Abstract
As receptor-targeted therapeutics become increasingly used in clinical oncology, the ability to quantify protein expression and pharmacokinetics in vivo is imperative to ensure successful individualized treatment plans. Current standards for receptor analysis are performed on extracted tissues. These measurements are static and often physiologically irrelevant, therefore, only a partial picture of available receptors for drug targeting in vivo is provided. Until recently, in vivo measurements were limited by the inability to separate delivery, binding, and retention effects but this can be circumvented by a dual-tracer approach for referencing the detected signal. We hypothesized that in vivo receptor concentration imaging (RCI) would be superior to ex vivo immunohistochemistry. Using multiple xenograft tumor models with varying epidermal growth factor receptor (EGFR) expression, we determined the EGFR concentration in each model using a novel targeted agent (anti-EGFR affibody-IRDye800CW conjugate) along with a simultaneously delivered reference agent (control affibody-IRDye680RD conjugate). The RCI-calculated in vivo receptor concentration was strongly correlated with ex vivo pathologist-scored immunohistochemistry and computer-quantified ex vivo immunofluorescence. In contrast, no correlation was observed with ex vivo Western blot or in vitro flow cytometry assays. Overall, our results argue that in vivo RCI provides a robust measure of receptor expression equivalent to ex vivo immuno-staining, with implications for use in non-invasive monitoring of therapy or therapeutic guidance during surgery.
Keywords: receptor concentration imaging, epidermal growth factor receptor, xenograft, fluorescence, infrared, carcinoma
Introduction
The ability to quantitatively measure the in vivo expression of extracellular proteins as well as the respective pharmacokinetics and receptor occupancy of any ligand-of-interest (i.e., therapeutics or imaging agents) in clinical oncology has long been recognized as an important key for personalized medicine including tumor detection, treatment stratification and therapeutic monitoring (1–5). The current practices for analyzing protein expression clinically and pre-clinically are performed on ex vivo tissues or in vitro cell culture; however, these tissue samples do not accurately represent the complexity of the in vivo environment and can grossly misinterpret the available receptor concentration for binding. Attempts at determining in vivo receptor concentration using a single targeted tracer have been studied, but are limited by the inability to distinguish actual binding events from vascular delivery and tumor retention. This paper examines the hypothesis that in vivo receptor concentration imaging (RCI) shows the accessible concentration of extracellular receptors when complemented with an appropriate reference tracer to negate the dominant effects of drug transport in vivo (6); and this value directly correlates to standard ex vivo tissue processing methods. This hypothesis may seem obvious, yet the concept of applying immunohistochemistry in vivo is largely undeveloped, and not validated in a panel of tumor lines. Moreover, the ability to dynamically quantify expression in vivo could be enormously important for evaluating the individual targeting of biological therapies and changes in multiple receptors during the course of a therapy.
Clinically, the most widely used method of determining the molecular expression in a tumor is biopsy followed by immunohistochemical (IHC) staining. Although IHC has been shown innumerable times to be correlative to the protein expression in a tumor, it suffers from a limited sampling of a potentially highly heterogeneous tissue, and is time consuming and invasive to the patient. Additionally, receptor staining in IHC is performed with all protein exposed on the cut surfaces, which is different than the in vivo situation where drug access to extracellular proteins is via interstitial transport mechanisms. In vivo, the available receptors for binding can potentially be fewer than those available on an ex vivo slide. In vivo imaging with positron emission tomography (PET) and single-photon emission computed tomography (SPECT) are two techniques that can be used to image the metabolic and molecular status of cancerous tissues; however, they cannot be interpreted as a direct receptor concentration image without accounting for the transport kinetics to the tumor, and so routinely are just calibrated into a standard uptake value (SUV).
Pre-clinically, molecular expression is most often measured by IHC, immunofluorescence (IF), Western blot, and Flow Cytometry (Figure 1). The most reliable and utilized immunostaining techniques, IHC and IF, can be used to compare relative expression when samples are stained simultaneously and under identical conditions but quantifying these techniques is limited by significant inter-operator errors, a low sensitivity range in pathologist scoring systems, and automatic analysis that relies on the linear optical properties of the stain (7–9). Western blot can be used to determine total relative protein expression in bulk samples, representing both intracellular and extracellular protein. However, detection is limited by optical density (OD) range in which absorption is linear and so is rarely used as a true quantitative measure but rather to monitor relative differences or changes in protein expression (10). Radiometric and fluorescence detection techniques for Western blots have been developed with greater sensitivity but are not as commonly used and can also suffer from signal saturation (10, 11). Flow cytometry is an extremely sensitive quantitative technique (using calibration methods that employ molecules of equivalent soluble fluorochrome, MESF, beads (12)) but is most often applied to in vitro cells or intracellular protein expression measurements in ex vivo tissues, as extracellular proteins can be compromised during cellular retrieval from in vivo samples. In addition, techniques studying gene expression using techniques such as fluorescent in situ hybridization (FISH) can determine whether a mutated gene is present and in some cases (e.g., Her-2/neu mutations in breast cancer) provide a measure of gene amplification that correlates to patient prognosis (13). However, protein expression is not always correlated with gene expression, especially in moderately expressing tumors, because multiple copies of a single protein can be made from a single mRNA strand or the mutation of interest is not causing the overexpression (13, 14). It is important to note that all of these techniques have a varying degree of tissue integrity preservation and in vivo relevance, which can be essential for predicting the accessible and therapeutically relevant receptor population.
Figure 1.

The standard methods of quantifying protein expression in tissue and cell samples are compared in terms of type of sample preparation, detection technique and sensitivity range. In each row, the relevance to in vivo tissues and tissue integrity decreases from left to right. IHC = immunohistochemistry, IF = immunofluoresence, EGFR = epidermal growth factor receptor, AR = amplification ratio, and OD = optical density. The sensitivity ranges for IHC and IF are reported as pathologist score (top) and computer analyzed (bottom).
