Abstract
The genetics of myoepithelial tumors (ME) of soft tissue and bone have recently been investigated, with EWSR1 related gene fusions being seen in approximately half of the tumors. The fusion partners of EWSR1 so far described include POU5F1, PBX1, ZNF444 and, in a rare case, ATF1. We investigated by RNA sequencing an index case of EWSR1-rearranged ME of the tibia, lacking a known fusion partner, and identified a novel EWSR1-PBX3 fusion. The fusion was further validated by reverse transcriptase polymerase chain reaction (RT-PCR) and Fluorescence In Situ hybridization (FISH). To evaluate if this is a recurrent event, an additional cohort of 22 EWSR1-rearranged ME cases lacking a fusion partner were screened by FISH for abnormalities in PBX3 gene. Thus 2 additional cases were identified showing an EWSR1-PBX3 gene fusion. One of them was also intra-osseous involving the ankle, while the other occurred in the soft tissue of the index finger. The morphology of the EWSR1-PBX3 fusion positive cases showed similar findings, with nests or sheets of epithelioid to spindle cells in a partially myxoid to collagenous matrix. All the 3 cases showed expression of S100 and EMA by immunohistochemistry. In summary, we report a novel EWSR1-PBX3 gene fusion in a small subset of ME, thereby expanding the spectrum of EWSR1-related gene fusions seen in these tumors. This gene fusion seems to occur preferentially in skeletal ME, with 2 of the 3 study cases occurring in intraosseous locations.
Keywords: Myoepithelial tumor, EWSR1, PBX3
INTRODUCTION
Myoepithelial tumors (ME) of soft tissue and bone are rare tumors that show morphologic similarity to myoepithelial tumors of salivary gland. They encompass a morphologically heterogeneous group of tumors, some of which were originally termed as parachordomas. Similar to salivary gland tumors, ME of soft tissue and bone can occasionally show gland formation leading to a biphasic morphology. While a subset of mixed tumors of skin and soft tissue often share PLAG1 rearrangement with salivary gland counterparts (Antonescu et al., 2013), other soft tissue ME tumors have been shown to harbor distinctly different molecular changes.
A t(1;22)(q23;q12) resulting in an EWSR1-PBX1 fusion was first described as a sole cytogenetic event in a soft tissue ME tumor, arising in the foot of a 59-year-old woman (Brandal et al., 2008), whereas a second case, of an occipital soft tissue ME carcinoma, arising in a 40-year-old woman, showed a t(19;22)(q13;q12), resulting in an EWSR1-ZNF444 fusion (Brandal et al., 2009). In addition, EWSR1 gene rearrangement by fluorescence in situ hybridization (FISH) has been reported in two ME tumors in the pediatric age group (Gleason and Fletcher 2007). A subsequent detailed molecular analysis of 66 ME cases (including 4 intra-osseous lesions) demonstrated EWSR1 rearrangement in 45% cases of ME tumors with fusion partners including PBX1, ZNF444, and POU5F1 (Antonescu et al., 2010). Another EWSR1 fusion partner was recently described as a case report in a soft tissue myoepithelioma showing EWSR1-ATF1 gene fusion (Flucke et al., 2012). In contrast, EWSR1-negative ME tumors are located superficially, with predilection for the skin and subcutaneous tissue, and often show ductal and glandular elements. It is in this latter subset of lesions that a pathogenetic link between skin and soft tissue ME and their salivary gland counterpart was established by the presence of PLAG1 gene rearrangements (Antonescu et al., 2013).
Despite the recent advances, there still remains a subset of ME tumors with unknown genetic alterations or a subset with EWSR1 rearrangements but no known fusion partner. In this study, we investigated one such EWSR1-rearranged ME tumor by RNA sequencing in an attempt to identify additional novel gene partners and determine its recurrent potential.
MATERIAL AND METHODS
Index Case and ME Extended Cohort
A 26 year-old male presenting with anteromedial knee and proximal tibial pain was noted to have a 1.0 cm lesion of the left tibia on CT scans. He was observed for 4 years by imaging when the lesion had grown to about 2.6 cm. He was treated with curetting, cementation and bone grafting. Twenty-two months later he developed local recurrence around the cemented area. The recurrence was treated once more with curetting, cementation and bone grafting. Tumor material from both primary and recurrent lesion was reviewed. Frozen tumor tissue and formalin fixed paraffin embedded (FFPE) tissue were available for molecular analysis. Additionally, FFPE sections from other EWSR1-rearranged ME tumors, some of which were reported in earlier studies (Antonescu et al., 2010; Jo et al., 2013), were reviewed and selected for the study. The study was approved by the Institutional Review Board 02-060.
