethers also have RNA-binding activity, a property that may be important for organization and movement of RNA around VP1 and VP3 [18].
Genome organization within the core remains poorly understood, although high-resolution structural studies indicate that en masse the 11 segments are arranged in concentric rings, reminiscent of other RNA viruses (Figure 1e) [12••, 13••, 19]. Given that the genome segments differ markedly in size, from 0.7 –3.1K base pairs, the contribution of individual segments to ring architecture must vary. The rings are interrupted at the five-fold axis by the presence of VP1 and VP3, and the VP2-A and VP2-B tethers become disordered upon reaching the outer most ring [12••, 13••].
Viral life cycle
During rotavirus entry, the virion VP4-VP7 outer capsid is disrupted, yielding a double-layered particle (DLP) [20, 21]. Through the activity of its VP1 and VP3 components, the DLP synthesizes 11 capped, nonpolyadenylated +RNAs. These are translated, giving rise to six structural proteins (VP1-VP4, VP6-VP7) and six nonstructural proteins (NSP1-NSP6). Transcriptionally-active DLPs become embedded within viral inclusions (viroplasms) formed by NSP2 and NSP5, and containing the core proteins VP1, VP2, and VP3, and the intermediate capsid protein VP6 (reviewed in [22]). Within viroplasms, the 11 viral +RNAs undergo assortment and interact with assembling progeny cores; the VP1 components of these cores then direct minus-strand synthesis to form dsRNA. The fact that the 11 viral dsRNAs are made in equimolar levels [2] but the +RNA transcripts are not, suggests that assortment is precise and RNA specific, and that the dsRNA genome is synthesized only after the complete complement of +RNAs is packaged into progeny cores. The association of VP6 with progeny cores occurs in association with viroplasms, forming DLPs that then migrate to the endoplasmic reticulum where virus particles mature into TLPs [23].
Polymerase activities
Several earlier studies provided evidence of a link between VP1 activity and the presence of the viral inner capsid proteins. Among these were results showing that rotavirus replicase and transcriptase activities were only detectable in isolated subviral particles that contained VP1 and VP2, or VP1, VP2, and VP6, respectively [3, 21, 24, 25]. Moreover, analysis of the rotavirus temperature-sensitive mutants, tsF and tsG, indicated that the formation of subviral particles with replicase activity requires VP2 (and not VP6), while the formation of particles with transcriptase activity is dependent on both VP2 and VP6 [26•, 27]. More recently, in vitro assays performed with recombinant core proteins have established that VP1 lacks replicase activity unless VP2 is present [5••, 6••]. Indeed, such assays have indicated that for optimal replicase activity, VP1 and VP2 must be present at a molar ratio of 1:10 [5••], suggesting that VP1 activity is triggered by its interaction with a VP2 decamer. Interestingly, DLPs lose transcriptase activity when their VP6 layer is removed, despite retaining a structurally intact core [26•]. The activity is restored when such cores are reconstituted with VP6, suggesting that VP6 and the integrity of the DLP has a critical role in the ability of VP1 to function as a transcriptase [3, 21, 25]. The need for VP6 may stem from its contribution to the formation and function of channels that help to translocate nucleotides and nascent transcripts into and out of the core.
