Abstract
Serine and Arginine-rich (SR) proteins play multiple roles in the eukaryotic gene expression pathway. Initially described as constitutive and alternative splicing factors, it is now clear that SR proteins are key determinants of exon identity and function as molecular adaptors, linking the pre-mRNA to the splicing machinery. In addition, SR proteins are now implicated in many aspects of mRNA and ncRNA processing well beyond splicing. These unexpected roles, including RNA transcription, export, translation and decay may prove to be the rule rather than the exception. To simply define this family of RNA binding proteins as splicing factors belies the broader roles of SR proteins in post-transcriptional gene expression.
Post-transcriptional regulation is critical to the accurate expression of human genes. This process is overseen in part by a large superfamily of RS-domain-containing proteins found throughout metazoans1, which contain SR and “SR-related” proteins (reviewed in2). Although it is often precarious to separate the two groups on a functional level, for the purposes of this review we will focus on the subfamily of “classic” SR proteins. SR proteins are structurally defined as a family of RNA binding proteins with a modular domain structure consisting of one to two amino- terminal RNA recognition motifs (RRMs) and a carboxyl-terminal domain rich in serine and arginine dipeptide repeats3. There are twelve canonical members of the SR protein family that share this characteristic domain structure (see Table 1). SR proteins are intimately involved in the gene expression pathway, influencing both nuclear pre-messenger RNA (pre-mRNA) processing as well as the cytoplasmic fate of the mature RNA (mRNA) message. Prominently known for their requirement in spliceosome assembly and regulation of alternative splicing decisions, important distinctions in SR protein biology have emerged over the years. For example, some SR proteins have a life in the cytoplasm whereas others remain confined in the nucleus. Here we will provide a summary of the nuclear roles of SR proteins as well as their emergent post-splicing functions in gene expression.
Table 1. The SR Protein Family.
Includes domain configuration of protein members, protein aliases, shuttling activities, reported molecular functions and biological processes.
| Gene Symbol | Domain Structure | Protein Aliases | Shuttling | Molecular Functions | Biological Processes | References |
|---|---|---|---|---|---|---|
| SRSF1 |
|
SF2, ASF, SRp30A | Yes | pre-mRNA splicing; mRNA export; translation; miRNA biogenesis; mRNA stability; NMD; transcriptional elongation | apoptosis; cell-cycle; senescence; cell growth proliferation; SUMOylation; Genomic Stability; cytoskeleton organization; embryogenesis; retinal development; cardiac development; cancer | 126, 132–141 |
| SRSF2 |
|
SC35, SRp30B | No | pre-mRNA splicing; Genomic Stability; transcriptional elongation | cell survival; cell cycle; cancer; metastasis; senescence; apoptosis; development; neural plasticity; metabolism | 37, 142–149 |
| SRSF3 |
|
SRp20 | Yes | pre-mRNA splicing; mRNA export; (viral) mRNA translation; transcriptional elongation | cell adhesion and migration; cell cycle; cell proliferation; cellular senescence; aerobic glycolysis; neuronal survival and growth; apoptosis; glucose and lipid metabolism; cholesterol homeostasis; LTM formation; development; neurological disorders; cancer | 150–160 |
| SRSF4 |
|
SRp75 | Yes | pre-mRNA splicing; | neural differentiation | 160 |
| SRSF5 |
|
SRp40 | No | pre-mRNA splicing; (viral) mRNA translation | insulin signaling; lipid transport; cell cycle; apoptosis; cancer; bipolar disorder | 161–164 |
| SRSF6 |
|
SRp55 | Yes | pre-mRNA splicing; (viral) mRNA translation | drosophilia development; cardiac development; eye development; apoptosis; wound healing; cell cycle; cytoskeleton organization; genomic integrity; angiogenesis; lipid transport; muscle development; calcium metabolism | 163, 168–178 |
| SRSF7 |
|
9G8 | Yes | pre-mRNA splicing; mRNA export; (viral) mRNA processing | microtubules stabilization; viral infection | 176, 177 |
| SRSF8 |
|
SRp46 | ND | pre-mRNA splicing | N/A | 190 |
| SRSF9 |
|
SRp30c | ND | pre-mRNA splicing mRNA translation | glucocorticoid signaling; apoptosis; cell-adhesion | 178–181 |
| SRSF10 |
|
SRp38, SRrp40 | Yes | pre-mRNA splicing; mRNA translation | stress response; neuronal differentiation; cholesterol biosynthesis; cell cycle | 73, 116, 182–186 |
| SRSF11 |
|
p54, NET2 | ND | pre-mRNA splicing; genomic stability | genomic integrity; ATP synthesis | 187, 188 |
| SRSF12 |
|
SRrp35 | ND | pre-mRNA splicing | cell cycle | 189 |
RRM, RNA recognition motif; RRMH, RNA recognition motif homology; RS, Arginine/Serine-rich motif; Zn, Zinc-binding domain. (References for presented data are included in table and text).
From the beginning, the functional characterization of SR proteins alluded to both diverse and redundant activities. The founding member of the SR protein family, SRSF1, was identified and characterized by concurrent studies using biochemical complementation assays. Not only was SRSF1 seen to preferentially enhance usage of the proximal authentic 5′ splice site of a β-globin splicing reporter 4, but it also altered splicing ratios of SV40 pre-mRNA, enhancing small T mRNA isoform production5. These data implicated the first “classic” SR protein as a regulator of both constitutive and alternative splicing. A second SR protein, SFRS6, was shown to complement splicing-deficient extracts to promote β-globin splicing, as well as alternative 5′ splice site usage in β-thalassemic pre-mRNA6, 7. At roughly the same time, a third SR protein, SRSF2 was shown to influence splice site selection. Using RNase T1 protection and immunoprecipitation assays, SRSF2 was seen to interact with both the 5′ and 3′ splice sites independently8. Furthermore, interactions between U1 and U2 snRNPs bound to 5′ and 3′ splice sites were shown to occur in an SRSF2-dependent fashion, implicating SR proteins in early spliceosome architecture. SRSF2 was also shown to have similar effects on splice site selection as SRSF1, alluding to functional redundancy during spliceosome assembly.
The SR protein family was rapidly expanded through clever biochemical fractionation by Zahler and colleagues, who co-purified a group of proteins, (including SRSF1, SRSF2, and SRSF6), by ammonium sulfate fractionation and precipitation with magnesium chloride9. This approach revealed five proteins of various molecular weights, which were selectively purified from both HeLa cell extract and calf thymus. These proteins presented reactivity to mAb104, an antibody previously shown to recognize phosphorylated SR proteins6, 10, 11. Furthermore, four of these proteins were shown to rescue splicing of β-globin and ftz splicing reporters in splicing-deficient extracts, providing evidence that, like SRSF1 and SRSF2, these proteins were splicing factors9. Finally, microsequencing of these proteins showed highly similar amino acid compositions, as well as an abundance of serine/arginine dipeptides, on which their family name is based9.
Regulation of SR proteins by post-translational modification
Post-translational modification plays critical roles in regulation of SR protein activity and localization. Phosphorylation of SR proteins is regulated by the SR-specific protein kinase (SRPK) family and other CMGC kinase family members, such as Clk/Sty (cdc2-like kinase/serine, threonine, and tyrosine kinase)12, 13. These kinases share similar abilities to phosphorylate serine residues throughout the RS domain but differ in their specificity and mechanism of phosphorylation14–17. Dynamic phosphorylation of SR proteins is vital to the initiation and progression of spliceosome assembly to catalysis18–22. Mechanistically, it is thought that phosphorylation of the RS domain increases RNA binding specificity23 and is also important for specific protein-protein interactions within the pre-spliceosome24. Structurally, phosphorylation results in entropic reduction of the intrinsically disordered RS domain to promote more ordered side chains for molecular recognition25. Together these data advocate that phosphorylation states of SR proteins act as “molecular switches” during spliceosome assembly.
RS domain phosphorylation also influences the dynamics of SR protein localization in the cell. Release of SR proteins from nuclear speckles requires phosphorylation by the RS domain kinase Clk/Sty13. Following spliceosome assembly, SR proteins encounter one of two potential paths: for a subset of SR proteins, re-phosphorylation by Clk/Sty and nuclear SRPKs will target the protein for nuclear recycling for further rounds of pre-mRNA splicing, whereas other SR proteins remain dephosphorylated and associated with spliced mRNAs9, 26–28. These studies suggest that dephosphorylated SR proteins retained on spliced mRNA may be a signal that an mRNA is ready for nuclear export. Following mRNA export and translation, SR proteins can then be rephosphorylated by cytoplasmic SRPKs, which facilitates interactions with transportin-SR and their import back into the nucleus29, 30.