RNA Sequencing
Total RNA was prepared for RNA sequencing in accordance with the standard Illumina mRNA sample preparation protocol (Illumina). Briefly, mRNA was isolated with oligo(dT) magnetic beads from total RNA (2 μg) extracted from case. The mRNA was fragmented by incubation at 94°C for 2.5 min in fragmentation buffer (Illumina). To reduce the inclusion of artifactual chimeric transcripts due to random priming of transcript fragments into the sequencing library because of inefficient A-tailing reactions that lead to self ligation of blunt-ended template molecules (Quail et al., 2008), an additional gel size-selection step (capturing 350-400 bp) was introduced prior to the adapter ligation step. The adaptor-ligated library was then enriched by PCR for 15 cycles and purified. The library was sized and quantified using DNA1000 kit (Agilent) on an Agilent 2100 Bioanalyzer according to the manufacturer’s instructions. Paired-end RNA-sequencing at read lengths of 50 or 51 bp was performed with the HiSeq 2000 (Illumina). A total of about 26.8 million paired-end reads were generated, corresponding to about 2.6 billion bases.
Analysis of RNA Sequencing Results with FusionSeq
All reads were independently aligned with the CASAVA 1.8 software provided by Illumina against the human genome sequence (hg19) and a splice junction library, simultaneously. The splice junction library was generated by considering all possible junctions between exons of each transcript. We used the University of California, Santa Cruz (UCSC) Known Genes annotation set (Hsu et al., 2006) to generate this library via RSEQtools, a computational method for processing RNA-seq data (Habegger et al., 2011). The mapped reads were converted into Mapped Read Format (Habegger et al., 2011) and analyzed with FusionSeq (Sboner et al., 2010) to identify potential fusion transcripts. FusionSeq is a computational method successfully applied to paired-end RNA-seq experiments for the identification of chimeric transcripts (Tanas et al., 2011; Pierron et al., 2012) (Mosquera et al., 2013). Briefly, paired-end reads mapped to different genes are first used to identify potential chimeric candidates. A cascade of filters, each taking into account different sources of noise in RNA-sequencing experiments, was then applied to remove spurious fusion transcript candidates. Once a confident list of fusion candidates was generated, they were ranked with several statistics to prioritize the experimental validation. In these cases, we used the DASPER score (Difference between the observed and Analytically calculated expected SPER): a higher DASPER score indicated a greater likelihood that the fusion candidate was authentic and did not occur randomly. See (Sboner et al., 2010) for further details about FusionSeq.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
An aliquot of the RNA extracted above from frozen tissue (Trizol Reagent, Invitrogen; Carlsbad, CA) was used to confirm the novel fusion transcript identified by FusionSeq. RNA quality was determined by Eukaryote Total RNA Nano Assay and cDNA quality was tested for PGK housekeeping gene (247 bp amplified product). Three microgram of total RNA was used for cDNA synthesis by SuperScript® III First-Strand Synthesis Kit (Invitrogen). RT-PCR was performed using the Advantage-2 PCR kit (Clontech, Mountain View, CA) for 30 cycles at a 64.5°C annealing temperature, using the following primers: EWSR1 Exon 7 Fwd: 5’– CTCTCAGCAGAACACCTATGG–3’ and PBX3 Exon 6 Rev: 5’– GGCAACAAACGAATCAGGTACAAG – 3’. Amplified products were purified and sequenced by Sanger method.
DNA PCR to investigate the intronic breakpoint
Genomic DNA was isolated from fresh frozen tissue, as described previously (Antonescu et al., 2003). PCR was performed using the following primers: EWSR1 Intron 8 Fwd: 5’–CCTCATTTGGCTCTCCCTTGG–3’ and PBX3 Intron 4 Rev: 5’–GCCTACTGACTCTGTGTTTTCTGACTG–3’, for 33 cycles at a 64.5°C annealing temperature.