Molecular imaging and quantification of protein expression has had a large impact on pre-clinical drug design and development (4, 5, 15), validation and study of xenograft cancer models (16–18), and advancement of surgical resection techniques (19, 20). In vivo targeted fluorescent imaging in both clinical and pre-clinical oncology settings has been limited in part by the extended times (24–48 hours) required to gain adequate contrast between tumor and normal tissues. Activatable fluorescent molecules are finding great success in tumor and metastases detection but they are generally not quantitative and like many other techniques can suffer from saturating signals (21). Receptor concentration imaging in PET and SPECT for applications in neurobiology has been successfully applied both clinically and pre-clinically for decades (3, 15, 17). However, these techniques do not simply transfer to oncology, even for brain tumors, because they rely on the simultaneous imaging of a single tracer in two structurally identical tissues where one tissue (the “reference tissue”) is devoid of any receptor (22). The hallmark of cancers is that they can be both physically and metabolically heterogeneous as compared to primary tissue, and so this two-tissue model fails to have value (23, 24). Multiple tracer imaging can be feasibly done with nuclear medicine methods, yet has been rarely attempted owing to the complexity of the signal and the complications of using multiple radiotracers in vivo.
The RCI method for fluorescence imaging of cancerous tissue overcomes the time delays and poor contrast of imaging a single fluorescent tracer and builds upon the receptor concentration imaging developed in the PET and SPECT neurobiology field (6). The RCI method enables in vivo quantification of receptor concentration within one-hour of fluorescent tracer administration by “normalizing” the uptake of the targeted tracer by the uptake of a second, untargeted tracer – that has similar physical and pharmacokinetic characteristics to the targeted tracer – to account and correct for nonspecific uptake and delivery kinetics of the targeted tracer.
This study employed clinically relevant targeted and untargeted Affibody® molecules (6 kDa antibody mimetics) labeled with near infrared fluorophores to determine the in vivo concentration of five tumor models known to express varying levels of epidermal growth factor receptor (EGFR). The in vivo receptor concentrations determined by RCI were then compared to standard clinical and pre-clinical methods of protein concentration determination: pathologist scored IHC, computer analyzed IF, Western blot, and flow cytometry. This study used EGFR as the model receptor system, but this technique is extendable to any extracellular protein receptor that has known and validated ligand or small molecule targeting agents.
Materials and Methods
Cell lines and culture methods
Four cell lines of varying origin and known EGFR expression where cultured for receptor quantification with in vitro flow cytometry and in vivo implantation: A431, a human epidermoid carcinoma (ATCC, Manassas, VA, USA) expresses a very high level of EGFR; AsPC-1, a human pancreatic adenocarcinoma (from author T. Hasan) and U251, a human neuronal glioblastoma (from Dr. Israel at Dartmouth College) have moderate levels of EGFR expression; and 9L (from Dr. Wheeler at Wake Forest University), a rat gliosarcoma known to be EGFR negative. Cell lines were cultured in either DMEM (A431, U251 and 9L) or RPMI (AsPC-1) with 10% fetal RPMI 10% (v/v) fetal bovine serum and 100 IU/mL penicillin-streptomycin.
Xenograft murine models
All animal procedures were conducted according to a protocol approved by the Dartmouth Institutional Animal Care and Use Committee (IACUC). A total of 30 female athymic nude mice were used with 6 mice per tumor group. The five tumor groups included subcutaneously implanted A431, AsPC-1, U251, and 9L, in addition to orthotopically implanted AsPC-1. For the subcutaneous tumor models, 1×106 cells were implanted in the right flank of the mouse using 50 µL of a 1:1 mixture of Matrigel (BD Biosciences, San Jose, CA, USA) and complete cell culture media. The orthotopic implantation of the AsPC-1 cells has been described elsewhere.(25) Briefly, 1×106 cells in 50 µL the 1:1 Matrigel to cell culture medium mixture is injected into the tail of the pancreas via a small incision in the mouse abdomen. All tumors were used for imaging and analysis when they reached 100–150 mm3 in volume.
Fluorescent labeling of anti-EGFR affibody proteins
The anti-EGFR affibody imaging agent (Affibody, Solna, Sweden) was labeled with IRDye800CW maleimide (LI-COR Biosciences, Lincoln, NE, USA) and used as the targeted imaging agent. The negative control affibody imaging agent was labeled with IRDye680RS maleimide (LI-COR Biosciences) and used as the non-targeted reference imaging agent. The affibody proteins were prepared for labeling as described by the manufacturer’s instructions and using a Pierce Polyacrylamide Desalting Column (6K MWCO, Thermo Scientific, Waltham, MA) to purify fluorescent labeling. The prepared affibody proteins were subsequently labeled with the maleimide formulation of fluorescent dyes according to LI-COR Biosciences instructions with the modification that the free dye was separated from protein using the above described desalting column, and the fluorescently labeled affibody proteins were concentrated with using a spin column (Vivaspin™ 2, MWCO 3K, GE Healthcare Life Sciences, Piscataway, NJ, USA) according to manufacturer’s instructions.
In vivo Receptor Concentration Imaging (RCI)
All animal procedures were conducted according to a protocol approved by the Dartmouth Institutional Animal Care and Use Committee (IACUC). The mice were prepared for imaging with an intraperatoneal injection ketamine-xylazine (100 mg/kg:10 mg/kg) and when a surgical plane of anesthesia was obtained (confirmed by toe-pinch), superficial tissue was removed to expose the tumor and thigh muscle. Each mouse was placed with the tumor and muscle facing down onto a glass side, to which the mice were loosely secured with surgical tape to reduce movement during injection. The mice were positioned on an Odyssey Scanner (LI-COR Biosciences) and pre-injection images were collected in both the 700 nm (excitation 685 nm, emission 700 nm LP) and 800 nm (excitation 785 nm, emission 800 nm LP) channels. The mixture of IRDye 800CW-AntiEGFR Affibody (0.2 nmol) and IRDye 680RD-control Affibody (0.2 nmol) was administered via tail vein injection and scanning was resumed with images taken every minute for the first 15 min and then every 2–5 min for a total of 40 min.
Estimates of EGFR binding potential were determined on a pixel-by-pixel basis by applying a multiple-time-point dual-tracer kinetic model based on a simplified reference tissue model (22) to the temporal uptake curves of the targeted and untargeted tracers. The model, which has been described in depth elsewhere (6, 26), was fitted to the data using the optimization algorithm, lsqcurvefit() in MATLAB (Natick, MA). Differences in the plasma pharmacokinetics were corrected using the deconvolution correction described previously (26). A region-of-interest was manually drawn around each tumor in the resulting binding potential parametric maps to calculate the mean tumor binding potential (BP) for each xenograft tumor, where BP is equal to the concentration of receptors available for binding multiplied by the binding affinity (kA) of the receptor-ligand pair (27). The mean receptor concentration of each xenograft tumor was determined by multiplying the BP by the dissociation constant (kD = 1/kA) determined by flow cytometry for each cell line (Supplementary Data).