SR protein phosphorylation is further modulated in response to a variety of different cellular conditions and signals. Changes in phosphorylation and subcellular distribution of SR proteins accompany the global regulation of RNA metabolism during early development31,32, viral infection33, and cell cycle progression12. One recent example demonstrates SR protein phosphorylation as a direct result of epidermal growth factor (EGF) signaling. EGF signaling is shown to increase AKT activation, which in turn activates SRPK and subsequent up-regulation of SR protein phosphorylation34. These data implicate SR proteins as integral players in propagating EGF signaling, which is linked to numerous human cancers.
SR proteins are modified by a variety of other marks including methylation and acetylation. The consequences of these modifications are less well-understood than phosphorylation but they appear to be functionally relevant. For example, acetylation of SRSF2 occurs in response to genotoxic stress. Acetylation within the RRM domain correlates with pre-mRNA alternative splicing regulation of caspase-8, a factor involved in apoptosis35. Arginine methylation also influences SR protein localization and activity37. Blocking methylation affects alternative splicing, translation, and mRNA decay, most likely due to mis-regulation of SR protein localization36–38.
The complex roles of SR proteins in pre-mRNA splicing
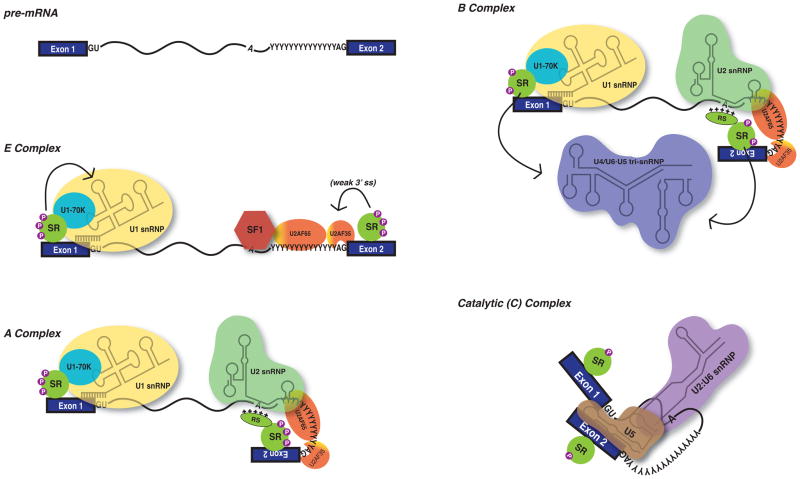
The spliceosome is assembled de novo on each and every intronic substrate. This dynamic process involves the sequential recruitment and rearrangement of Uracil-rich small nuclear ribonucleoprotein particles (U snRNPs). SR proteins contribute to spliceosome assembly primarily through the recognition of exonic splicing enhancers (ESEs)39–41. These interactions are particularly important during formation and stabilization of the Early (E) Complex (Fig. 1)8, 42–45. E complex is defined by association of both the U1 snRNP and the heterodimeric splicing factor U2 snRNP auxiliary factor (U2AF) with the 5′ and 3′ splice sites, respectively. This step is mediated by phosphorylation-dependent interactions between the RS and RRM domains of ESE-bound SR proteins and the U1-70K at the 5′ splice site21, 46 and the small subunit of U2AF (U2AF35) at the 3′ splice site 47–50. E-complex is assumed to form on either end of an intron, however, the same interaction network can occur across exons, in a process called exon definition (see below).
Figure 1. SR proteins regulate spliceosome assembly.
Spliceosome assembly onto the pre-mRNA occurs in a coordinated, stepwise manner. In E complex, SR proteins regulate U1 snRNP recruitment to the 5′ splice site GU, and U2AF35/65 bound to the pyrimidine tract and 3′ splice site AG. In the A complex, SR proteins may facilitate U2 snRNP binding at the branchpoint by neutralizing the negative phosphodiester backbone charge. SR proteins can also recruit U4/U6•U5 tri-snRNP during B complex. Molecular rearrangements and dephosphorylation of SR proteins occurs to form the catalytically active C complex, in which U2 and U6 interact, and U6 replaces U1 snRNP, and U5 coordinates exons prior to splicing and ligation. SF1, splicing factor 1; snRNP, small nuclear ribonucleoprotein, SR, SR protein; RS, Arginine/Serine motif; 5′ and 3′ splice sites are indicated by GU and AG dinucleotides, respectively; (Y)n, polypyrimidine tract; P, phosphate moiety](References for presented data are included in text)
E complex is converted to A complex by the addition of the U2 snRNP. During A complex formation, SR proteins are thought to promote interactions of U2 snRNP with the branch point sequence through non-specific interactions of the RS domain with the phosphodiester backbone, possibly neutralizing its negative charge and enhancing base pairing51. Additionally, SR proteins are implicated in recruitment of the U4/U6.U5 tri-snRNP52, forming a cross-exon “B-like” complex, which can ultimately rearrange into cross-intron B complexes53. The RS domain of SR proteins (presumably not associated with ESEs) are also hypothesized to associate with the phosphodiester backbone near the 5′ ss to promote U6 binding54. Finally, extensive remodeling and rearrangement of RNA-RNA and RNA-protein interactions, coupled with dephosphorylation of SR proteins, results in formation of the catalytically active C complex18, 55, 56. In summary, SR proteins promote recruitment of multiple factors throughout spliceosome assembly, and are critical in formation of the final catalytic core.
SR proteins also play important roles in establishing exon-intron boundaries in large metazoan genes. The process of “exon definition” is hypothesized to solve a significant problem related to finding relatively short exons within the context of long intronic sequences57. Exon definition occurs through a complex interaction network that links the 3′ss at the 5′ end of the exon with the 5′ss at the 3′ end of the exon (reviewed in 58). In metazoans exon definition precedes intron-definition in which 5′ and 3′ splice sites are paired during spliceosome assembly53. SR proteins also contribute to intron-definition through a series of protein-protein interactions, mediated by the RS domain linking U1 snRNP at the 5′ss to U2AF35 at the 3′ss45. Intron bridging has been alluded to through protein-protein interaction studies, and observed on splicing substrates using electron microscopy59, but the precise role of SR proteins is not well understood.
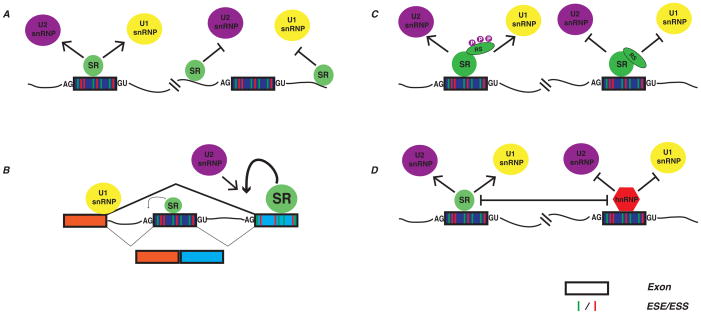
The mechanisms described above not only contribute to the roles of SR proteins in constitutive splicing, but similarly in alternative splicing. The distinction between the two processes is simply the context in which SR proteins engage the pre-mRNA50 (see Fig. 2A). A general theme emerging from both in vitro and in vivo assays is that SR proteins act as enhancers of splicing when associated with exonic sequences, but function as silencers while binding to intronic sequences downstream of the 5′ splice site39, 60. However, this simplistic perspective belies the complex cis-regulatory landscape of most regulated exons. Several features distinguishing alternative exons from constitutive exons, including their shorter length and weaker 5′ splice sites, respectively. Exonic regulatory sequences (ESRs) are also more strongly conserved in the context of alternative exons as compared to their counterparts in constitutive exons61, reflecting requirements for ESRs in definition of sub-optimal exons50. Remarkably, the same ESR sequence can have opposing affects on splicing when placed into distinct positions within the same alternative exon61. These studies suggest that the regulatory roles of SR proteins in alternative splicing are most likely highly position- and context-dependent.
Figure 2. SR proteins regulate alternative splicing.
A, SR proteins have been shown to promote or inhibit U1/U2 snRNP recruitment with respect to their orientation to 5′ and 3′ splice sites. B, SR proteins bound to adjacent exons can compete for U2 snRNP recruitment to their respective 3′ splice sites, likely depending on the “strength” of the SR protein to recruit spliceosomal factors. C, Phosphorylation states of the RS domain can influence SR protein-dependent recruitment of U1 and U2 snRNPs. D, Antagonistic relationships of SR proteins and hnRNP proteins often influence recruitment of spliceosomal factors. ESE/ESS, exonic splicing enhancers/exonic splicing silencers. (References for presented data are included in text).