Fluorescence In Situ Hybridization (FISH)
FISH on interphase nuclei from paraffin-embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BAC), covering and flanking genes that were identified as potential fusion partners in the RNA-seq experiment. BAC clones were chosen according to USCS genome browser (http://genome.uscs.edu), see Supplementary Table 1. The BAC clones were obtained from BACPAC sources of Children's Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (Antonescu et al., 2010). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
RESULTS
Radiologic and Pathologic Features of Index Case (ME1)
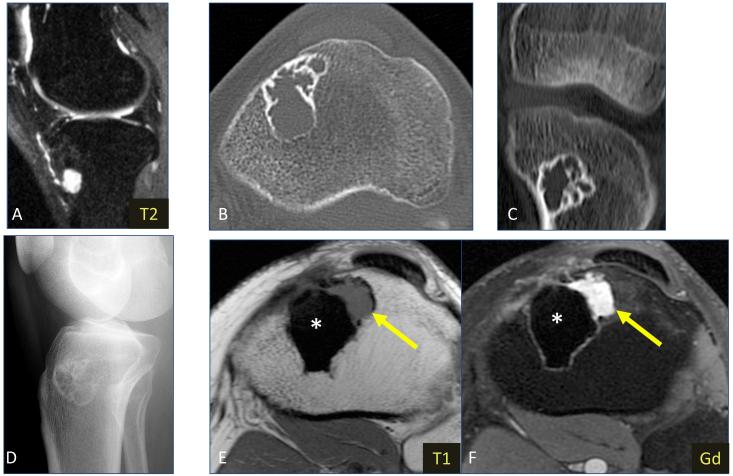
Radiographs of the ME1 index case, a 26 year-old male presenting with anteromedial knee and proximal tibial pain, showed a lytic lesion in the proximal metaphysis of the left tibia. T2-weighted MR images revealed a hyperintense lesion in the anterior subcortical region. No surrounding marrow edema pattern or extraosseous extension was noted. CT images showed a multilobulated lesion with multiple internal septations, a thin sclerotic margin and no calcified matrix (Fig. 1) Histologic findings showed epithelioid to oval cells arranged in short fascicles, embedded in a myxoid to sclerotic stroma. Focal areas of clear cell change were identified. (Fig. 2) Nuclei showed vesicular chromatin with occasional prominent nucleoli. No significant cytologic atypia was noted. Mitotic activity was low (1 per 10 high power fields) and no necrosis was noted. Immunohistochemical stains showed that the tumor cells were positive for S100 protein, EMA and HHF35, in keeping with myoepithelial differentiation. FISH studies performed showed an EWSR1 gene rearrangement, while no gene abnormalities noted in POU5F1, PBX1 and ZNF444.
Figure 1.
Imaging for index case ME1 (A) Sagittal fat-suppressed T2-weighted MR image of the primary 1.4 cm lesion showing lesion with hyperintense signal, located in the anterior subcortical region of proximal tibial metaphysis. No surrounding marrow edema pattern or soft tissue extension is evident. (B) Axial and (C) coronal CT images without intravenous contrast obtained 44 months later demonstrate increase in size (3.8 cm) of the lytic lesion. The lesion is multilobulated, and contains multiple internal septations, a thin sclerotic margin and no calcified matrix. (D) Lateral radiograph obtained 3 months later shows the peripheral sclerotic margin and septations. Axial (E) T1-weighted and (F) post-gadolinium fat-suppressed T1-weighted MR images obtained 33 months later show the low-signal, non-enhancing surgical cement (*), as well as a small focus of recurrent tumor (arrow) that has developed along the anterolateral margin of cement, with intermediate T1 signal and avid enhancement.
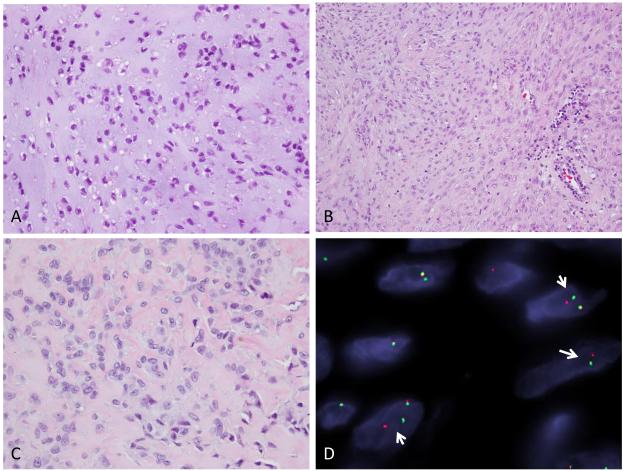
Figure 2.
ME tumor involving the tibial metaphysis in a 26 year-old male (ME1). Low power image from the first curettings showing epithelioid cells in a myxoid stroma (A, 200x), while other areas displaying spindle cells in short fascicles. (B, 100x) Recurrent lesion showing epithelioid cells in a partially sclerotic stroma (C, 100x); FISH break-apart assay for PBX3 gene showing split signal indicating rearrangement (D).