VP1 structure and function
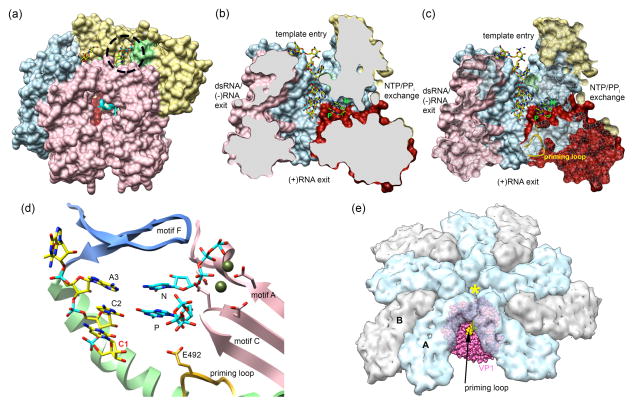
Studies reporting atomic structures for VP1, both in its apo form and in complex with short viral RNAs, have provided important insights into the mechanism by which VP2 affects rotavirus polymerase activity. These analyses revealed that VP1 has a closed cage-like structure consisting of a central polymerase domain that is sandwiched in between large N-terminal and C-terminal (bracelet) domains (Figure 2) [28••]. The polymerase domain has the classical “right-handed” polymerase architecture that includes fingers, palm, and thumb subdomains and the canonical polymerase motifs, A–F [29]. Four tunnels lead to the catalytic center of the polymerase; one operates as the template entry tunnel, another as the nucleotide/pyrophosphate (NTP/PPi) exchange tunnel, and two others as RNA exit tunnels (Figure 2) [28••, 30••]. Only one RNA exit tunnel is believed to operate during replication; this tunnel extends through the bracelet domain and represents the pathway for release of newly made dsRNA (Figure 3a) to the core interior. During transcription, both RNA exit tunnels of the polymerase are expected to function. One is used for release of the minus-strand template RNA from the polymerase and is the same tunnel used for release of the dsRNA product during replication (dsRNA/-RNA exit tunnel). The other is used for release of newly made +RNAs (Figure 3b) and represents a conduit that directs nascent transcripts out of the core. An unusual feature of VP1 is the presence of a shallow cleft near the template entry tunnel that represents an RNA cap-binding site (Figure 2a) [28••]. This site may help to recruit capped +RNA templates during replication and/or provide an anchoring point for the capped 5′-end of the +RNA of viral genome segments during multiple rounds of transcription.
Figure 2. Features of VP1 that contribute to RNA initiation.
(a) A surface representation of VP1 is shown colored by domain: N-terminal (khaki), fingers (blue), palm (red), thumb (light green), and C-terminal bracelet (light pink) (PDB: 2R7R, 2R7W). The C-terminal plug (cyan) can be seen inside the tunnel formed by the C-terminal bracelet. The cap-binding site on VP1-located to the right of the template entry tunnel-is indicated by a dashed circle. (b) A cutaway representation of VP1 in complex with a ribonucleotide representing the 3′ consensus sequence (3’CS) of rotavirus +RNA and nucleotides modeled into the N and P sites. This cutaway representation reveals the four tunnels to the active site: template entry, dsRNA/-RNA exit, NTP/PPi exchange, and +RNA exit (PDB: 2R7R). (c) Similar to panel b, with a transparent view of the clipped VP1 surface. The priming loop (yellow) is extended away from the P site, unable to support initiation (PDB: 2R7R). (d) Magnification of the polymerase active site reveals details of initiation (polymerase domains colored as in panel a). The active site residues from motifs A and C (red) in the palm domain are represented as sticks and motif F (dark blue) encloses the top of the active site. In the absence of VP2, VP1 exists in an inactive conformation. The priming loop (yellow ribbon) is retracted away from the priming nucleotide (P) and the template RNA is one nucleotide past the initiation register (C1) [PDB: 2R7R (VP1), 1N1H (nucleotides)]. (e) Estrozi and colleagues determined the orientation of VP1 (pink) within the DLP [12••], finding that the polymerase is offset from the five-fold axis (yellow asterisk) at the center of the VP2 decamer (blue and gray). The position of VP1 revealed that the priming loop (yellow surface on VP1) and neighboring regions of VP1 likely interact directly with the VP2 decamer. It is probable that the interaction between VP1 and VP2 within the core particle forces the priming loop further into the active site to support a priming nucleotide for initiation.