SR proteins can also have long-range effects on regulation of alternative exons. Several studies described a new mode for SR protein-mediated splicing regulation that occurs through their association with constitutive exons that are adjacent to alternative exons62–64 (see Fig. 2B). For example, SRSF1 has been shown to promote skipping of exon 16 in CamKIIδ through its association with downstream constitutive exon 1763. A similar mechanism influences splicing of the receptor tyrosine kinase MET, a key driver of malignant breast cancer65. In this context, elevated SRSF1 levels lead to increased skipping of exon 11, an effect mediated by ESEs located within exon 1266. These data suggest an intriguing model in which SR proteins may alter the competition between 3′ splice sites of adjacent exons with a common upstream 5′ splice site 61,62,81,82.
Another important aspect of the mechanisms through which SR proteins influence alternative splicing involves their interplay with members of the hnRNP family. The hnRNPs include several well-established splicing repressors, which mediate the repressive effects of exonic splicing silencers (ESSs). The functional antagonism of SR proteins and hnRNP proteins was first observed between SRSF1 and hnRNP A1 on several different alternative splicing modalities67 (see Fig. 2D). Not surprisingly, the underlying molecular mechanisms can be quite distinct. In the case of competing splice site donors, SRSF1 promoted selection of the proximal 5′ splice site (closest to the 3′ss) whereas hnRNP A1 promoted usage of more distal sites68 by reducing binding of U1 snRNP at the proximal site. This functional antagonism also extends to alternative cassette exons. In an elegant series of experiments, Zhu and Krainer demonstrated that binding of an SR protein to an ESE inhibits the repressive affects of hnRNPs bound to adjacent silencers69. Because the relative expression levels of SR proteins and hnRNPs can vary dramatically across tissues and during tumorigenesis70, 71, the complex functional interplay between hnRNPs and SR proteins are likely to play important roles in regulating patterns of alternate splicing across a wide array of conditions.
While often thought of as general splicing enhancers, there are also instances where SR proteins can inhibit splicing (see Fig. 2C). For example SRSF9 promotes skipping of exon 7B in the hnRNP A1 pre-mRNA72. This activity requires an intronic splicing silencer element located upstream of the exon 7B 3′ ss. Likewise, the poorly characterized SRSF11 is reported to promote skipping of exon 10 of the Tau pre-mRNA by binding an exonic splicing silencer73. By contrast to these transcript specific affects, SRSF10 functions as an inducible, global repressor of splicing74. SRSF10 activity is inhibited by phosphorylation-dependent interactions with 14-3-3 proteins. Conditions that promote activation of protein phosphatase 1, including heat shock and mitosis, leads to dephosphorylation of SRSF10, liberation from 14-3-3 proteins and activation of splicing repressor activity75. Although the mechanisms of splicing inhibition are likely to be very different for each of theses SR proteins, it is nonetheless intriguing that SR proteins are capable of having potentially opposite affects on splicing depending on their phosphorylation state76 or the context in which they engage the pre-mRNA.
Global analysis of SR protein RNA binding specificity
SR proteins are sequence-specific RNA binding proteins. For most SR proteins, a putative consensus motif has been identified (reviewed in 77) but the challenge now is to determine how these elements function within different sequence contexts. Additionally, it is clear that there is significant functional redundancy in binding specificity78–80. These data imply that SR proteins may compete with each other for binding to closely related sites.
The first clues for understanding how this competition plays out on a global scale emerged from studies of SR proteins distribution on fixed insect polytene chromosomes and amphibian oocyte lampbrush chromosomes10, 81. Imaging of nascent transcripts on Chironomus tentans polytene chromosomes revealed that SR proteins are distributed across the genome in a non-random pattern. Distinct SR protein staining patterns were observed at different loci, suggesting that different combinations of SR proteins associated with nascent transcripts. High-resolution analysis of the Balbiani Ring (BR) locus revealed that the BR mRNP is extensively remodeled as it is matured. Many of the SR proteins are replaced between the steps of mRNA export and translation, such that only SRSF1 is bound to polyribosome-associated mRNPs82. This observation is consistent with work from mammalian cells, which demonstrate that SR proteins are sorted on nascent transcripts through a phosphorylation-dependent mechanism83.
The high throughout sequencing and crosslinking immunoprecipitation (HITS-CLIP) method allowed for global analysis of in situ protein-RNA interactions and provides key information such as consensus binding motifs, genome-wide binding site distribution, gene ontology of RNA targets, etc.84. HITS-CLIP analysis of SRSF1 revealed a diverse pool of RNA transcripts, including mRNA, miRNAs, snoRNAs, and intergenic transcripts of unknown function, advocating roles for SR proteins beyond pre-mRNA processing62, 85. Subsequent studies confirmed many of these hypotheses, including interactions with long non-coding RNAs MALAT1 and Xist, as well as pre-cursors of miRNA processing86–89. Furthermore, gene ontology analysis of SRSF1 mRNA targets showed an enrichment for RNA processing factors, suggesting a broad, highly integrated post-transcriptional network that governs splicing factor levels and ultimately global gene expression85. Like SRSF1, SRSF3 and SRSF4 engage a functionally diverse pool of RNA transcripts. However, their consensus binding sites and their CLIP tag distribution across transcripts is distinct90. Most intriguing was that CLIP tags for both SRSF3 and SRSF4 were enriched in ncRNAs, many of which have yet to be prescribed functions within the cell. Finally, SRSF3, but not SRSF4, was seen to regulate splicing of other splicing factors, further supporting the hypothesis of regulatory cascades that may extend the roles of SR proteins beyond their known targets85, 90, 91.
Global studies of SR protein RNA target specificity demonstrate that most exons are bound by at least one SR protein64, 82, 85, 90. A major challenge now is to understand how different combinations of SR proteins influence splicing of specific exons. Recent work from Pandit et al. suggests that this is likely to be a complex problem64. An initial comparison of SRSF1 and -2 in situ binding sites in mouse embryo fibroblasts (MEFs) demonstrated that both proteins have considerable overlap in their binding specificity, suggesting that there may be competition for binding to related exon sequences. Interestingly, depletion of SRSF2 resulted in complex changes in SRSF1 binding site occupancy. In some cases SRSF1 binding increased in the absence of SRSF2, whereas the opposite pattern was observed at other locations. Together these data suggest that SR proteins can play both redundant roles and cooperative roles in exon recognition.
Emerging roles for SR proteins in gene expression
While it is generally accepted that the majority of splicing occurs in a co-transcriptional manner, only recently have these two processes been observed to directly regulate one another92, 93. Live cell imaging initially showed dynamic recruitment of various splicing factors from nuclear speckles to sites of transcription activation94. Indeed, SR proteins co-localize with RNA polymerase II in nuclear speckles, an interaction mediated by the Pol II C-terminal domain (CTD)95 in a serine phosphorylation-dependent manner22, 96. Truncation of the CTD prevents targeting of splicing factors to sites of transcription and markedly inhibits pre-mRNA splicing97. Also, selective mutations in the CTD cause diffusion of SR proteins away from nuclear speckles, and accumulation of unspliced β-globin transcripts98. These data indicate that interactions between SR proteins and Pol II are involved in splicing regulation. Using a minigene spicing reporter, de la Mata and colleagues showed that the CTD was required for SRSF3 recruitment and subsequent exon exclusion. Furthermore, the affect of the CTD on SRSF3-regualted alternative splicing was independent of transcription kinetics99. Together, these data imply that the CTD may play a direct role in spliceosome assembly through SR protein recruitment. Conversely, recent data imply that the association of SR proteins and RNA Pol II may only occur after initiation of transcription. In the context of nascent FOS RNA, association of various SR proteins with Pol II was seen to be RNA-dependent100, suggesting that recruitment of SR proteins to actively transcribed genes may not occur during initiation in all contexts. Additional experiments are needed to determine the mechanisms through which SR proteins regulate co-transcriptional alternative splicing. Regardless, these data provide a functional link between the processes of Pol II transcription and alternative splicing decisions mediated by SR proteins.
SR proteins may also directly regulate elongation rates of RNAPII. In general, depletion of either SRSF1 or SRSF2 have global effects on Pol II transcription in cells, and SRSF2 levels have been shown to affect the accumulation of Pol II at gene loci101. Mechanistically, SRSF2 is thought to enhance the release of transcriptional regulator TEFb from 7SK RNA due to emergence of an SRSF2-recognized ESE following initial Pol II elongation. This may induce SRSF2 to switch from the 7SK RNA to nascent RNA, triggering the coordinated release of P-TEFb from the 7SK complex, and subsequent phosphorylation and un-pausing of Pol II102. This suggests that some SR proteins may have direct effects in recruitment of Pol II factors to initiated Pol II complexes to facilitate elongation.