A Novel EWSR1-PBX3 Gene Fusion is Identified by FusionSeq
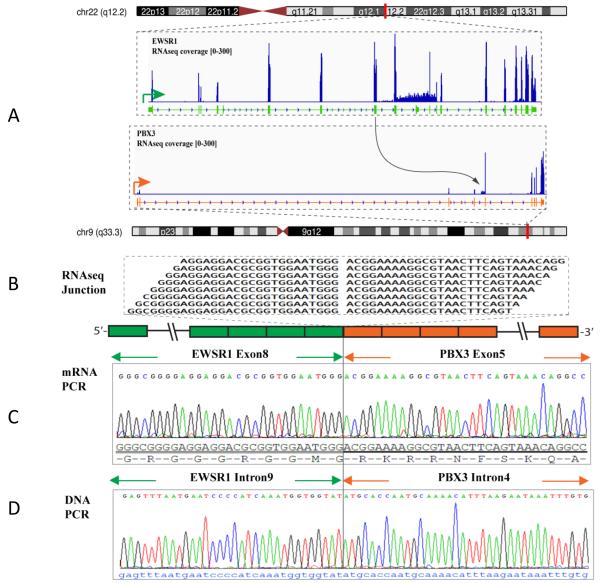
The top fusion candidate identified by FusionSeq in the index case was the EWSR1-PBX3. Reads alignment suggested a fusion of EWSR1 exon 8 to PBX3 exon 5. Experimental validation confirmed this fusion transcript sequence by RT-PCR (Fig. 3). Subsequent FISH analysis, using a break-apart assay, showed rearrangements in both EWSR1 and PBX3 genes (Fig. 2). The intronic break was further confirmed by DNA PCR, showing the fusion of EWSR1 intron 9 to PBX3 intron 4 (Fig. 3).
Figure 3.
EWSR1-PBX3 gene fusion in ME tumor (ME1). (A) Schematic representation of the EWSR1-PBX3 fusion indicating the chromosomal loci joint together; EWSR1 exon 8 being fused to PBX3 exon 5; (B) RNA reads covering the fusion junction were isolated independent to FusionSeq analysis work flow, supporting the EWSR1-PBX3 fusion candidate; (C) RT-PCR experimental validation of the fusion shows the EWSR1 exon 8 fused to PBX3 exon 5; (D) DNA PCR further confirming the genomic breakpoint, showing the fusion of intron 9 of EWSR1 to the intron 4 of PBX3.
Further PBX3 screening by FISH identifies two additional ME with EWSR1-PBX3 Fusions
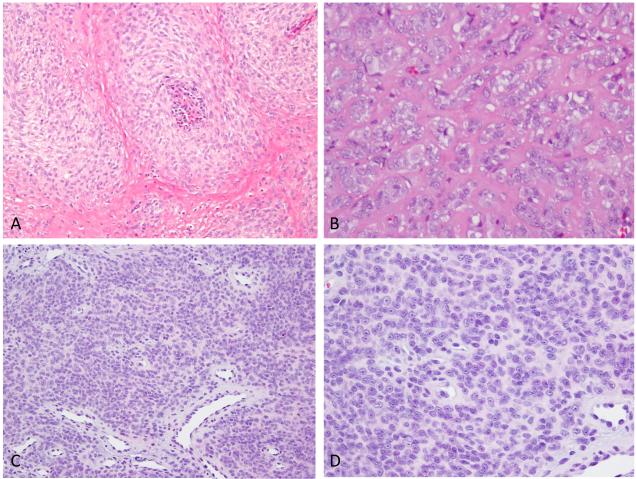
FISH analysis was performed in 22 additional EWSR1-rearranged ME cases, lacking a known fusion gene partner. The 22 cases included 17 soft tissue, 3 cutaneous and 2 intraosseous ME tumors. Thirteen of the 22 cases were previously reported by Antonescu et al., 2010 (10 cases) and Jo et al., 2013 (3 cases). Two of 22 tumors tested showed similar PBX3 gene rearrangements in keeping with an EWSR1-PBX3 fusion. One of these two cases presented as an intra-osseous ME tumor, involving the fibula of a 16 year-old girl. (ME2, Fig. 4) Morphologically, the tumor was composed of nests of spindle to epithelioid cells separated by a collagenous matrix. Focal areas of clear cell change were also noted. Mitotic activity was low (1 per 10 high power fields) and no necrosis was identified. (Fig. 4) Immunohistochemical stains showed reactivity of the tumor cells for S100 protein and EMA.