Immunohistochemistry (ICH, ex vivo)
All tissue was fixed in 10% buffered formalin (Biochemical Science Inc, Swedesboro, NJ). Tissue sections were cut at (4 µm), mounted on Leica Bond Plus Slides Cat # 00270) and air-dried at room temperature. Using the automated protocol of the Leica Bond Rx Automated Stainer (Leica Products/Equipment, Leica Microsystems, Inc., Buffalo Groove, IL), the slides were baked for 30 minutes, and dewaxed with Leica Bond Dewax solution (Cat #AR9222). The antigen retrieval was Bond Epitope Retrieval 2 (Cat #ar9640), carried out in a pH 9.0 solution for 20 minutes. The EGFR primary antibody dilution was 1:300 for 15 minutes (Abcam Cat # ab52894; Abcam Inc., 1 Kendall Sq Suite B2304 Cambridge, MA 02139-1517). Primary antibody binding was visualized using Leica Bond Refine Detection kit (Cat # DS9800) with a diaminobenzidine (DAB) chromogen and a hematoxylin counterstain. The tissue sections were scored by a pathologist (author W. Wells) according to the following criteria: 0 = no staining; 1+ = partial membranous staining around cells; 2+ = weaker membranous staining completely around cells; and 3+ = strong membranous staining completely around cells.
Immunofluorescence (IF, ex vivo)
Half of the extracted tumor was covered in optimum cutting temperature (OCT) compound (TissueTek®, Torrance, CA, USA) and flash frozen in a mixture of methylbutane and dry ice. The tissues were prepared in 10 µm frozen sections, with three sections per slide, each 100 µm apart. Slides were fixed in pre-cooled (-20°C) 1:1 acetone:methanol for 15 minutes at −20°C, air dried for 30 min, washed two times (5min/wash) in PBS, and then blocked with prepared blocking solution (5% FBS, 1% BSA, 0.3% Triton™ X-100 in PBS) for 30 min at room temperature. Slides were then incubated overnight at 4°C with Anti-EGFR (1:50, Cell Signaling Technologies, Danvers, MA, USA) in the blocking solution. Slides were washed 3 times for 5 min in PBS and incubated for 1 h with Goat Anti-Rabbit Alexa Fluor 555 secondary (1:400, Invitrogen, Grand Island, NY, USA) in blocking solution and prior to a further 3 washes PBS. Slides were prepared for imaging by adding a coverslip with mounting medium (SlowFad Gold with DAPI, Invitrogen). Ten fluorescent images were obtained per tumor line using identical camera and microscope settings (Olympus BX51 microscope fitted with a Q-Color3 CCD camera). Fluorescent images using U-MF2 and U-MWIG2 filter cubes (Chroma, Bellows falls, VT), for DAPI and Alexa Fluor 555, respectively, collected for each tumor line were read into, and analyzed identically using the NIH freeware ImageJ (http://rsb.info.nih.gov/ij/) by measuring the average pixel intensity of the entire image using pre-determined thresholding parameters.
Western Blot (ex vivo)
Tissue was flash frozen in liquid nitrogen, pulverized with a morter and pestle, and solubilized in RIPA lysis buffer (Thermo Scientific, Rockford, IL), 1-mM phenylmethylsulfonyl fluoride, and Complete Protease Inhibitor (Roche, Madison, WI). Proteins were subjected to SDS-polyacrylamide gel electrophoresis and transferred to PVDF. Membranes were blocked with 5% milk/PBS/0.1% Tween (Milk/PBT) for 1 hour, incubated overnight with anti-EGFR antibody (1:1000, Santa Cruz Biotechnologies, Dallas, TX) or Anti-ERK2 (1:5000, Cell Signaling Technologies), washed 3 times in PBT, incubated for 1 hour with Goat Anti-Rabbit HRP-conjugated secondary (BioRad, Hercules, CA), and washed 3 times in PBT. Bound antibodies were visualized using enhanced chemiluminescence (Pierce, Rockford, IL). Densitrometry was performed and normalized to β-actin 1 for equal loading of lanes (Supplementary Data).
Flow cytometry (in vitro)
The in vitro number of receptors per cell for each tumor cell line was determined as described previously (6). Briefly, when 80% confluence was reached, each cell line was labeled with 4 mg/mL of EGF Biotin (Molecular Probes, Invitrogen, Camarillo, CA, USA), and secondarily labeled with a 1:25 dilution of Cy5-Streptavidin (Invitrogen). Control cells were stained only with the Cy-5 Streptavidin to account for autofluorescence and non-specific staining. Quantification was performed by comparison to Quantum Cy5 MESF beads, used as described by the manufacturer (Bangs Laboratory, Fisher, IN, USA). Flow cytometric data was acquired with a FACSCalibur analysis system equipped with a FACStation, and Cell Quest Acquisition software (Becton Dickinson, San Jose, CA, USA). The average fluorescence measured for the control cells was subtracted from the EGF stained cells and the number of receptors per cell was calculated assuming one molecule of biotin per EGF and three Cy5 fluorophores per Streptavidin molelecule, as specified by Invitrogen.
Statistical analysis
All statistical analyses and linear regressions were performed using OriginPro 8 (OriginLab, Northhampton, MA). The in vivo receptor concentration for each tumor group and control muscle tissue were compared using a one-way ANOVA with Bonferroni correction and the tissue-type as the between-subjects factor. Linear regressions were employed to compare the in vivo receptor concentration determined with affibody molecules to pathologist determined immunohistochemistry scores, computer-analyzed immunofluorescence image intensities, relative Western blot staining intensities and receptor concentration per cell determined by flow cytometry. The results of the linear regressions are reported as Pearson’s Product Correlation Coefficient (r), ANOVA for regression (p) with Bonferroni correction, slope of the line (m), and the y-intercept (b). Statistical significance was based on p < 0.05 and all data are reported in mean ± SD.