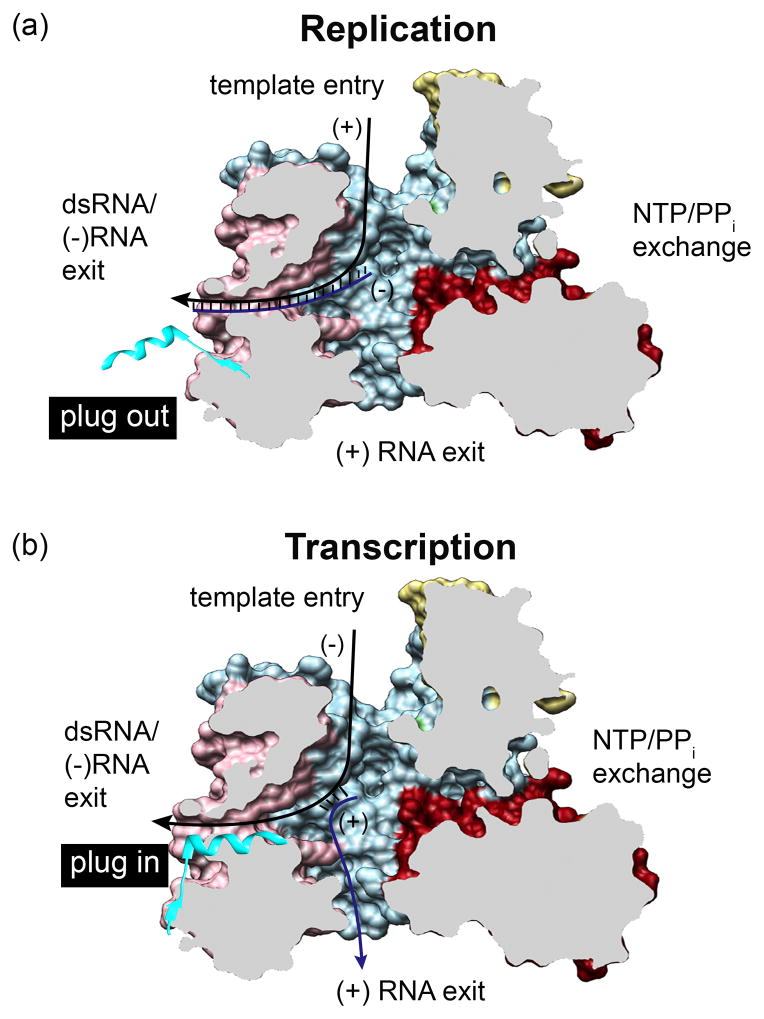
Figure 3. Regulation of replication versus transcription in VP1.
(a–b) A cutaway of VP1 (PDB: 2R7R), colored as in Figure 2, reveals the four tunnels to the active site. Template −RNA/+RNA and the C-terminal plug were modeled on top of VP1. (a) During replication, +RNA templates enter the polymerase through the template entry tunnel, the complementary −RNA is synthesized, and the resulting dsRNA product is thought to exit through the dsRNA/−RNA exit tunnel into the virion core. The C-terminal plug (cyan) must extend out (“plug out”), allowing dsRNA to pass through. (b) During transcription, −RNA templates enter the polymerase through the template entry tunnel, and +RNA is synthesized. In this reaction, the template −RNA is thought to exit through the dsRNA/−RNA exit tunnel. The C-terminal plug may be stabilized by interactions with the surface of the tunnel (“plug in”), reducing its diameter. +RNA products are thought to exit through the fourth tunnel and out of the DLP.
Regulation of replication
Even when incubated with NTPs and viral +RNAs, rotavirus VP1 lacks polymerase activity unless VP2 is present [5••, 6••]. However, VP1 alone can specifically bind +RNAs [6••, 31•], an interaction mediated by an extensive network of hydrogen bonds formed between residues lining the template entry tunnel of VP1 and the conserved 3′-terminal UGUGACC element of the RNA [28••]. The polymerase forms hydrogen bonds with the bases of the UGUG residues and the ribose-phosphate backbone of the ACC residues [32, 33]. Importantly, the UGUG interaction allows the polymerase to specifically recognize viral +RNAs. However, analysis of complexes formed by VP1 and a UGUGACC polyribonucleotide revealed the 3′-end of the RNA to be stabilized in a position that is one nucleotide past initiation register and, thus, not opposite of the catalytic priming (P) site of the polymerase (Figure 2c) [28••]. The 1+ overshot register suggests that the presence of VP2 induces conformational changes within the polymerase that repositions the 3′-end of +RNA relative to the P site. Such re-positioning would ensure that dsRNA synthesis begins at the very first residue of the template strand.