Figure 4.
Pathologic spectrum of EWSR1-PBX3 fusion positive ME tumors. ME tumor involving the fibula in a 16 year-old female (ME2) showing nests of ovoid cells (A, 100x), with focally more epithelioid cells having clear cytoplasm (B, 400x); ME lesion involving the index finger soft tissue in a 53 year-old female (ME3) showing sheets of monomorphic cells (C, 100x), which at higher power reveal ovoid or epithelioid cells with prominent nucleoli in a loose matrix (D, 400x).
The other EWSR1-PBX3 positive case occurred as a right index finger soft tissue lesion in a 53 year-old woman. (ME3, Fig. 4) Microscopically, it revealed sheets of monomorphic round to ovoid cells with round nuclei and prominent nucleoli, in a partially sclerotic background. (Fig 4) Mitotic activity was low (0-1 per 10 high power fields) and no necrosis was identified. Immunohistochemical stains showed reactivity of the tumor cells for S100 protein, Cytokeratin and EMA.
Discussion
ME neoplasms of soft tissue represent a heterogeneous group of tumors, ranging from benign to highly aggressive lesions, which, based on their anatomical location, are sometimes designated under different terminologies: soft tissue myoepithelioma, parachordoma and cutaneous mixed tumor. (Kilpatrick et al., 1997; Hornick and Fletcher 2003). Despite the wide spectrum of morphologies and clinical presentations, the defining denominator among these different lesions is the consistent immunoexpression for S100 protein and EMA/cytokeratin, in keeping with a myoepithelial lineage. EWSR1 related gene fusions are seen in roughly half of ME tumors, with a known fusion partner being identified in 17% cases (Antonescu et al., 2010). The most common fusions include EWSR1-POU5F1 and EWSR1-PBX1 seen in 15% of cases, with only rare cases described harboring either EWSR1-ZNF444 or EWSR1-ATF1 (Brandal et al., 2009; Antonescu et al., 2010; Flucke et al., 2012). Despite the heterogeneous morphology of the ME neoplasms, there are clues to suggest that variant gene fusions correlate with a specific phenotype. All 5 tumors showing the EWSR1-POU5F1 gene fusion, in the study by Antonescu et al. showed striking similarities in clinical presentation and with morphologic features of predominantly epithelioid cells with abundant clear cytoplasm, arranged in a nested growth pattern (Antonescu et al., 2010). Furthermore, tumors with EWSR1-PBX1 fusion showed a deceptively bland spindle cell proliferation within a fibrotic stroma, reminiscent of fibromatosis (Brandal et al., 2008; Antonescu et al., 2010). The rare tumors harboring EWSR1-ZNF444 gene fusion (Brandal et al., 2009; Antonescu et al., 2010) have been described with an epithelioid, nested appearance. Recently, in a study of 38 cutaneous syncytial myoepitheliomas, tumors showed similar morphologic features of uniform, ovoid to spindle or histiocytoid cells with eosinophilic syncytial cytoplasm arranged in solid sheets (Jo et al., 2013). Fourteen of the 17 (82%) tumors tested in that study showed EWSR1 gene rearrangement, but no fusion partner was detected. In the current study, 3 of the EWSR1 rearranged cutaneous syncytial myoepitheliomas, included in the prior study by Jo et al., were tested for PBX3 abnormalities by FISH but no abnormalities were detected.
The three EWSR1-PBX3 fusion positive ME tumors identified in this study showed similar morphologic features, of spindle to epithelioid cells, arranged in fascicles, in a myxoid to sclerotic stroma. EWSR1-PBX3 fusion positive tumors seem to preferentially arise in intra-osseous locations with 2 of the 3 cases in our study occurring in the bone. Primary ME tumors of bone are rare, with 21 cases reported in the literature (Table 1)(de Pinieux et al., 2001; Ferretti et al., 2003; Alberghini et al., 2007; Cuesta Gil et al., 2008; Antonescu et al., 2010; Park et al., 2010; Rekhi et al., 2011; Kurzawa et al., 2013) Based on the reported cases, intra-osseous ME tend to involve flat and long bones with common locations including maxilla, ilium, tibia and fibula. Morphologically, most of them appear to share a similar histology, with epithelioid to spindle cells arranged in cords and nests, in a myxoid to collagenous stroma. In the series by Antonescu et al. (Antonescu et al., 2010), 5 intra-osseous lesions were included, 4 showing EWSR1 rearrangement, with one having EWSR1-PBX1 fusion. Kurzawa et al. reported the largest series of 8 skeletal ME tumors cases, including 2 cases reported previously by Antonescu et al, of which 5 (64%) showed rearrangement of the EWSR1 gene (Kurzawa et al., 2013). Two of the 3 intra-osseous ME reported in this study were previously included in Antonescu et al., 2010. Including the additional case reported in this study, the incidence of EWSR1 gene rearrangement in intra-osseous ME is 67% of the cases tested.