Results
The targeted anti-EGFR Affibody and untargeted Affibody imaging agent control display similar uptake trends at short time periods after injection (Figure 2, columns 1 and 2, respectively) and fluorescence intensity in the tumor region does not reflect EGFR expression levels (Figure 2). The predicted trend in EGFR concentration (A431 > AsPC-1(Or) > AsPC-1(SQ) > U251 > 9L) was validated by the RCI method as seen in Figure 2, column 3 and correlates with the previously determined concentrations using the native human EGF ligand (Supplemental Data). The kinetic curves of the targeted and untargeted tracers (Figure 2, column 4) display greater separation in tissues that have high EGFR concentrations and almost identical curves in the 9L model, which is an EGFR negative tumor (6, 28). The calculated in vivo receptor concentration (Figure 3) in A431, AsPC-1(Or), AsPC-1(SQ) and U251 tumor models were statistically significant from the control muscle tissue (p < 0.05), known to lack endogenous EGFR, as well as the EGFR negative 9L tumor (p < 0.05). In addition, the orthotopic and subcutaneous AsPC-1 tumor models were not significantly different from each other.
Figure 2.

The concentration of in vivo EGFR is determined by RCI in five different tumor models (rows). The tumor lines are presented in the order of predicted EGFR concentration (fading purple triangle on far left). The first column shows the fluorescence uptake of the EGFR targeted tracer in the tumor (boundary designated by white dashed line) and surrounding tissues at 40 minutes post-injection. The second column is the corresponding untargeted tracer uptake in the same tissue at the same time point. The third column presents maps of the EGFR concentration (nM), determined using the RCI method. The forth column show the time trajectories of the average fluorescence within the tumor region of the targeted (red) and untargeted (green) tracers. The curves are normalized to the first time point.
Figure 3.

A box plot of the RCI determined in vivo receptor concentration for each tumor model is shown. Each box represents the 25–75% percentile range, the filled square is the mean, the solid line dividing the box is the median, and the open circles are the individual tumors within each group. The A431, AsPC-1(Or), AsPC-1(SQ) and U251 tumor models were found to be statistically different than muscle (p < 0.05, denoted by *) and the EGFR-negative 9L tumor (p < 0.05, denoted by #). The 9L tumor model was not significantly different than the muscle.
The anti-EGFR Affibody in vivo receptor concentration was compared to standard techniques of receptor concentration determination including ex vivo IHC, IF, and Western blots in addition to in vitro flow cytometry (Figure 4). The in vivo receptor concentration had a strong linear correlation to the EGFR concentration determined by ex vivo IHC (r = 0.69, p = 2×10−5, m = 0.29 ± 0.06, b = 0.8 ± 0.2) and IF (r = 0.62, p = 5×10−4, m = 1.0 ± 0.2, b = 2.1 ± 0.9). There was no significant correlation between in vivo receptor concentration and ex vivo Western blot (r = 0.35, p = 0.08, m = 0.04 ± 0.03, b = 0.06 ± 0.08), or in vivo receptor concentration and in vitro flow cytometry (r = 0.43, p = 0.017, m = 9×104 ± 3×104, intercept = 4×104 ± 1.3×105,) methods of receptor concentration determination. The IHC and IF EGFR staining trends for each tumor model are shown in Figure 5.
Figure 4.

The receptor concentration determined using anti-EGFR affibody RCI is compared to four standard methods used to determine protein concentration: A) pathologist scored ex vivo immunohistochemistry (IHC, r = 0.69, p = 2×10−5, m = 0.29 ± 0.06, b = 0.8 ± 0.2), B) ex vivo immunofluorescence (r = 0.62, p = 5×10−4, m = 1.0 ± 0.2, b = 2.1 ± 0.9), C) ex vivo Western blot (r = 0.35, p = 0.08, m = 0.04 ± 0.03, b = 0.06 ± 0.08), and D) in vitro flow cytometry (r = 0.43, p = 0.017, m = 9×104 ± 3×104, intercept = 4×104 ± 1.3×105). The linear regression (solid line) is displayed as well as the 95% confidence bands (dashed lines) and the 95% prediction bands (dotted lines).
Figure 5.
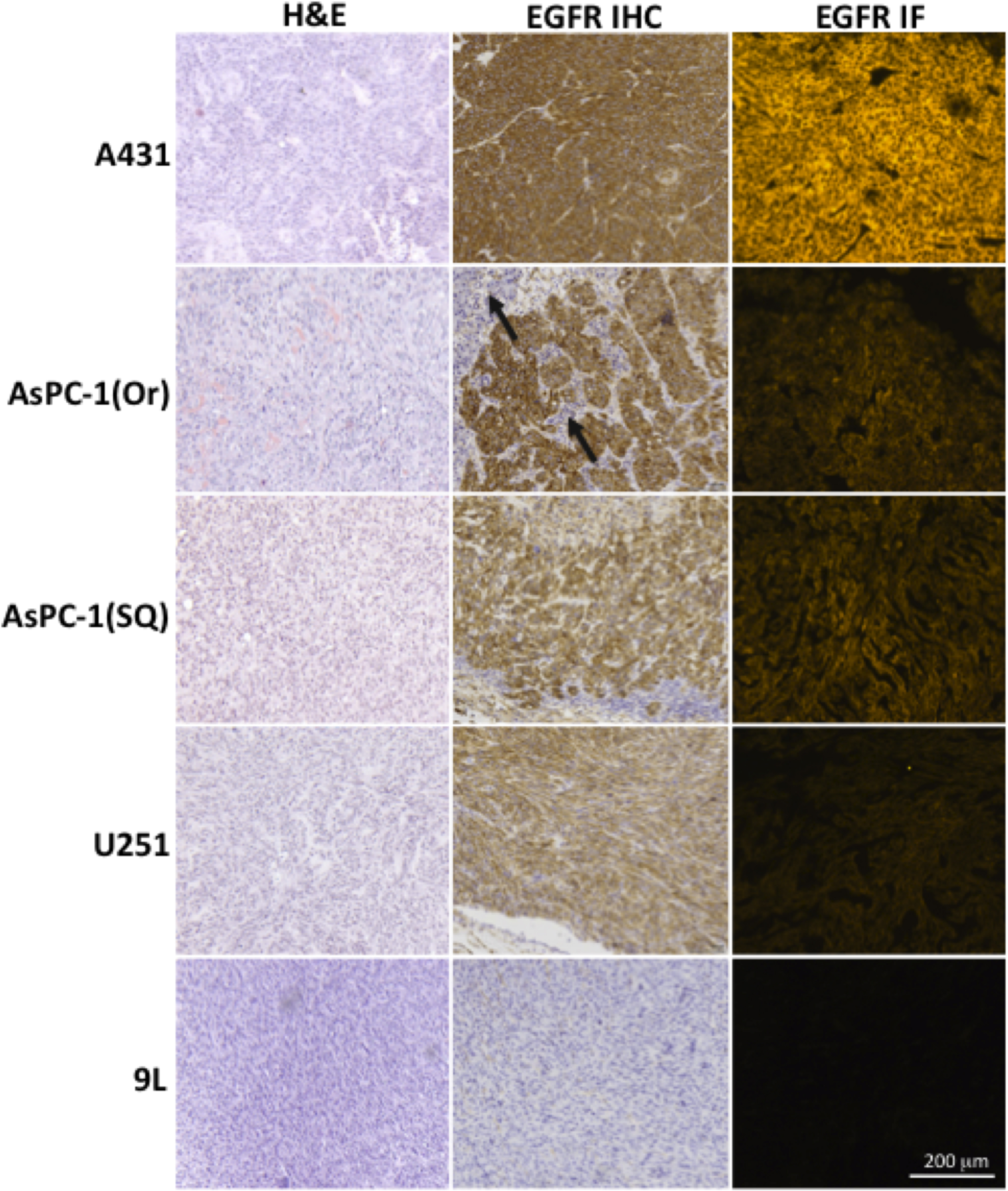
Ex vivo tissue sections of the five tumor models with varying expression of EGFR studied. Column 1: hemotoxylin and eosin (H&E, column 1), Column 2: EGFR immunohistochemistry (IHC, column 2), and Column 3: EGFR immunofluorescence (IF, column 1). The images are ordered from highest expressing (top row) to lowest expressing (bottom row). Note in the AsPC-1(Or) model, the tumor cells are interspersed and surrounded by normal pancreas acinar cells that are EGFR negative (arrow).
Discussion
The successful delivery of molecular targeted agents to tumors depends on many biological and physiological factors, including hemodynamics, vascular permeability, density and diffusivity of the interstitial tissue, and non-specific binding. These parameters can not only vary greatly from one region of a tumor to another but individual patients with tumors of the same origin can have vastly different biological and physiological parameters. In vivo single tracer studies of receptor concentration have shown some success in correlating EGFR, or EGFR family receptors, concentrations (29–32); however, these studies look at target accumulation after a minimum of 24 hrs where tumor to normal tissue contrast is high but the enhanced permability and retention effect dominates tracer accumulation. In fact at these late time points, high contrast of non-targeted tracers can be observed (33); thus, imaging with a single tracer lacks the ability to distinguish true binding events (available receptor concentrations) from the physiological characteristics of the tumor. The currently accepted methods of protein quantification and assessment in tumors occur either ex vivo or in vitro and as such, these dynamic, heterogeneous biological factors are ignored or it is difficult to obtain a global sample of the tissue.