Rotavirus VP1 supports the initiation of dsRNA synthesis through a process that is independent of a priming RNA or a nucleotidylated protein. Based on earlier studies of the structurally similar reovirus RdRP (λ3), initiation of dsRNA synthesis by VP1 is likely supported by the activity of a specialized flexible element, termed a priming loop (Figure 2c–e) [30••]. This loop (10 aa) is located at the junction of the fingers and palm subdomains and roughly extends from the base of the polymerase cage in a direction towards the P site. As shown for λ3, the priming loop probably exists in multiple conformations, including retracted and extended forms. In its extended form, the loop stabilizes a priming nucleotide into the P site. In its retracted form, the loop is too far removed from the P site to support such a function, yielding a polymerase that is catalytically inactive. Analysis of VP1 crystals soaked with a UGUGACC oligoribonucleotide and NTPs have revealed that both P and N sites are unoccupied [28••]. Such VP1-UGUGACC complexes are characterized by a retracted priming loop, probably explaining the lack of VP1 polymerase activity. Given that the polymerase becomes catalytic active in the presence of VP2, the core capsid protein is expected to trigger the extension of the priming loop to support a priming GTP in the P site, thereby allowing a another GTP to occupy the N site and, ultimately, the formation of the initial phosphodiester bond of the dsRNA product. Subsequently, the priming loop retracts, allowing passage of the elongating dsRNA product out of the polymerase.
Based on structural analysis of DLPs, Estrozi et al [13••] established that the surface region of VP1 that comes in contact with the VP2 decamer includes residues located at or near the retracted priming loop (Figure 2e). Thus, VP1-VP2 contacts may induce electrostatic changes that promote a conformational shift in the priming loop to an extended form. Based on cell-free replication assays, VP2-mediated activation of VP1 polymerase activity represents a specific process that is only possible when VP1 and VP2 of the same rotavirus species are combined. For example, VP2 proteins of group C rotaviruses cannot activate the VP1 polymerase of group A rotaviruses [16••, 34], a result that may explain the failure of group A and C rotaviruses to successfully reassort with one another. Mutational studies have determined that both the scaffold and tether domains of VP2 are required for optimal polymerase activity [16••]. However, such studies have shown that it is the scaffold domain, and not the tether, that is responsible for the species specificity of VP2-dependent polymerase activation [16••].
Regulation of transcription
Structural analysis has revealed the presence of a flexible element (plug) at the C-terminus of VP1 that extends from the surface of the bracelet domain into the dsRNA/-RNA exit tunnel (Figure 3) [28••]. Although the plug was observed within this tunnel in crystal structures of VP1, successful purification of VP1 using a C-terminal polyhistidine affinity tag indicates that the plug can also extend outside the tunnel [28••]. Deletion mutagenesis has indicated that while the plug is not required for VP1 catalytic activity [28••], its affect on the diameter of the dsRNA/-RNA exit tunnel suggests that the plug may regulate the polymerase’s function as a replicase or a transcriptase. Specifically, when the plug is out of the tunnel, the tunnel’s diameter is sufficiently wide to accommodate dsRNA. However, when the plug is present, the tunnel’s diameter is reduced to the point that only single-stranded RNA can be accommodated [28••]. Thus, the presence of the plug may cause separation of the duplex product formed by the minus-strand template and nascent +RNA during transcription, resulting in the release of the two RNAs from different tunnels of the polymerase. How the position of the plug is controlled is unknown. However, as the predicted heterodimeric complex of VP1 and VP3 is bound to the interior face of VP2 decamers within the DLP, VP3 may prevent the release of the plug from the dsRNA/-RNA exit tunnel, promoting transcription by VP1. When VP3 is not present, or is not stably bound to VP1, the nascent dsRNA product likely pushes the plug out of the exit tunnel, allowing the polymerase to operate as a replicase.