Table 1.
Clinical, radiologic and genetic abnormalities of intra-osseous ME cases reported in the literature.
| Study | Number of cases |
Sites of involvement |
Radiology | Histology | Genetic abnormality |
|---|---|---|---|---|---|
| de Pinieux et al. 2001 | 1 | cuboid | lytic lesion | epithelioid cells in a myxo- chondroid stroma, with associated keratinous cysts |
NA |
| Ferretti et al. 2003 | 5 | Maxilla (5) | lucency in maxillary alveolus |
epithelioid / plasmacytoid cells in abundant myxoid stroma |
NA |
| Alberghini et al. 2007 | 1 | femur | lytic lesion with extra- cortical extension |
epithelioid and spindle cells arranged in cords, in a hyalinized to chondromyxoid matrix |
NA |
| Cuesta Gil et al. 2008 | 1 | maxilla | circumscribed and encapsulated mass involving the left side of the maxilla and extending superiorly at the maxillary sinus |
epithelioid cells arranged in cords |
NA |
| Park et al. 2010 | 1 | humerus | lytic lesion with focal cortical erosion |
epithelioid cells with round- to-ovoid nuclei arranged in sheets in a cord-like fashion with squamous differentiation |
NA |
| Antonescu et al. 2010 | 5 | Ilium, L1 vertebra, hip, humerus, fibula |
NA | spindle to epithelioid cells in fascicles in a collagenous or myxoid matrix |
EWSR1-PBX1 fusion - 1 case; EWSR1 rearrangement - 3 cases; No EWSR1 rearrangement - 1 case |
| Rekhi et al. 2011 | 1 | ilium | lytic lesion with extra- cortical extension |
epithelioid and spindle cells arranged in cords and nests, in a sclerotic and myxo- hyaline stroma, with associated squamous differentiation |
EWSR1 rearrangement - negative |
| Kurzawa et al. 2013 | 6 | tibia (2), maxilla, ilium(2), sacrum |
radiolucent lesions | spindle to epithelioid cells in fascicles in a hyaline, collagenous or myxoid matrix |
EWSR1 rearranged - 3 cases; No EWSR1 rearrangement - 2 cases |
| Agaram et al. (current study) | 3 | Tibia, fibula*and L1 vertebra* |
lytic lesions | epithelioid to oval cells arranged in fascicles, focal myxoid to sclerotic stroma |
EWSR1-PBX3 fusion - 2 cases; EWSR1 rearrangement - 1 case |
previously reported in Antonescu et al. 2010; NA, not assessed
PBX3 (Pre-B cell leukemia homeobox 3) is encoded by a gene located at 9q33.3 locus. The pre-B-cell leukemia transcription factors (PBX) are members of the TALE (three amino acid loop extension) homeobox gene family, which are involved in regulation of developmental gene expression, differentiation of urogenital organs and steroidogenesis through their abilities to form hetero-oligomeric DNA complexes (Schnabel et al., 2001; Laurent et al., 2008). PBX proteins interact with a subset of HOX proteins and with the Meinox subfamily of TALE class proteins to enhance their DNA-binding affinities and specificities. Human PBX1 was originally identified as a proto-oncogene in pre-B cell acute lymphoblastic leukemia, where it is expressed as a fusion protein with E2A after a chromosomal translocation (Kamps et al., 1990; Nourse et al., 1990). Subsequently, PBX2, PBX3 and PBX4 were identified as other members of the PBX family based on their high degree of sequence homology within and flanking their DNA-binding homeodomains (Monica et al., 1991; Wagner et al., 2001). Biochemical studies and expression profiling of PBX proteins suggest both overlapping and specific functions (Laurent et al., 2008). Both PBX1 and PBX3 are expressed in the cortex of developing adrenal glands where they play a significant role in regulation of steroidogenesis (Di Giacomo et al., 2006; Lichtenauer et al., 2007). No previous reports have implicated PBX3 in a fusion gene event.