The in vivo RCI method uses targeted and non-targeted tracers to simultaneously experience the same hemodynamics, vascular permeability and nonspecific uptake in the tissue of interest (6) and as such, overcomes the difficulties of the ex vivo and in vitro methods. The binding activity of the targeted tracer is used to elucidate the concentration of the receptor of interest using the RCI model. Imaging for the RCI model is performed immediately after injection and therefore can separate true binding events from the physiological characteristics of the tumor. In fact, the fluorescent contrast observed for either of the imaging agents (Figure 2, columns 1 and 2) is negligible at these time points and only when the RCI model is applied, does significant contrast occur (Figure 2, column 3). Previously, the BP was determined for EGFR using the native human EGF ligand (6), and here is shown to have a strong linear correlation to the BP determined here with the anti-EGFR targeting agent (Supplementary Data). This is important for two reasons: 1) anti-EGFR affibody is a clinically relevant inhibitor of EGFR, whereas EGF is a stimulant; and 2) it demonstrates that the RCI model first presented by Tichauer et al. (2012) holds for various tracers.
The in vivo receptor concentration of each of the five tumor models (A431, AsPC-1(Or), AsPC-1(SQ), U251 and 9L) was determined and mapped using the RCI method and an anti-EGFR and control affibody imaging pair (Figure 1). The in vivo receptor concentration for each tumor model was compared to standard methods of measuring receptor concentration in clinical and pre-clinical oncology using excised tissues or cells grown in culture (IHC, IF, Western blot, and flow cytometry; Figure 4). The RCI calculated receptor concentration had a high linear correlation and statistical significance with IHC scored by a trained pathologist and digitally analyzed IF. These results are a key indication that in vivo RCI can determine receptor concentration with very high accuracy and is equivalent to ex vivo methods of immunohistochemistry. In addition, the RCI method has the distinct advantage of imaging the tumor in its entirety, unlike IHC and IF that require small biopsy samples or thinly sliced tissue sections, and also has the potential to be imaged repeatedly without the necessity of excising tissue multiple times.
Although there was a moderate linear correlation determined using Pearson’s Product Correlation Coefficient (defined as 0.3 > r > 0.5) between the receptor concentrations determined by ex vivo Western blot and in vitro flow cytometry with in vivo RCI, the correlation was not statistically significant. The largest discrepancy in both techniques can be attributed to the large overestimation of EGFR receptors in A431, a tumor line that expresses an order of magnitude more EGFR than the others tested. There are several possible explanations for this: 1) there was no interfering biological physiology to limit deliver of molecular targets, such as tumor interstitium, restricted vascular flow or permeability, or varying cellular density; or 2) both ex vivo Western blot and in vitro flow cytometry use receptor saturating concentrations for EGFR labeling, thus providing full and uninhibited access to all receptors (intracellular and extracellular). The inconsistencies between ex vivo Western blot and the in vivo uptake of EGFR targeted agents in A431 tumors have been observed before (6, 31). Both Tichauer et al (6) and Aerts et al (31) found that A431 tumors had significantly less delivery of an EGFR targeted agent in vivo than was expected from ex vivo or in vitro quantification of receptor concentration. In fact, delivery of a single targeted agent in A431 tumors was far lower in than tumor lines that were known to produce as much as an order of magnitude less EGFR. Similarly, McLarty et al (32) have demonstrated previously in breast cancer cell lines that highly overexpress HER2, that retention of a single agent is less than in cell lines that moderately overexpress HER2. Aerts et al. (31) attributes this discrepancy to physiological parameters, such as vascular density and permeability, as well as cellular viability, which is likely true when considering the retention of an any single agent in a tumor (targeted or non-targeted). However, the RCI model accounts for these factors, suggesting that indeed there is a non-proportional discrepancy between in vivo receptor concentration (RCI, IHC and IF) and total protein content (Western blot and flow cytometry) with respect to the concentration of receptors physiologically available for targeted binding.