Compartmentalization of genetic material in the nucleus allows for separation of mRNA transcription from its fate as a template for protein synthesis. The discovery that a subset of SR proteins shuttle between the nucleus and cytoplasm (reviewed in 103) immediately suggested that SR proteins might remain bound with their mRNA targets beyond pre-mRNA splicing (Fig. 3). At least one non-canonical function for shuttling SR proteins appears to be in mRNA export pathways. Specific roles for the SR proteins SRSF3 and SRSF7 in intronless histone mRNA export provided the first direct evidence for SR protein moonlighting104. Shuttling SR proteins can interact with the canonical mRNA export factor nuclear RNA export factor 1 (NXF1; also known as TAP)105, 106. These data suggest that SR proteins may function broadly in the export of both spliced and unspliced mRNAs104, 107, 108. Interactions between NXF1 and SR proteins require dephosphorylation of the RS domain, suggesting an elegant mechanism for signaling the completion of an export-ready mRNA106. Surprisingly, depletion of specific SR proteins does not induce general defects in mRNA export83, suggesting that SR proteins may function redundantly or play roles in nuclear export of specific mRNAs.
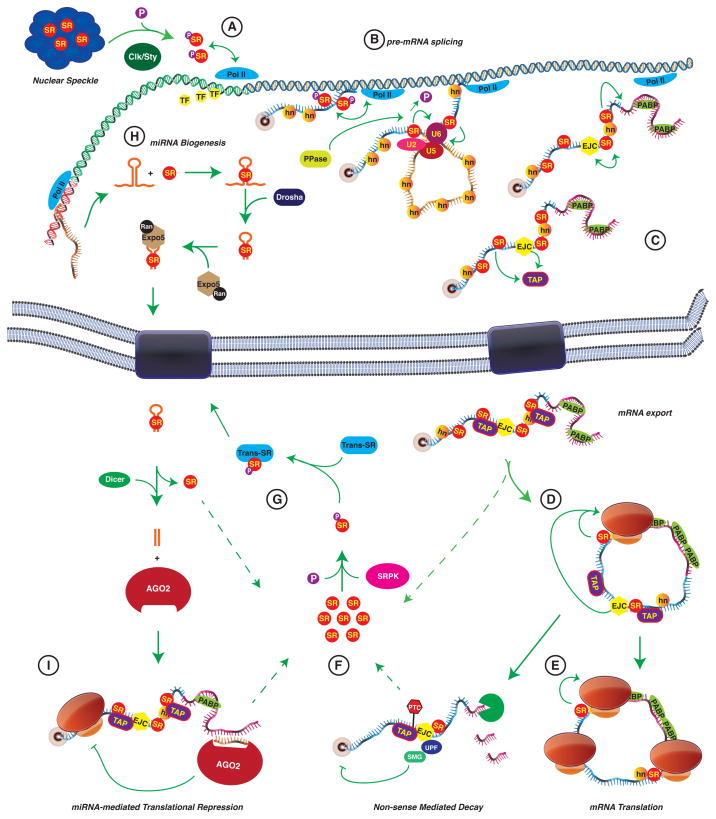
Figure 3. The life cycle of an SR protein.
A, SR proteins remain localized to nuclear speckles until they are phosphorylated by Clk/Sty. At this point they can be recruited to areas of active transcription, possibly in Pol II-dependent manner. B, SR proteins can then bind to splicing enhancers in the nascent pre-mRNA transcript to facilitate spliceosome assembly co-transcriptionally in phosphorylation dependent manner. C, Following maturation of the mRNA transcript, SR proteins, along with other factors (e.g. EJC proteins) can facilitate TAP binding to the mRNP and subsequent nuclear export. D, After export, SR proteins can enhance the pioneering round of mRNA translation and send the transcript down one of two pathways: E, the ribosome encounters no pre-termination codons (PTC) and continues with steady-state translation, or F, a PTC is encountered and nonsense-mediated decay proceeds. G, Released SR proteins can then be phosphorylated by cytoplasmic SRPK, which triggers binding of Transportin-SRs and nuclear import of SR proteins for storage or further rounds of splicing. H, SR proteins may also play roles in miRNA biogenesis by facilitating export of pre-microRNAs to the cytoplasm for further processing and use in RNA-induced silencing (I). SR, SR protein. P, phosphate moiety. TF, transcription factor. Pol II, RNA polymerase II. hn, hnRNP proteins. U2, U5, U6, U snRNPs. PPase, protein phosphatase. EJC, exon junction complex. PABP, poly-A binding protein. TAP, TAP/nuclear export factor 1. Exo, exosome. SRPK, SR protein Kinase. Trans-SR, Transportin-SR. Expo5, Exportin 5. AGO2, Argonaute 2. (References for presented data are included in text).
The nucleocytoplasmic shuttling SR protein SRSF1 is readily detectable in the polyribosome fraction of cultured human cells suggesting that SR proteins may be involved in mRNA translation109. This hypothesis was confirmed through three different functional assays. Perhaps most importantly, non-shuttling mutants of SRSF1 failed to enhance mRNA translation28, 109. Subsequent experiments demonstrated that SRSF1 stimulates translation initiation through a novel mechanism involving the mechanistic target of rapamycin complex (mTOR)110. These data support an intriguing model whereby SRSF1 functions as an adaptor protein linking specific mRNA transcripts to translational control by mTOR (see Fig. 4A). Recent work from the Caceres laboratory dramatically extends this model with the identification of >500 mRNAs that are likely to be translationally controlled by SRSF1 and mTOR111.
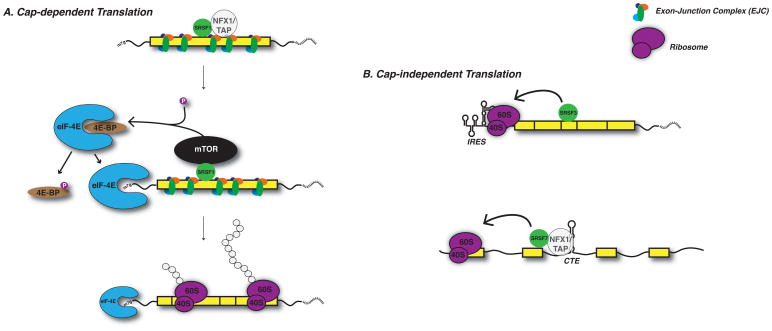
Figure 4. SR proteins function in translation initiation.
SRSF1 bound to exported mRNAs can associate with mTOR kinase and recruit it to cytoplasmic mRNP complexes. This facilitates phosphorylation of 4E-BP, causing dissociation from eIF4E, and increasing efficiency of cap-dependent translation initiation. SR proteins have also been shown to enhance cap-independent translation initiation of viral RNAs that contain internal ribosome entry site (IRES) elements and constitutive transport elements (CTEs). (References for presented data are included in text).
Moreover, other shuttling SR proteins including SRSF3 and SRSF7 are implicated in translational control. SRSF3 and SRSF7 mediate the effects of two distinct cis-regulatory elements including a viral IRES and cellular constitutive transport elements (CTEs)108, 112 (see Fig. 4B). Likewise, SRSF5 and SRSF6 enhance translation of gag protein from unspliced HIV-1 RNA, an activity that depends on their ability to shuttle from the nucleus to the cytoplasm113. SR proteins also have the potential to repress translation. During Xenopus development SRSF10 has been shown to interact with the peptidyltransferase center of 28S rRNA in undifferentiated neural cells114. Furthermore, this mechanism may help neuronal stem cells to maintain an undifferentiated state. These data paint a larger role for SR proteins in translation through regulating interactions with their respective RNA targets and translational machinery.
The roles of SRSF1 in mRNA translation suggest an intriguing hypothesis, that the fates of mRNA isoforms generated by alternative splicing may be subject to differential translation109, 115, 116. This idea is supported by the recent observation that >30% of alternative mRNA isoforms exhibit differential polyribosome association117. Shuttling SR proteins, such as SRSF1, which remain associated with its’ targets throughout the gene expression pathway, are likely to contribute to this mechanism62, 83. Over-expression of SRSF1 results in isoform-specific recruitment of mRNAs to polyribosomes, suggesting a direct role in coordinating the alternative fates of mRNA isoforms111.
In addition to splicing, export and translation, SR proteins also influence mRNA stability118. This activity can occur through several different mechanisms. First, SR proteins regulate alternative splicing and in many cases, such as post-transcriptional control of splicing factor levels, alternative splicing generates isoforms that are inherently unstable91, 119. Unstable isoforms contain pre-mature termination codons that trigger the nonsense mediated decay (NMD) RNA surveillance pathway. By contrast to this splicing based mode of gene regulation, SR proteins have also been shown to directly enhance NMD120. Intriguingly, this study showed the RS domain is required for augmentation of NMD, but not SR protein shuttling activity. These data suggest that SR proteins stimulate a rate-limiting step in the nucleus or that SR proteins may regulate the expression of NMD factors. Alternatively, SRSF1 may stimulate the pioneer round of mRNA translation leading to more efficient NMD121. Taken together there is little doubt that SR proteins have complex effects on transcript stability.