In summary, we have identified a novel gene fusion, EWSR1-PBX3, in a small subset (7.5%) of EWSR1-rearranged ME tumors, thereby expanding the spectrum of EWSR1-related gene fusions seen in these lesions. Although this gene fusion seems to occur preferentially in intra-osseous ME tumors, additional studies with larger numbers of cases are required to confirm this finding.
Supplementary Material
Acknowledgments
Supported in part by: P01CA47179 (CRA), P50 CA 140146-01 (CRA), Cycle for Survival
Footnotes
Conflict of interest: none
REFERENCES
- Alberghini M, Pasquinelli G, Zanella L, Pignatti G, Benini S, Bacchini P, Bertoni F. Primary malignant myoepithelioma of the distal femur. APMIS. 2007;115(4):376–380. doi: 10.1111/j.1600-0463.2007.apm_569.x. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49(12):1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonescu CR, Zhang L, Shao SY, Mosquera JM, Weinreb I, Katabi N, Fletcher CD. Frequent PLAG1 gene rearrangements in skin and soft tissue myoepithelioma with ductal differentiation. Genes Chromosomes Cancer. 2013;52(7):675–682. doi: 10.1002/gcc.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandal P, Panagopoulos I, Bjerkehagen B, Gorunova L, Skjeldal S, Micci F, Heim S. Detection of a t(1;22)(q23;q12) translocation leading to an EWSR1-PBX1 fusion gene in a myoepithelioma. Genes Chromosomes Cancer. 2008;47(7):558–564. doi: 10.1002/gcc.20559. [DOI] [PubMed] [Google Scholar]
- Brandal P, Panagopoulos I, Bjerkehagen B, Heim S. t(19;22)(q13;q12) Translocation leading to the novel fusion gene EWSR1-ZNF444 in soft tissue myoepithelial carcinoma. Genes Chromosomes Cancer. 2009;48(12):1051–1056. doi: 10.1002/gcc.20706. [DOI] [PubMed] [Google Scholar]
- Cuesta Gil M, Bucci T, Navarro Cuellar C, Duarte Ruiz B, Pannone G, Bufo P, Navarro Vila C. Intraosseous myoepithelioma of the maxilla: clinicopathologic features and therapeutic considerations. J Oral Maxillofac Surg. 2008;66(4):800–803. doi: 10.1016/j.joms.2007.06.002. [DOI] [PubMed] [Google Scholar]
- de Pinieux G, Beabout JW, Unni KK, Sim FH. Primary mixed tumor of bone. Skeletal Radiol. 2001;30(9):534–536. doi: 10.1007/s002560100384. [DOI] [PubMed] [Google Scholar]
- Di Giacomo G, Koss M, Capellini TD, Brendolan A, Popperl H, Selleri L. Spatio-temporal expression of Pbx3 during mouse organogenesis. Gene Expr Patterns. 2006;6(7):747–757. doi: 10.1016/j.modgep.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Ferretti C, Coleman H, Altini M, Meer S. Intraosseous myoepithelial neoplasms of the maxilla: diagnostic and therapeutic considerations in 5 South African patients. J Oral Maxillofac Surg. 2003;61(3):379–386. doi: 10.1053/joms.2003.50075. [DOI] [PubMed] [Google Scholar]
- Flucke U, Mentzel T, Verdijk MA, Slootweg PJ, Creytens DH, Suurmeijer AJ, Tops BB. EWSR1-ATF1 chimeric transcript in a myoepithelial tumor of soft tissue: a case report. Hum Pathol. 2012;43(5):764–768. doi: 10.1016/j.humpath.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Gleason BC, Fletcher CD. Myoepithelial carcinoma of soft tissue in children: an aggressive neoplasm analyzed in a series of 29 cases. Am J Surg Pathol. 2007;31(12):1813–1824. doi: 10.1097/PAS.0b013e31805f6775. [DOI] [PubMed] [Google Scholar]
- Habegger L, Sboner A, Gianoulis TA, Rozowsky J, Agarwal A, Snyder M, Gerstein M. RSEQtools: a modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics. 2011;27(2):281–283. doi: 10.1093/bioinformatics/btq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornick JL, Fletcher CD. Myoepithelial tumors of soft tissue. A clinicopathologic and immunohistochemical study of 101cases with evaluation of prognostic parameters. Am J Surg Pathol. 2003;27:1183–1196. doi: 10.1097/00000478-200309000-00001. [DOI] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22(9):1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Jo VY, Antonescu CR, Zhang L, Dal Cin P, Hornick JL, Fletcher CD. Cutaneous syncytial myoepithelioma: clinicopathologic characterization in a series of 38 cases. Am J Surg Pathol. 2013;37(5):710–718. doi: 10.1097/PAS.0b013e3182772bba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamps MP, Murre C, Sun XH, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990;60(4):547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- Kilpatrick SE, Hitchcock MG, Kraus MD, Calonje E, Fletcher CD. Mixed tumors and myoepitheliomas of soft tissue: a clinicopathologic study of 19 cases with a unifying concept. Am J Surg Pathol. 1997;21(1):13–22. doi: 10.1097/00000478-199701000-00002. [DOI] [PubMed] [Google Scholar]