The in vivo receptor concentration determined for the five tumor models demonstrated expected trends when compared to the control tissues: muscle and 9L tumor model. The receptor concentration of the 9L tumor model was not significantly different from the muscle (Figure 3), which was expected as the tumor line has been shown to have no apparent EGFR expression (6, 28) and was confirmed here (Figures 3 and 5). The in vivo receptor concentrations of the EGFR positive tumor models were all significantly different than the control muscle tissue and the 9L tumor line (p < 0.05, Figure 3). It was hypothesized that EGFR expression would be different between the subcutaneous and orthotopic tumor models owing to the variations in physical environments, blood supply and distribution, and cellular content (34). Interestingly, the receptor concentration of A431 was not significantly different from AsPC-1(Or), which has been determined previously using EGF as the targeted tracer (6). Although the available concentration determined by the RCI method of EGFR in the AsPC-1(SQ) model was lower than that of the AsPC-1(Or) model, the difference was not significant: indeed the two tumor models both received a pathologist score of 2+. However, the EGFR staining patterns were visually different, as shown in Figure 5, with the AsPC-1(Or) staining being slightly darker. This staining difference is potentially attributable to a combination of tumor and normal pancreas tissue in the orthotopic tissues, and thus our calculated receptor concentration could also include benign EGFR negative acinar cells. The significant contrast observed between the AsPC-1 tumor and the normal pancreas, illustrates the potential significance of this technique for surgical tissue resection or tumor detection in vivo.
Here, the experimentally determined in vivo BP was converted to receptor concentration using the dissociation constants found by in vitro flow cytometry binding studies. This requires the assumption that the in vivo and in vitro dissociation constants are the same. In neuroreceptor ligand studies, it has been demonstrated that in vivo rate of dissociation is much slower than that in vivo, but the dissociation constant itself stays relatively constant implying that ligand association is also decreased.(27, 35, 36) However, this may be attributed to binding events occurring in the confined space of the synaptic barrier.(37) Further studies on the apparent in vivo dissociation constant need to be performed to confirm ligand-receptor affinity but is beyond the scope of the work presented here.
The results presented here highlight an important concept for therapeutic and imaging agent delivery to tumors: in vitro or ex vivo total protein measures are inaccurate methods of estimating the available receptor population for binding in vivo. Although both IHC and IF predicted the number of receptors in vivo, they: 1) require invasive measures to acquire tissue; 2) are not realistic techniques to monitor receptor expression changes over time; and 3) do not account for dynamic biological and physiological parameters that influence vascular delivery. Here, the correlation of RCI determined in vivo receptor concentration to ex vivo immunohistochemistry techniques was established for the first time with clinically relevant affibody molecules using minimally invasive techniques. Development of clinical non-invasive imaging techniques requires thorough analysis of native EGFR levels in the skin and other high EGFR expressing tissues. In Figure S2, it is shown that mouse skin has far less EGFR than the A431 and AsPC-1 tumors but could significantly contribute to the signal in U251 or 9L and as such appropriate receptor concentration thresholds need to be determined. However, non-invasive RCI has recently been demonstrated in mice for lymph node imaging of breast tumor metastases using fluorescence alone (38) and glioma tumors using MRI- and CT-guided fluorescence tomography (39, 40). Additionally, RCI has potential for imaging other primary cancers such as breast tumors (41–43) where multiple measurements could be taken over a course antibody therapy to monitor treatment progress. Surface tumors such as skin cancers (melanomas, squamous and basal cell carcinomas) and some head and neck cancers, as well as hollow organ tumors, such as esophageal and bladder cancers, would also be prime targets as optical or fluorescence imaging is often used preclinically in these instances. For instance, RCI of melanomas has recently been demonstrated using an intravascular compartment model similar to the one presented here and a vascular targeting agent (44). Currently, the use of RCI for fluorescence-guided surgical resection is being tested in our laboratories.
In vivo receptor concentration imaging provides a robust, quantitative measure of extracellular receptors available for binding with a targeted imaging or therapeutic agent. For the first time, it has been demonstrated that in vivo RCI determined receptor concentration is equivalent to ex vivo immuno-staining techniques but does not correlate with ex vivo and in vitro techniques such as Western blot and flow cytometry. RCI will likely be most successfully applied clinically for guidance during surgical resection or for treatment monitoring of tumors accessible for light imaging such as breast cancer, hollow organ tumors and skin cancer.
Supplementary Material
Acknowledgements
This work has been supported by NIH grants R01CA156177 (KSS, BWP, TH), U54CA151662 (BWP, JRG, KSS), and P01CA84201 (KSS, JRG, BWP). The authors would like to thank Jonathan Park from the Department of Pathology at Dartmouth-Hitchcock Medical Center for the fluorescence in situ hybridization image in Figure 1, Pathology Translational Research Program (a shared resource of the Geisel School of Medicine at Dartmouth, the Dartmouth Hitchcock Medical Center and the Norris Cotton Cancer Center), and histotechnologist Rebecca O'Meara ASCP (HT), who performed the immunohistochemical studies.
Footnotes
Conflicts of Interest: The authors have no conflicts to disclose.