By contrast to their roles in pre-mRNA splicing, relatively little is known concerning the molecular mechanisms through which SR proteins influence post-splicing steps of gene expression. One hypothesis is that shuttling SR proteins work in concert with the exon junction complex (EJC) to influence mRNA export105, 122, stability118 and translation109, 110, 123. The EJC is deposited near exon-exon junctions as a result of pre-mRNA splicing and regulates post-transcriptional control of mature mRNAs. Proteomic analysis of EJC factors revealed numerous RNA-independent interactions with SR proteins. This observation is in good agreement with analysis of EJC RNA footprints, which revealed a myriad of non-canonical binding sites (i.e. those not centered 24 nucleotides upstream of an exon-exon junction). A significant proportion of these footprints overlapped with exonic splicing enhancers (ESEs), which are often occupied by SR proteins. EJC-SR protein interactions appear to be functionally significant as SRSF1 and -3 exhibit reduced mRNA binding activity when the EJC factor eIF4AIII is depleted from cells 124. Together, these data reveal extensive, cooperative associations between SR proteins and the EJC in mRNP biogenesis and may explain their functional redundancy in regulation of mRNA export105, 122, translation109, 110, 123, and decay121, 122, as well as maintenance of genomic stability125, 126.
The roles of SR proteins in gene regulation extend beyond mRNA processing. SR proteins have recently been attributed functional roles in microRNA biogenesis. Specifically, SRSF1 has been found to associate with primary-miR-7 transcript thorough a putative SRSF1 binding site in the stem loop. This interaction promotes cleavage of the pre-miR by the microprocessor complex protein Drosha89. The role for SRSF1 in microRNA biogenesis appears to be direct, as it is independent of its role in splicing. HITS-CLIP analysis of SRSF1, -3 and -4 suggests that most SR proteins interact with a small but distinct group of miRNAs, suggesting that shuttling SR proteins are involved in miRNA biogenesis on a more general level85, 90. Overall, these data highlight a potential coordination between splicing regulation and miRNA-mediated transcriptome regulation.
Conclusion
The characteristics of SR proteins mirror another general regulator of nucleic acid structure and function: histones. Like histones, SR proteins control the accessibility of their nucleic acid targets to the gene expression machinery. The similarities extend to their biochemical properties as well. Both are highly alkaline, associate with and regulate the use of their respective bound nucleic acids, and can form homo- and heteroligomers to package DNA/RNA within the cell. Also, both histones and SR proteins are extensively post-translationally modified, which can control the functionality of the nucleic acid to which they are bound. Furthermore, both sets of proteins are used as the foundational basis for recruitment of additional factors to their respective nucleic acids to accomplish biochemical work, whether it be DNA-binding complexes that regulate and catalyze transcription or RNA-binding complexes that regulate pre-mRNA splicing, mRNA export, translation, and degradation. Similar comparisons have been made of hnRNP oligomers that have the ability to wrap up RNA species to form mRNPs that share some resemblance to nucleosomes127, 128. Clearly, this hypothesis warrants further investigation given the intimate association of SR proteins with virtually every aspect of post-transcriptional gene regulation.
The coming years will undoubtedly see an explosion in data utilizing high-throughput assays (e.g. HITS-CLIP, iCLIP, etc.) to determine the transcriptome-wide RNA-interactions networks of SR proteins. These studies will provide a more general overview as to what RNAs SR proteins associate with and how they bind to them. This will certainly solidify the notion that SR proteins function in all aspects of RNA metabolism and gene expression rather than just splicing. The challenge for future work is to begin to determine how fluctuations in the levels of SR proteins influence the binding specificity of other SR proteins and splicing factors globally 64, 129–131. These types of experiments will elucidate the context-specific interactions that determine how exon identity is established in living cells as well as other RBP “codes” as they function in downstream steps of gene expression. Overall, the near future holds a greater understanding of how SR proteins govern the RNA world.
Acknowledgments
The authors wish to thank current and past Sanford Lab member and Alan Zahler for comments on the manuscript and acknowledge support from the NIH (GM109146 and AG042003) and the Ellison Medical Research Foundation New Scholar Award to J.R.S.
Contributor Information
Jonathan M. Howard, Department of Molecular, Cellular and Developmental Biology, University of California Santa Cruz, Santa Cruz, California 95064, USA
Jeremy R. Sanford, Email: jsanfor2@ucsc.edu, Department of Molecular, Cellular and Developmental Biology, University of California Santa Cruz, Santa Cruz, California 95064, USA
References
- 1.Boucher L, Ouzounis CA, Enright AJ, Blencowe BJ. A genome-wide survey of RS domain proteins. RNA. 2001;7:1693–1701. [PMC free article] [PubMed] [Google Scholar]
- 2.Blencowe BJ, Bowman JA, McCracken S, Rosonina E. SR-related proteins and the processing of messenger RNA precursors. Biochem Cell Biol. 1999;77:277–291. [PubMed] [Google Scholar]
- 3.Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24:1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krainer AR, Conway GC, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 5.Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 6.Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayeda A, Zahler AM, Krainer AR, Roth MB. Two members of a conserved family of nuclear phosphoproteins are involved in pre-mRNA splicing. Proc Natl Acad Sci U S A. 1992;89:1301–1304. doi: 10.1073/pnas.89.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu XD, Maniatis T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc Natl Acad Sci U S A. 1992;89:1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 10.Roth MB, Murphy C, Gall JG. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roth MB, Gall JG. Monoclonal antibodies that recognize transcription unit proteins on newt lampbrush chromosomes. J Cell Biol. 1987;105:1047–1054. doi: 10.1083/jcb.105.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- 13.Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- 14.Aubol BE, Plocinik RM, Hagopian JC, Ma CT, McGlone ML, Bandyopadhyay R, Fu XD, Adams JA. Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases. J Mol Biol. 2013;425:2894–2909. doi: 10.1016/j.jmb.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma CT, Hagopian JC, Ghosh G, Fu XD, Adams JA. Regiospecific phosphorylation control of the SR protein ASF/SF2 by SRPK1. J Mol Biol. 2009;390:618–634. doi: 10.1016/j.jmb.2009.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colwill K, Feng LL, Yeakley JM, Gish GD, Caceres JF, Pawson T, Fu XD. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J Biol Chem. 1996;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- 17.Huynh N, Ma CT, Giang N, Hagopian J, Ngo J, Adams J, Ghosh G. Allosteric interactions direct binding and phosphorylation of ASF/SF2 by SRPK1. Biochemistry. 2009;48:11432–11440. doi: 10.1021/bi901107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mermoud JE, Cohen PT, Lamond AI. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanopka A, Muhlemann O, Petersen-Mahrt S, Estmer C, Ohrmalm C, Akusjarvi G. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature. 1998;393:185–187. doi: 10.1038/30277. [DOI] [PubMed] [Google Scholar]
- 20.Ma CT, Ghosh G, Fu XD, Adams JA. Mechanism of dephosphorylation of the SR protein ASF/SF2 by protein phosphatase 1. J Mol Biol. 2010;403:386–404. doi: 10.1016/j.jmb.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1–70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci U S A. 2011;108:8233–8238. doi: 10.1073/pnas.1017700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misteli T, Caceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci U S A. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 25.Xiang S, Gapsys V, Kim HY, Bessonov S, Hsiao HH, Mohlmann S, Klaukien V, Ficner R, Becker S, Urlaub H, et al. Phosphorylation drives a dynamic switch in serine/arginine-rich proteins. Structure. 2013;21:2162–2174. doi: 10.1016/j.str.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol Cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 28.Sanford JR, Ellis JD, Cazalla D, Caceres JF. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc Natl Acad Sci U S A. 2005;102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kataoka N, Bachorik JL, Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J Cell Biol. 1999;145:1145–1152. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7957. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- 31.Sanford JR, Bruzik JP. Developmental regulation of SR protein phosphorylation and activity. Genes Dev. 1999;13:1513–1518. doi: 10.1101/gad.13.12.