- Kurzawa P, Kattapuram S, Hornicek FJ, Antonescu CR, Rosenberg AE, Nielsen GP. Primary myoepithelioma of bone: a report of 8 cases. Am J Surg Pathol. 2013;37(7):960–968. doi: 10.1097/PAS.0b013e3182858a0e. [DOI] [PubMed] [Google Scholar]
- Laurent A, Bihan R, Omilli F, Deschamps S, Pellerin I. PBX proteins: much more than Hox cofactors. Int J Dev Biol. 2008;52(1):9–20. doi: 10.1387/ijdb.072304al. [DOI] [PubMed] [Google Scholar]
- Lichtenauer UD, Duchniewicz M, Kolanczyk M, Hoeflich A, Hahner S, Else T, Bicknell AB, Zemojtel T, Stallings NR, Schulte DM, Kamps MP, Hammer GD, Scheele JS, Beuschlein F. Pre-B-cell transcription factor 1 and steroidogenic factor 1 synergistically regulate adrenocortical growth and steroidogenesis. Endocrinology. 2007;148(2):693–704. doi: 10.1210/en.2006-0681. [DOI] [PubMed] [Google Scholar]
- Monica K, Galili N, Nourse J, Saltman D, Cleary ML. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol Cell Biol. 1991;11(12):6149–6157. doi: 10.1128/mcb.11.12.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera JM, Sboner A, Zhang L, Kitabayashi N, Chen CL, Sung YS, Wexler LH, LaQuaglia MP, Edelman M, Sreekantaiah C, Rubin MA, Antonescu CR. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013;52(6):538–550. doi: 10.1002/gcc.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourse J, Mellentin JD, Galili N, Wilkinson J, Stanbridge E, Smith SD, Cleary ML. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60(4):535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- Park JS, Ryu KN, Han CS, Park YK. Malignant myoepithelioma of the humerus with a satellite lesion: a case report and literature review. Br J Radiol. 2010;83(991):e161–164. doi: 10.1259/bjr/64670838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierron G, Tirode F, Lucchesi C, Reynaud S, Ballet S, Cohen-Gogo S, Perrin V, Coindre JM, Delattre O. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44(4):461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5(12):1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhi B, Amare P, Gulia A, Baisane C, Patil A, Agarwal S, Puri A, Jambhekar NA. Primary intraosseous myoepithelioma arising in the iliac bone and displaying trisomies of 11, 15, 17 with del (16q) and del (22q11)--A rare case report with review of literature. Pathol Res Pract. 2011;207(12):780–785. doi: 10.1016/j.prp.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS, Demichelis F, Rubin MA, Gerstein MB. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11(10):R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel CA, Selleri L, Jacobs Y, Warnke R, Cleary ML. Expression of Pbx1b during mammalian organogenesis. Mech Dev. 2001;100(1):131–135. doi: 10.1016/s0925-4773(00)00516-5. [DOI] [PubMed] [Google Scholar]
- Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, Flanagan J, Luo Y, Fenwick K, Natrajan R, Mitsopoulos C, Zvelebil M, Hoch BL, Weiss SW, Debiec-Rychter M, Sciot R, West RB, Lazar AJ, Ashworth A, Reis-Filho JS, Lord CJ, Gerstein MB, Rubin MA, Rubin BP. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. 2011;3(98):98–82. doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- Wagner K, Mincheva A, Korn B, Lichter P, Popperl H. Pbx4, a new Pbx family member on mouse chromosome 8, is expressed during spermatogenesis. Mech Dev. 2001;103(1-2):127–131. doi: 10.1016/s0925-4773(01)00349-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.