References
- 1.Mankoff DA, Link JM, Linden HM, Sundararajan L, Krohn KA. Tumor Receptor Imaging. Journal of Nuclear Medicine. 2008;49:149S–163S. doi: 10.2967/jnumed.107.045963. [DOI] [PubMed] [Google Scholar]
- 2.Ntziachristos V. Fluorescence Molecular Imaging. Annual Review of Biomedical Engineering. 2006;8:1–33. doi: 10.1146/annurev.bioeng.8.061505.095831. [DOI] [PubMed] [Google Scholar]
- 3.Oyen WJG, Van Der Graaf WTA. Molecular imaging of solid tumors: exploiting the potential. Nature Reviews Clinical Oncology. 2009;6:609–611. doi: 10.1038/nrclinonc.2009.139. [DOI] [PubMed] [Google Scholar]
- 4.Willmann JK, van Bruggen N, Dinkelborg LM, Gambhir SS. Molecular imaging in drug development. Nat Rev Drug Discov. 2008;7:591–607. doi: 10.1038/nrd2290. [DOI] [PubMed] [Google Scholar]
- 5.Rudin M, Weissleder R. Molecular imaging in drug discovery and development. Nat Rev Drug Discov. 2003;2:123–131. doi: 10.1038/nrd1007. [DOI] [PubMed] [Google Scholar]
- 6.Tichauer K, Samkoe K, Sexton K, Hextrum S, Yang H, Klubben WS, et al. In Vivo Quantification of Tumor Receptor Binding Potential with Dual-Reporter Molecular Imaging. Molecular Imaging and Biology. 2012;14:584–592. doi: 10.1007/s11307-011-0534-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor CR, Levenson RM. Quantification of immunohistochemistry—issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49:411–424. doi: 10.1111/j.1365-2559.2006.02513.x. [DOI] [PubMed] [Google Scholar]
- 8.Walker RA. Quantification of immunohistochemistry—issues concerning methods, utility and semiquantitative assessment I. Histopathology. 2006;49:406–410. doi: 10.1111/j.1365-2559.2006.02514.x. [DOI] [PubMed] [Google Scholar]
- 9.Cregger M, Berger AJ, Rimm DL. Immunohistochemistry and Quantitative Analysis of Protein Expression. Archives of Pathology & Laboratory Medicine. 2006;130:1026–1030. doi: 10.5858/2006-130-1026-IAQAOP. [DOI] [PubMed] [Google Scholar]
- 10.Dennis-Sykes CA, Miller WJ, McAleer WJ. A quantitative Western Blot method for protein measurement. Journal of Biological Standardization. 1985;13:309–314. doi: 10.1016/s0092-1157(85)80044-5. [DOI] [PubMed] [Google Scholar]
- 11.Diamandis EP, Christopoulos TK, Bean CC. Quantitative Western blot analysis and spot immunodetection using time-resolved fluorometry. Journal of Immunological Methods. 1992;147:251–259. doi: 10.1016/s0022-1759(12)80015-9. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Gaigalas AK, Ahbasi F, Marti GE, Vogt RF, Schwartz A. Quantitating Fluorescence Intensity from Fluorophores: Practical Use of MESF Values. J Res Natl Inst Stand Technol. 2002;107:339–353. doi: 10.6028/jres.107.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez E, Press M, Dueck A, Jenkins R, Kim C, Chen B, et al. Immunohistochemistry and fluorescence in situ hybridization assessment of HER2 in clinical trials of adjuvant therapy for breast cancer (NCCTG N9831, BCIRG 006, and BCIRG 005) Breast Cancer Res Treat. 2013;138:99–108. doi: 10.1007/s10549-013-2444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammock L, Lewis M, Phillips C, Cohen C. Strong HER-2/neu protein overexpression by immunohistochemistry often does not predict oncogene amplification by fluorescence in situ hybridization. Human Pathology. 2003;34:1043–1047. doi: 10.1053/s0046-8177(03)00409-x. [DOI] [PubMed] [Google Scholar]
- 15.Hargreaves RJ. The Role of Molecular Imaging in Drug Discovery and Development. Clin Pharmacol Ther. 2008;83:349–353. doi: 10.1038/sj.clpt.6100467. [DOI] [PubMed] [Google Scholar]
- 16.Leblond F, Davis SC, Valdés PA, Pogue BW. Pre-clinical whole-body fluorescence imaging: Review of instruments, methods and applications. Journal of Photochemistry and Photobiology B: Biology. 2010;98:77–94. doi: 10.1016/j.jphotobiol.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pysz MA, Gambhir SS, Willmann JK. Molecular imaging: current status and emerging strategies. Clinical Radiology. 2010;65:500–516. doi: 10.1016/j.crad.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herschman HR. Molecular Imaging: Looking at Problems, Seeing Solutions. Science. 2003;302:605–608. doi: 10.1126/science.1090585. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen QT, Olson ES, Aguilera TA, Jiang T, Scadeng M, Ellies LG, et al. Surgery with molecular fluorescence imaging using activatable cell-penetrating peptides decreases residual cancer and improves survival. Proceedings of the National Academy of Sciences. 2010;107:4317–4322. doi: 10.1073/pnas.0910261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hama Y, Urano Y, Koyama Y, Choyke PL, Kobayashi H. Activatable Fluorescent Molecular Imaging of Peritoneal Metastases following Pretargeting with a Biotinylated Monoclonal Antibody. Cancer Research. 2007;67:3809–3817. doi: 10.1158/0008-5472.CAN-06-3794. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi H, Ogawa M, Alford R, Choyke PL, Urano Y. New strategies for fluorescent probe design in medical diagnostic imaging. Chemical Reviews. 2009;110:2620–2640. doi: 10.1021/cr900263j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lammerstsma A, Hume H. Simplified reference tissue model for PET receptor studies. Neuroimage. 1996;4:153–158. doi: 10.1006/nimg.1996.0066. [DOI] [PubMed] [Google Scholar]
- 23.Tichauer K, Samkoe K, Klubben W, Hasan T, Pogue B. Advantages of a dual-tracer model over reference tissue models for binding potential measurement in tumors. Physics in Medicine and Biology. 2012;57:6647. doi: 10.1088/0031-9155/57/20/6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomasi G, Turkheimer F, Aboagye E. Importance of Quantification for the Analysis of PET Data in Oncology: Review of Current Methods and Trends for the Future. Molecular Imaging and Biology. 2012;14:131–146. doi: 10.1007/s11307-011-0514-2. [DOI] [PubMed] [Google Scholar]