Conclusion
Rotavirus polymerase activity requires the inner capsid proteins, a connection that links genome replication with core assembly and transcription with DLP assembly. For either replicase or transcriptase activity, VP1 likely requires the movement of the polymerase’s priming loop from a retracted form to an extended form, thereby allowing RNA initiation. Because such movement is dependent on assembled VP2, VP1 alone lacks polymerase activity, even though it can bind viral template RNA. Whether VP1 functions as a replicase or transcriptase may be regulated through the polymerase’s C-terminal plug, an element affecting the diameter of the dsRNA/-RNA exit tunnel. The interaction of VP3 with VP1 in the core may stabilize the plug in the dsRNA/-RNA exit tunnel, causing the polymerase to operate as transcriptase and not as a replicase. The VP6 capsid shell is also required for transcriptase activity, probably supporting in part the activities of channels that translocate +RNAs out of the core. Further insights into the importance of the VP1 priming loop and plug await determination of atomic structures for VP1 in enzymatically-active states. A model incorporating our current knowledge and ideas into the assembly of the VP1-VP2-VP3 decamer complex and its functions is illustrated in Figure 4.
Figure 4. Formation of core particles.
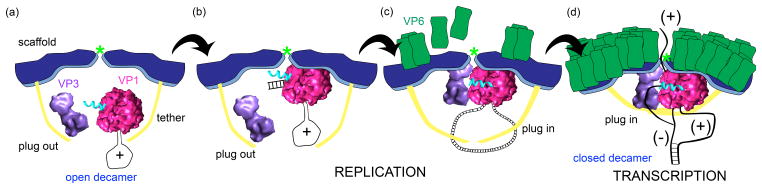
(a) VP2 decamers (blue) with open tethers (yellow) associate with VP1/+RNA complexes (pink) and VP3 (purple). The five-fold axis is indicated with a light green asterisk. (b) The specific interaction of the VP1/+RNA complex with the VP2 scaffold domain triggers RNA initiation. Elongation of dsRNA displaces the plug (cyan) from the exit tunnel and releases dsRNA into the core interior. (c) Interaction of VP3 with VP1 and VP2, along with closure of the tethers, stabilizes the plug within the exit tunnel. The intermediate VP6 layer (green) begins to assemble on top of VP2. (d) Assembly of the VP6 capsid shell promotes formation of channels that allow transcription and release of nascent transcripts. The positive-strand of the dsRNA segment likely remains associated with VP1, bound at the cap-binding site, to facilitate multiple rounds of transcription within the DLP.
Highlights.
Rotavirus RNA polymerase activity is strictly particle associated.
Rotavirus replicase activity requires the core shell protein VP2.
VP2 may induce conformational changes in the polymerase priming loop.
Transcriptase activity requires VP2 and VP6 capsid shell proteins.
C-terminal plug of VP1 may regulate replicase versus transcriptase activity.
Acknowledgments
We thank Marco Morelli and Kristen Ogden for their help in preparing the manuscript. The authors are supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stacy-Phipps S, Patton JT. Synthesis of plus- and minus-strand RNA in rotavirus-infected cells. J Virol. 1987;61:3479–84. doi: 10.1128/jvi.61.11.3479-3484.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patton JT. Evidence for equimolar synthesis of double-stranded and minus-stranded RNA in rotavirus-infected cells. Virus Res. 1990;17:199–208. doi: 10.1016/0168-1702(90)90065-j. [DOI] [PubMed] [Google Scholar]
- 3.Bican P, Cohen J, Charpilienne A, Scherrer R. Purification and characterization of bovine rotavirus cores. J Virol. 1982;43:1113–1117. doi: 10.1128/jvi.43.3.1113-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helmberger-Jones M, Patton JT. Characterization of subviral particles in cells infected with simian rotavirus SA11. Virology. 1986;155:655–665. doi: 10.1016/0042-6822(86)90225-4. [DOI] [PubMed] [Google Scholar]