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanford JR, Bruzik JP. Regulation of SR protein localization during development. Proc Natl Acad Sci U S A. 2001;98:10184–10189. doi: 10.1073/pnas.181340498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bridge E, Xia DX, Carmo-Fonseca M, Cardinali B, Lamond AI, Pettersson U. Dynamic organization of splicing factors in adenovirus-infected cells. J Virol. 1995;69:281–290. doi: 10.1128/jvi.69.1.281-290.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, Hu Q, Ghosh G, Adams JA, Rosenfeld MG, et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47:422–433. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edmond V, Moysan E, Khochbin S, Matthias P, Brambilla C, Brambilla E, Gazzeri S, Eymin B. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 2011;30:510–523. doi: 10.1038/emboj.2010.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bressan GC, Moraes EC, Manfiolli AO, Kuniyoshi TM, Passos DO, Gomes MD, Kobarg J. Arginine methylation analysis of the splicing-associated SR protein SFRS9/SRP30C. Cell Mol Biol Lett. 2009;14:657–669. doi: 10.2478/s11658-009-0024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinha R, Allemand E, Zhang Z, Karni R, Myers MP, Krainer AR. Arginine methylation controls the subcellular localization and functions of the oncoprotein splicing factor SF2/ASF. Mol Cell Biol. 2010;30:2762–2774. doi: 10.1128/MCB.01270-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen YC, Milliman EJ, Goulet I, Cote J, Jackson CA, Vollbracht JA, Yu MC. Protein arginine methylation facilitates cotranscriptional recruitment of pre-mRNA splicing factors. Mol Cell Biol. 2010;30:5245–5256. doi: 10.1128/MCB.00359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavigueur A, La Branche H, Kornblihtt AR, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 40.Sun Q, Mayeda A, Hampson RK, Krainer AR, Rottman FM. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 41.Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 42.Chiara MD, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Identification of proteins that interact with exon sequences, splice sites, and the branchpoint sequence during each stage of spliceosome assembly. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staknis D, Reed R. SR proteins promote the first specific recognition of Pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 45.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 46.Xiao SH, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu J, Krainer AR. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 2000;14:3166–3178. doi: 10.1101/gad.189500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Hoffmann HM, Grabowski PJ. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 49.Guth S, Tange TO, Kellenberger E, Valcarcel J. Dual function for U2AF(35) in AG-dependent pre-mRNA splicing. Mol Cell Biol. 2001;21:7673–7681. doi: 10.1128/MCB.21.22.7673-7681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graveley BR, Hertel KJ, Maniatis T. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA. 2001;7:806–818. doi: 10.1017/s1355838201010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 52.Roscigno RF, Garcia-Blanco MA. SR proteins escort the U4/U6. U5 tri-snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider M, Will CL, Anokhina M, Tazi J, Urlaub H, Luhrmann R. Exon definition complexes contain the tri-snRNP and can be directly converted into B-like precatalytic splicing complexes. Mol Cell. 2010;38:223–235. doi: 10.1016/j.molcel.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 54.Shen H, Green MR. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 55.Cao W, Jamison SF, Garcia-Blanco MA. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 56.Amin EM, Oltean S, Hua J, Gammons MV, Hamdollah-Zadeh M, Welsh GI, Cheung MK, Ni L, Kase S, Rennel ES, et al. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell. 2011;20:768–780. doi: 10.1016/j.ccr.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 58.De Conti L, Baralle M, Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA. 2013;4:49–60. doi: 10.1002/wrna.1140. [DOI] [PubMed] [Google Scholar]
- 59.Stark JM, Bazett-Jones DP, Herfort M, Roth MB. SR proteins are sufficient for exon bridging across an intron. Proc Natl Acad Sci U S A. 1998;95:2163–2168. doi: 10.1073/pnas.95.5.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erkelenz S, Mueller WF, Evans MS, Busch A, Schoneweis K, Hertel KJ, Schaal H. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013;19:96–102. doi: 10.1261/rna.037044.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences--The complex definition of enhancers and silencers. Mol Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 62.Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan W, Caceres JF. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One. 2008;3:e3369. doi: 10.1371/journal.pone.0003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han J, Ding JH, Byeon CW, Kim JH, Hertel KJ, Jeong S, Fu XD. SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons. Mol Cell Biol. 2011;31:793–802. doi: 10.1128/MCB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M, Jr, Fu XD. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell. 2013;50:223–235. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maggiora P, Marchio S, Stella MC, Giai M, Belfiore A, De Bortoli M, Di Renzo MF, Costantino A, Sismondi P, Comoglio PM. Overexpression of the RON gene in human breast carcinoma. Oncogene. 1998;16:2927–2933. doi: 10.1038/sj.onc.1201812. [DOI] [PubMed] [Google Scholar]
- 66.Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 67.Caceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 68.Eperon IC, Makarova OV, Mayeda A, Munroe SH, Caceres JF, Hayward DG, Krainer AR. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol Cell Biol. 2000;20:8303–8318. doi: 10.1128/mcb.20.22.8303-8318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 70.Hanamura A, Caceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 71.Zerbe LK, Pino I, Pio R, Cosper PF, Dwyer-Nield LD, Meyer AM, Port JD, Montuenga LM, Malkinson AM. Relative amounts of antagonistic splicing factors, hnRNP A1 and ASF/SF2, change during neoplastic lung growth: implications for pre-mRNA processing. Mol Carcinog. 2004;41:187–196. doi: 10.1002/mc.20053. [DOI] [PubMed] [Google Scholar]
- 72.Simard MJ, Chabot B. SRp30c is a repressor of 3′ splice site utilization. Mol Cell Biol. 2002;22:4001–4010. doi: 10.1128/MCB.22.12.4001-4010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu JY, Kar A, Kuo D, Yu B, Havlioglu N. SRp54 (SFRS11), a regulator for tau exon 10 alternative splicing identified by an expression cloning strategy. Mol Cell Biol. 2006;26:6739–6747. doi: 10.1128/MCB.00739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- 75.Shi Y, Manley JL. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol Cell. 2007;28:79–90. doi: 10.1016/j.molcel.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 76.Feng Y, Chen M, Manley JL. Phosphorylation switches the general splicing repressor SRp38 to a sequence-specific activator. Nat Struct Mol Biol. 2008;15:1040–1048. doi: 10.1038/nsmb.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 78.Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu XD. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- 81.Bauren G, Jiang WQ, Bernholm K, Gu F, Wieslander L. Demonstration of a dynamic, transcription-dependent organization of pre-mRNA splicing factors in polytene nuclei. J Cell Biol. 1996;133:929–941. doi: 10.1083/jcb.133.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bjork P, Jin S, Zhao J, Singh OP, Persson JO, Hellman U, Wieslander L. Specific combinations of SR proteins associate with single pre-messenger RNAs in vivo and contribute different functions. J Cell Biol. 2009;184:555–568. doi: 10.1083/jcb.200806156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin S, Xiao R, Sun P, Xu X, Fu XD. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol Cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 84.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 85.Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Royce-Tolland ME, Andersen AA, Koyfman HR, Talbot DJ, Wutz A, Tonks ID, Kay GF, Panning B. The A-repeat links ASF/SF2-dependent Xist RNA processing with random choice during X inactivation. Nat Struct Mol Biol. 2010;17:948–954. doi: 10.1038/nsmb.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martins SB, Rino J, Carvalho T, Carvalho C, Yoshida M, Klose JM, de Almeida SF, Carmo-Fonseca M. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat Struct Mol Biol. 2011;18:1115–1123. doi: 10.1038/nsmb.2124. [DOI] [PubMed] [Google Scholar]
- 93.Lee KM, Tarn WY. Coupling pre-mRNA processing to transcription on the RNA factory assembly line. RNA Biol. 2013;10:380–390. doi: 10.4161/rna.23697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 95.Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci U S A. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Du L, Warren SL. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 99.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 100.Sapra AK, Anko ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol Cell. 2009;34:179–190. doi: 10.1016/j.molcel.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 101.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Twyffels L, Gueydan C, Kruys V. Shuttling SR proteins: more than splicing factors. FEBS J. 2011;278:3246–3255. doi: 10.1111/j.1742-4658.2011.08274.x. [DOI] [PubMed] [Google Scholar]