- 25.Samkoe KS, Chen A, Rizvi I, O'Hara JA, Hoopes PJ, Pereira SP, et al. Imaging Tumor Variation in Response to Photodynamic Therapy in Pancreatic Cancer Xenograft Models. International Journal of Radiation Oncology*Biology*Physics. 2010;76:251–259. doi: 10.1016/j.ijrobp.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tichauer KM, Diop M, Elliott JT, Samkoe KS, Hasan T, Lawrence KS, et al. Accounting for pharmacokinetic differences in dual-tracer receptor density imaging. Physics in Medicine and Biology. 2014;59:2341. doi: 10.1088/0031-9155/59/10/2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, et al. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab. 2007;27:1533–1539. doi: 10.1038/sj.jcbfm.9600493. [DOI] [PubMed] [Google Scholar]
- 28.Gibbs-Strauss SL, Samkoe KS, O'Hara JA, Davis SC, Hoopes PJ, Hasan T, et al. Detecting Epidermal Growth Factor Receptor Tumor Activity In Vivo During Cetuximab Therapy of Murine Gliomas. Academic Radiology. 2010;17:7–17. doi: 10.1016/j.acra.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Dijk LK, Hoeben BAW, Kaanders JHAM, Franssen GM, Boerman OC, Bussink J. Imaging of Epidermal Growth Factor Receptor Expression in Head and Neck Cancer with SPECT/CT and 111In-Labeled Cetuximab-F(ab')2. Journal of Nuclear Medicine. 2013;54:2118–2124. doi: 10.2967/jnumed.113.123612. [DOI] [PubMed] [Google Scholar]
- 30.Ardeshirpour Y, Chernomordik V, Capala J, Hassan M, Zielinsky R, Griffiths G, et al. Using in-vivo fluorescence imaging in personalized cancer diagnostics and therapy, an image and treat paradigm. Technology in cancer research & treatment. 2011;10:549. doi: 10.1177/153303461101000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aerts HJWL, Dubois L, Perk L, Vermaelen P, van Dongen GAMS, Wouters BG, et al. Disparity Between In Vivo EGFR Expression and 89Zr-Labeled Cetuximab Uptake Assessed with PET . Journal of Nuclear Medicine. 2009;50:123–131. doi: 10.2967/jnumed.108.054312. [DOI] [PubMed] [Google Scholar]
- 32.McLarty K, Cornelissen B, Scollard D, Done S, Chun K, Reilly R. Associations between the uptake of 111In-DTPA-trastuzumab, HER2 density and response to trastuzumab (Herceptin) in athymic mice bearing subcutaneous human tumour xenografts. Eur J Nucl Med Mol Imaging. 2009;36:81–93. doi: 10.1007/s00259-008-0923-x. [DOI] [PubMed] [Google Scholar]
- 33.Tichauer KM, Samkoe KS, Sexton KJ, Gunn JR, Hasan T, Pogue BW. Improved tumor contrast achieved by single time point dual-reporter fluorescence imaging. JBO. 2012;17:0660011–06600110. doi: 10.1117/1.JBO.17.6.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vakoc BJ, Lanning RM, Tyrrell JA, Padera TP, Bartlett LA, Stylianopoulos T, et al. Three-dimensional microscopy of the tumor microenvironment in vivo using optical frequency domain imaging. Nat Med. 2009;15:1219–1223. doi: 10.1038/nm.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laruelle M, Abi-Dargham A, Al-Tikriti MS, Baldwin RM, Zea-Ponce Y, Zoghbi SS, et al. SPECT Quantification of [123I]Iomazenil Binding to Benzodiazepine Receptors in Nonhuman Primates: II. Equilibrium Analysis of Constant Infusion Experiments and Correlation with In Vitro Parameters. J Cereb Blood Flow Metab. 1994;14:453–465. doi: 10.1038/jcbfm.1994.56. [DOI] [PubMed] [Google Scholar]
- 36.Robertson MW, Leslie CA, Bennett JP., Jr Apparent synaptic dopamine deficiency induced by withdrawal from chronic cocaine treatment. Brain Research. 1991;538:337–339. doi: 10.1016/0006-8993(91)90451-z. [DOI] [PubMed] [Google Scholar]
- 37.Delforge J, Syrota A, Lançon JP, Nakajima K, Loc'h C, Janier M, et al. Cardiac beta-adrenergic receptor density measured in vivo using PET, CGP 12177, and a new graphical method. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 1991;32:739–748. [PubMed] [Google Scholar]
- 38.Tichauer KM, Samkoe KS, Gunn JR, Kanick SC, Hoopes PJ, Barth RJ, et al. Quantitative imaging of cancer receptors in lymph nodes with a dual-tracer technique reveals microscopic burden level with macroscopic measurement. Nature Medicine. 2014 doi: 10.1038/nm.3732. Accepted for publication (NMED-NT66028C1): July 3, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis SC, Samkoe KS, Tichauer KM, Sexton KJ, Gunn JR, Deharvengt SJ, et al. Dynamic dual-tracer MRI-guided fluorescence tomography to quantify receptor density in vivo. Proceedings of the National Academy of Sciences. 2013;110:9025–9030. doi: 10.1073/pnas.1213490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tichauer KM, Holt RW, El-Ghussein F, Davis SC, Samkoe KS, Gunn JR, et al. Dual-tracer background subtraction approach for fluorescent molecular tomography. JBO. 2013;18:016003. doi: 10.1117/1.JBO.18.1.016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leff D, Warren O, Enfield L, Gibson A, Athanasiou T, Patten D, et al. Diffuse optical imaging of the healthy and diseased breast: A systematic review. Breast Cancer Res Treat. 2008;108:9–22. doi: 10.1007/s10549-007-9582-z. [DOI] [PubMed] [Google Scholar]
- 42.Tromberg BJ, Pogue BW, Paulsen KD, Yodh AG, Boas DA, Cerussi AE. Assessing the future of diffuse optical imaging technologies for breast cancer management. Medical Physics. 2008;35:2443–2451. doi: 10.1118/1.2919078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ntziachristos V, Chance B. Breast imaging technology: Probing physiology and molecular function using optical imaging - applications to breast cancer. Breast Cancer Res. 2001;3:41–46. doi: 10.1186/bcr269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tichauer K, Deharvengt S, Samkoe K, Gunn J, Bosenberg M, Turk M-J, et al. Tumor Endothelial Marker Imaging in Melanomas Using Dual-Tracer Fluorescence Molecular Imaging. Molecular Imaging and Biology. 2013:1–11. doi: 10.1007/s11307-013-0692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