- 5••.Patton JT, Jones MT, Kalbach AN, He YW, Xiaobo J. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J Virol. 1997;71:9618–26. doi: 10.1128/jvi.71.12.9618-9626.1997. Initial study demonstrating that rotavirus VP1 and VP2 were required at a 1:10 molar ratio for polymerase activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6••.Tortorici MA, Broering TJ, Nibert ML, Patton JT. Template recognition and formation of initiation complexes by the replicase of a segmented double-stranded RNA virus. J Biol Chem. 2003;278:32673–82. doi: 10.1074/jbc.M305358200. Critical study describing the requirement of VP2 for initiation of minus-strand synthesis by VP1. [DOI] [PubMed] [Google Scholar]
- 7.Patton JT, Vasquez-Del Carpio R, Spencer E. Replication and transcription of the rotavirus genome. Curr Pharm Des. 2004;10:3769–77. doi: 10.2174/1381612043382620. [DOI] [PubMed] [Google Scholar]
- 8.Mathieu M, Petitpas I, Navaza J, Lepault J, Kohli E, Pothier P, Prasad BV, Cohen J, Rey FA. Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion. EMBO J. 2001;20:1485–97. doi: 10.1093/emboj/20.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Baker ML, Jiang W, Estes MK, Prasad BV. Rotavirus architecture at subnanometer resolution. J Virol. 2009;83:1754–66. doi: 10.1128/JVI.01855-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawton JA, Zeng CQ-Y, Mukherjee SK, Cohen J, Estes MK, Prasad BV. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer. J Virol. 1997;71:7353–7360. doi: 10.1128/jvi.71.10.7353-7360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasad BV, Wang GJ, Clerx JP, Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988;199:269–75. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 12••.McClain B, Settembre E, Temple BR, Bellamy AR, Harrison SC. X-ray crystal structure of the rotavirus inner capsid particle at 3.8 resolution. J Mol Biol. 2010;397:587–99. doi: 10.1016/j.jmb.2010.01.055. Describes the first high-resolution crystal structure of the rotavirus DLP components VP2 and VP6. Tshe authors proposed specific interactions between VP2 and VP1 and/or RNA that facilitate packaging and polymerase activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Estrozi LF, Settembre EC, Goret G, McClain B, Zhang X, Chen JZ, Grigorieff N, Harrison SC. Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles. J Mol Biol. 2013;425:124–132. doi: 10.1016/j.jmb.2012.10.011. This study re-examines the X-ray map of the DLP to discern the location of VP1 within the core. The orientation of the polymerase relative to the VP2 decamer and the five-fold axis provides insight into the mechanism by which the core capsid protein activates polymerase activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogden KM, Snyder MJ, Dennis AF, Patton JT. Predicted structure and domain organization of rotavirus capping enzyme and innate immune antagonist VP3. J Virol. 2014;88:9072–9085. doi: 10.1128/JVI.00923-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boudreaux CE, Vile DC, Gilmore BL, Tanner JR, Kelly DF, McDonald SM. Rotavirus core shell subdomains involved in polymerase encapsidation into virus-like particles. J Gen Virol. 2013;94:1818–26. doi: 10.1099/vir.0.052951-0. [DOI] [PubMed] [Google Scholar]
- 16••.McDonald SM, Patton JT. Rotavirus VP2 core shell regions critical for viral polymerase activation. J Virol. 2011;85:3095–105. doi: 10.1128/JVI.02360-10. Study showing that the VP2 tether and scaffold domains were both required for maximal VP1 polymerase activity, but the role of the scaffold domains was specific. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng CQ-Y, Estes MK, Charpilienne A, Cohen J. The N terminus of rotavirus VP2 is necessary for encapsidation of VP1 and VP3. J Virol. 1998;72:201–208. doi: 10.1128/jvi.72.1.201-208.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labbe M, Baudoux P, Charpilienne A, Poncet D, Cohen J. Identification of the nucleic acid binding domain of the rotavirus VP2 protein. J Gen Virol. 1994;75:3423–30. doi: 10.1099/0022-1317-75-12-3423. [DOI] [PubMed] [Google Scholar]