- 104.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 105.Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 106.Lai MC, Tarn WY. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J Biol Chem. 2004;279:31745–31749. doi: 10.1074/jbc.C400173200. [DOI] [PubMed] [Google Scholar]
- 107.Escudero-Paunetto L, Li L, Hernandez FP, Sandri-Goldin RM. SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs. Virology. 2010;401:155–164. doi: 10.1016/j.virol.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Swartz JE, Bor YC, Misawa Y, Rekosh D, Hammarskjold ML. The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J Biol Chem. 2007;282:19844–19853. doi: 10.1074/jbc.M701660200. [DOI] [PubMed] [Google Scholar]
- 109.Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 111.Maslon MM, Heras SR, Bellora N, Eyras E, Caceres JF. The translational landscape of the splicing factor SRSF1 and its role in mitosis. Elife. 2014:e02028. doi: 10.7554/eLife.02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Swanson CM, Sherer NM, Malim MH. SRp40 and SRp55 promote the translation of unspliced human immunodeficiency virus type 1 RNA. J Virol. 2010;84:6748–6759. doi: 10.1128/JVI.02526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu KJ, Harland RM. Inhibition of neurogenesis by SRp38, a neuroD-regulated RNA-binding protein. Development. 2005;132:1511–1523. doi: 10.1242/dev.01703. [DOI] [PubMed] [Google Scholar]
- 115.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 116.Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Caceres JF, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 117.Sterne-Weiler T, Martinez-Nunez RT, Howard JM, Cvitovik I, Katzman S, Tariq MA, Pourmand N, Sanford JR. Frac-seq reveals isoform-specific recruitment to polyribosomes. Genome Res. 2013;23:1615–1623. doi: 10.1101/gr.148585.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lemaire R, Prasad J, Kashima T, Gustafson J, Manley JL, Lafyatis R. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev. 2002;16:594–607. doi: 10.1101/gad.939502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 120.Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 121.Sato H, Hosoda N, Maquat LE. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol Cell. 2008;29:255–262. doi: 10.1016/j.molcel.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 122.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ, Liaw HJ, Myung K, Walsh CA, Gaiano N, et al. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci. 2010;13:551–558. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 127.Huang M, Rech JE, Northington SJ, Flicker PF, Mayeda A, Krainer AR, LeStourgeon WM. The C-protein tetramer binds 230 to 240 nucleotides of pre-mRNA and nucleates the assembly of 40S heterogeneous nuclear ribonucleoprotein particles. Mol Cell Biol. 1994;14:518–533. doi: 10.1128/mcb.14.1.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, Reyes A, Anders S, Luscombe NM, Ule J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 131.Schueler M, Munschauer M, Gregersen LH, Finzel A, Loewer A, Chen W, Landthaler M, Dieterich C. Differential protein occupancy profiling of the mRNA transcriptome. Genome Biol. 2014;15:R15. doi: 10.1186/gb-2014-15-1-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pelisch F, Gerez J, Druker J, Schor IE, Munoz MJ, Risso G, Petrillo E, Westman BJ, Lamond AI, Arzt E, et al. The serine/arginine-rich protein SF2/ASF regulates protein sumoylation. Proceedings of the National Academy of Sciences of the United States of America. 2010;37:16119–16124. doi: 10.1073/pnas.1004653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shimoni-Sebag A, Lebenthal-Loinger I, Zender L, Karni R. RRM1 domain of the splicing oncoprotein SRSF1 is required for MEK1-MAPK-ERK activation and cellular transformation. Carcinogenesis. 2013;11:2498–2504. doi: 10.1093/carcin/bgt247. [DOI] [PubMed] [Google Scholar]
- 134.Fregoso OI, Das S, Akerman M, Krainer AR. Splicing-factor oncoprotein SRSF1 stabilizes p53 via RPL5 and induces cellular senescence. Molecular cell. 2013;1:56–66. doi: 10.1016/j.molcel.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Das S, Anczukow O, Akerman M, Krainer AR. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell reports. 2012;2:110–117. doi: 10.1016/j.celrep.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nature structural & molecular biology. 2007;3:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kanadia RN, Clark VE, Punzo C, Trimarchi JM, Cepko CL. Temporal requirement of the alternative-splicing factor Sfrs1 for the survival of retinal neurons. Development. 2008;23:3923–3933. doi: 10.1242/dev.024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gabut M, Dejardin J, Tazi J, Soret J. The SR family proteins B52 and dASF/SF2 modulate development of the Drosophila visual system by regulating specific RNA targets. Molecular and cellular biology. 2007;28:3087–3097. doi: 10.1128/MCB.01876-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes & development. 2005;22:2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR, Jr, Ye Z, Liu F, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;1:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 141.Longman D, Johnstone IL, Caceres JF. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. The EMBO journal. 2000;7:1625–1637. doi: 10.1093/emboj/19.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Edmond V, Merdzhanova G, Gout S, Brambilla E, Gazzeri S, Eymin B. A new function of the splicing factor SRSF2 in the control of E2F1-mediated cell cycle progression in neuroendocrine lung tumors. Cell cycle. 2013;12(8):1267–1278. doi: 10.4161/cc.24363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Apostolatos A, Song S, Acosta S, Peart M, Watson JE, Bickford P, Cooper DR, Patel NA. Insulin promotes neuronal survival via the alternatively spliced protein kinase CdeltaII isoform. The Journal of biological chemistry. 2012;12:9299–9310. doi: 10.1074/jbc.M111.313080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sharma S, Liao W, Zhou X, Wong DT, Lichtenstein A. Exon 11 skipping of E-cadherin RNA downregulates its expression in head and neck cancer cells. Molecular cancer therapeutics. 2011;9:1751–1759. doi: 10.1158/1535-7163.MCT-11-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Edmond V, Brambilla C, Brambilla E, Gazzeri S, Eymin B. SRSF2 is required for sodium butyrate-mediated p21(WAF1) induction and premature senescence in human lung carcinoma cell lines. Cell cycle. 2011;12:1968–1977. doi: 10.4161/cc.10.12.15825. [DOI] [PubMed] [Google Scholar]
- 146.Xiao R, Sun Y, Ding JH, Lin S, Rose DW, Rosenfeld MG, Fu XD, Li X. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Molecular and cellular biology. 2007;15:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Meshorer E, Bryk B, Toiber D, Cohen J, Podoly E, Dori A, Soreq H. SC35 promotes sustainable stress-induced alternative splicing of neuronal acetylcholinesterase mRNA. Molecular psychiatry. 2005;11:985–997. doi: 10.1038/sj.mp.4001735. [DOI] [PubMed] [Google Scholar]
- 148.Gabut M, Mine M, Marsac C, Brivet M, Tazi J, Soret J. The SR protein SC35 is responsible for aberrant splicing of the E1alpha pyruvate dehydrogenase mRNA in a case of mental retardation with lactic acidosis. Molecular and cellular biology. 2005;8:3286–3294. doi: 10.1128/MCB.25.8.3286-3294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Crovato TE, Egebjerg J. ASF/SF2 and SC35 regulate the glutamate receptor subunit 2 alternative flip/flop splicing. FEBS letters. 2005;19:4138–4144. doi: 10.1016/j.febslet.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 150.Kim J, Park RY, Chen JK, Kim J, Jeong S, Ohn T. Splicing factor SRSF3 represses the translation of programmed cell death 4 mRNA by associating with the 5′-UTR region. Cell death and differentiation. 2014;3:481–490. doi: 10.1038/cdd.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kano S, Nishida K, Nishiyama C, Akaike Y, Kajita K, Kurokawa K, Masuda K, Kuwano Y, Tanahashi T, Rokutan K. Truncated serine/arginine-rich splicing factor 3 accelerates cell growth through up-regulating c-Jun expression. The journal of medical investigation : JMI. 2013;3–4:228–235. doi: 10.2152/jmi.60.228. [DOI] [PubMed] [Google Scholar]
- 152.Sen S, Jumaa H, Webster NJ. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nature communications. 2013;4:1336. doi: 10.1038/ncomms2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang Z, Chatterjee D, Jeon HY, Akerman M, Vander Heiden MG, Cantley LC, Krainer AR. Exon-centric regulation of pyruvate kinase M alternative splicing via mutually exclusive exons. Journal of molecular cell biology. 2012;2:79–87. doi: 10.1093/jmcb/mjr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jia R, Li C, McCoy JP, Deng CX, Zheng ZM. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. International journal of biological sciences. 2010;6(7):806–826. doi: 10.7150/ijbs.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Goncalves V, Matos P, Jordan P. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Human molecular genetics. 2009;18(19):3696–3707. doi: 10.1093/hmg/ddp317. [DOI] [PubMed] [Google Scholar]