- 19.Prasad BV, Rothnagel R, Zeng CQ, Jakana J, Lawton JA, Chiu W, Estes MK. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature. 1996;382:471–3. doi: 10.1038/382471a0. [DOI] [PubMed] [Google Scholar]
- 20.Clark SM, Spendlove RS, Barnett BB. Role of two particle types in bovine rotavirus morphogenesis. J Virol. 1980;34:272–6. doi: 10.1128/jvi.34.1.272-276.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen J, Laporte J, Charpilienne A, Scherrer R. Activation of rotavirus RNA polymerase by calcium chelation. Archives of Virology. 1979;60:177–186. doi: 10.1007/BF01317489. [DOI] [PubMed] [Google Scholar]
- 22.Patton JT, Silvestri LS, Tortorici MA, Vasquez-Del Carpio R, Taraporewala ZF. Rotavirus genome replication and morphogenesis: role of the viroplasm. Curr Top Microbiol Immunol. 2006;309:169–87. doi: 10.1007/3-540-30773-7_6. [DOI] [PubMed] [Google Scholar]
- 23.Gallegos CO, Patton JT. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology. 1989;172:616–27. doi: 10.1016/0042-6822(89)90204-3. [DOI] [PubMed] [Google Scholar]
- 24.Patton JT, Gallegos CO. Structure and protein composition of the rotavirus replicase particle. Virology. 1988;166:358–65. doi: 10.1016/0042-6822(88)90506-5. [DOI] [PubMed] [Google Scholar]
- 25.Sandino AM, Jashes M, Faundez G, Spencer E. Role of the inner protein capsid on in vitro human rotavirus transcription. J Virol. 1986;60:797–802. doi: 10.1128/jvi.60.2.797-802.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Mansell EA, Patton JT. Rotavirus RNA replication: VP2, but not VP6, is necessary for viral replicase activity. J Virol. 1990;64:4988–96. doi: 10.1128/jvi.64.10.4988-4996.1990. Early report revealing the essential roles of VP2 and VP6 in the formation of rotavirus particles with replicase activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gombold JL, Ramig RF. Assignment of simian rotavirus SA11 temperature-sensitive mutant groups A, C, F, and G to genome segments. Virology. 1987;161:463–73. doi: 10.1016/0042-6822(87)90140-1. [DOI] [PubMed] [Google Scholar]
- 28••.Lu X, McDonald SM, Tortorici MA, Tao YJ, Vasquez-Del Carpio R, Nibert ML, Patton JT, Harrison SC. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–88. doi: 10.1016/j.str.2008.09.006. The first high-resolution structures of the rotavirus RdRP provided significant insights into the mechanism of polymerase activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Te Velthuis AJ. Common and unique features of viral RNA-dependent polymerases. Cell Mol Life Sci. 2014 doi: 10.1007/s00018-014-1695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage–structural studies of reovirus polymerase λ3. Cell. 2002;111:733–45. doi: 10.1016/S0092-8674(02)01110-8. First publication describing the structure of aReoviridae RNA polymerase and function of its priming loop. [DOI] [PubMed] [Google Scholar]
- 31•.Patton JT. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA, but the interaction is not sufficient to initiate minus-strand synthesis. J Virol. 1996;70:7940–7. doi: 10.1128/jvi.70.11.7940-7947.1996. Publication demonstrating that purified recombinant VP1 alone lacked polymerase activity but retained viral-specific RNA binding activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogden KM, Ramanathan HN, Patton JT. Residues of the rotavirus RNA-dependent RNA polymerase template entry tunnel that mediate RNA recognition and genome replication. J Virol. 2011;85:1958–69. doi: 10.1128/JVI.01689-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patton JT, Vasquez-Del Carpio R, Tortorici MA, Taraporewala ZF. Coupling of rotavirus genome replication and capsid assembly. Adv Virus Res. 2007;69:167–201. doi: 10.1016/S0065-3527(06)69004-0. [DOI] [PubMed] [Google Scholar]
- 34.McDonald SM, Patton JT. Assortment and packaging of the segmented rotavirus genome. Trends Microbiol. 2011;19:136–44. doi: 10.1016/j.tim.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]