- 156.Watanuki T, Funato H, Uchida S, Matsubara T, Kobayashi A, Wakabayashi Y, Otsuki K, Nishida A, Watanabe Y. Increased expression of splicing factor SRp20 mRNA in bipolar disorder patients. Journal of affective disorders. 2008;1–2:62–69. doi: 10.1016/j.jad.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 157.Jumaa H, Wei G, Nielsen PJ. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Current biology : CB. 1999;16:899–902. doi: 10.1016/s0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- 158.Antunes-Martins A, Mizuno K, Irvine EE, Lepicard EM, Giese KP. Sex-dependent up-regulation of two splicing factors, Psf and Srp20, during hippocampal memory formation. Learning & memory. 2007;10:693–702. doi: 10.1101/lm.640307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Corbo C, Orru S, Salvatore F. SRp20: an overview of its role in human diseases. Biochemical and biophysical research communications. 2013;1:1–5. doi: 10.1016/j.bbrc.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 160.Anko ML, Morales L, Henry I, Beyer A, Neugebauer KM. Global analysis reveals SRp20- and SRp75-specific mRNPs in cycling and neural cells. Nature structural & molecular biology. 2010;8:962–970. doi: 10.1038/nsmb.1862. [DOI] [PubMed] [Google Scholar]
- 161.Lu C, Li JY, Ge Z, Zhang L, Zhou GP. Par-4/THAP1 complex and Notch3 competitively regulated pre-mRNA splicing of CCAR1 and affected inversely the survival of T-cell acute lymphoblastic leukemia cells. Oncogene. 2013;50:5602–5613. doi: 10.1038/onc.2013.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Akula N, Barb J, Jiang X, Wendland JR, Choi KH, Sen SK, Hou L, Chen DT, Laje G, Johnson K, et al. RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Molecular psychiatry. 2014 doi: 10.1038/mp.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dance GS, Sowden MP, Cartegni L, Cooper E, Krainer AR, Smith HC. Two proteins essential for apolipoprotein B mRNA editing are expressed from a single gene through alternative splicing. The Journal of biological chemistry. 2002;15:12703–12709. doi: 10.1074/jbc.M111337200. [DOI] [PubMed] [Google Scholar]
- 164.Patel NA, Kaneko S, Apostolatos HS, Bae SS, Watson JE, Davidowitz K, Chappell DS, Birnbaum MJ, Cheng JQ, Cooper DR. Molecular and genetic studies imply Akt-mediated signaling promotes protein kinase CbetaII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. The Journal of biological chemistry. 2005;14:14302–14309. doi: 10.1074/jbc.M411485200. [DOI] [PubMed] [Google Scholar]
- 165.Fic W, Juge F, Soret J, Tazi J. Eye development under the control of SRp55/B52-mediated alternative splicing of eyeless. PloS one. 2007;2:e253. doi: 10.1371/journal.pone.0000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hara H, Takeda T, Yamamoto N, Furuya K, Hirose K, Kamiya T, Adachi T. Zinc-induced modulation of SRSF6 activity alters Bim splicing to promote generation of the most potent apoptotic isoform BimS. The FEBS journal. 2013;14:3313–3327. doi: 10.1111/febs.12318. [DOI] [PubMed] [Google Scholar]
- 167.Jensen MA, Wilkinson JE, Krainer AR. Splicing factor SRSF6 promotes hyperplasia of sensitized skin. Nature structural & molecular biology. 2014;2:189–197. doi: 10.1038/nsmb.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jin W, Cote GJ. Enhancer-dependent splicing of FGFR1 alpha-exon is repressed by RNA interference-mediated down-regulation of SRp55. Cancer research. 2004;24:8901–8905. doi: 10.1158/0008-5472.CAN-04-0716. [DOI] [PubMed] [Google Scholar]
- 169.Juge F, Fernando C, Fic W, Tazi J. The SR protein B52/SRp55 is required for DNA topoisomerase I recruitment to chromatin, mRNA release and transcription shutdown. PLoS genetics. 2010;9:e1001124. doi: 10.1371/journal.pgen.1001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lal S, Allan A, Markovic D, Walker R, Macartney J, Europe-Finner N, Tyson-Capper A, Grammatopoulos DK. Estrogen alters the splicing of type 1 corticotropin-releasing hormone receptor in breast cancer cells. Science signaling. 2013;282:ra53. doi: 10.1126/scisignal.2003926. [DOI] [PubMed] [Google Scholar]
- 171.Ring HZ, Lis JT. The SR protein B52/SRp55 is essential for Drosophila development. Molecular and cellular biology. 1994;11:7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shukla S, Dirksen WP, Joyce KM, Le Guiner-Blanvillain C, Breathnach R, Fisher SA. TIA proteins are necessary but not sufficient for the tissue-specific splicing of the myosin phosphatase targeting subunit 1. The Journal of biological chemistry. 2004;14:13668–13676. doi: 10.1074/jbc.M314138200. [DOI] [PubMed] [Google Scholar]
- 173.Tran Q, Coleman TP, Roesser JR. Human transformer 2beta and SRp55 interact with a calcitonin-specific splice enhancer. Biochimica et biophysica acta. 2003;2:141–152. doi: 10.1016/s0167-4781(02)00600-0. [DOI] [PubMed] [Google Scholar]
- 174.Tran Q, Roesser JR. SRp55 is a regulator of calcitonin/CGRP alternative RNA splicing. Biochemistry. 2003;4:951–957. doi: 10.1021/bi026753a. [DOI] [PubMed] [Google Scholar]
- 175.Ramchatesingh J, Zahler AM, Neugebauer KM, Roth MB, Cooper TA. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Molecular and cellular biology. 1995;9:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lopez-Mejia IC, Vautrot V, De Toledo M, Behm-Ansmant I, Bourgeois CF, Navarro CL, Osorio FG, Freije JM, Stevenin J, De Sandre-Giovannoli A, et al. A conserved splicing mechanism of the LMNA gene controls premature aging. Human molecular genetics. 2011;23:4540–4555. doi: 10.1093/hmg/ddr385. [DOI] [PubMed] [Google Scholar]
- 177.Valente ST, Gilmartin GM, Venkatarama K, Arriagada G, Goff SP. HIV-1 mRNA 3′ end processing is distinctively regulated by eIF3f, CDK11, and splice factor 9G8. Molecular cell. 2009;2:279–289. doi: 10.1016/j.molcel.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jain A, Wordinger RJ, Yorio T, Clark AF. Spliceosome protein (SRp) regulation of glucocorticoid receptor isoforms and glucocorticoid response in human trabecular meshwork cells. Investigative ophthalmology & visual science. 2012;2:857–866. doi: 10.1167/iovs.11-8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Xu Q, Leung DY, Kisich KO. Serine-arginine-rich protein p30 directs alternative splicing of glucocorticoid receptor pre-mRNA to glucocorticoid receptor beta in neutrophils. The Journal of biological chemistry. 2003;29:27112–27118. doi: 10.1074/jbc.M300824200. [DOI] [PubMed] [Google Scholar]
- 180.Fu Y, Huang B, Shi Z, Han J, Wang Y, Huangfu J, Wu W. SRSF1 and SRSF9 RNA binding proteins promote Wnt signalling-mediated tumorigenesis by enhancing beta-catenin biosynthesis. EMBO molecular medicine. 2013;5:737–750. doi: 10.1002/emmm.201202218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cloutier P, Toutant J, Shkreta L, Goekjian S, Revil T, Chabot B. Antagonistic effects of the SRp30c protein and cryptic 5′ splice sites on the alternative splicing of the apoptotic regulator Bcl-x. The Journal of biological chemistry. 2008;31:21315–21324. doi: 10.1074/jbc.M800353200. [DOI] [PubMed] [Google Scholar]
- 182.Shi Y, Manley JL. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Molecular cell. 2007;1:79–90. doi: 10.1016/j.molcel.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 183.Shi Y, Nishida K, Campigli Di Giammartino D, Manley JL. Heat shock-induced SRSF10 dephosphorylation displays thermotolerance mediated by Hsp27. Molecular and cellular biology. 2011;3:458–465. doi: 10.1128/MCB.01123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ling IF, Estus S. Role of SFRS13A in low-density lipoprotein receptor splicing. Human mutation. 2010;6:702–709. doi: 10.1002/humu.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Komatsu M, Kominami E, Arahata K, Tsukahara T. Cloning and characterization of two neural-salient serine/arginine-rich (NSSR) proteins involved in the regulation of alternative splicing in neurones. Genes to cells : devoted to molecular & cellular mechanisms. 1999;10:593–606. doi: 10.1046/j.1365-2443.1999.00286.x. [DOI] [PubMed] [Google Scholar]
- 186.Feng Y, Valley MT, Lazar J, Yang AL, Bronson RT, Firestein S, Coetzee WA, Manley JL. SRp38 regulates alternative splicing and is required for Ca(2+) handling in the embryonic heart. Developmental cell. 2009;4:528–538. doi: 10.1016/j.devcel.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Straub T, Grue P, Uhse A, Lisby M, Knudsen BR, Tange TO, Westergaard O, Boege F. The RNA-splicing factor PSF/p54 controls DNA-topoisomerase I activity by a direct interaction. The Journal of biological chemistry. 1998;41:26261–26264. doi: 10.1074/jbc.273.41.26261. [DOI] [PubMed] [Google Scholar]
- 188.Sakashita E, Tatsumi S, Werner D, Endo H, Mayeda A. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Molecular and cellular biology. 2004;3:1174–1187. doi: 10.1128/MCB.24.3.1174-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cowper AE, Caceres JF, Mayeda A, Screaton GR. Serine-arginine (SR) protein-like factors that antagonize authentic SR proteins and regulate alternative splicing. The Journal of biological chemistry. 2001;52:48908–48914. doi: 10.1074/jbc.M103967200. [DOI] [PubMed] [Google Scholar]
- 190.Soret J, Gattoni R, Guyon C, Sureau A, Popielarz M, Le Rouzic E, Dumon S, Apiou F, Dutrillaux B, Voss H, Ansorge W, Stevenin J, Perbal B. Characterization of SRp46, a novel human SR splicing factor encoded by a PR264/SC35 retropseudogene. Molecular and cellular biology. 1998;8:4924–34. doi: 10.1128/mcb.18.8.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]