Abstract
Sialyltransferase structures fall into either GT-A or GT-B glycosyltransferase fold. Some sialyltransferases from the Photobacterium genus have been shown to contain an additional N-terminal immunoglobulin (Ig)-like domain. Photobacterium damselae α2–6-sialyltransferase has been used efficiently in enzymatic and chemoenzymatic synthesis of α2–6-linked sialosides. Here we report three crystal structures of this enzyme. Two structures with and without a donor substrate analogue CMP-3F(a)Neu5Ac contain an immunoglobulin (Ig)-like domain and adopt the GT-B sialyltransferase fold. The binary structure reveals a non-productive pre-Michaelis complex, which are caused by crystal lattice contacts that prevent the large conformational changes. The third structure lacks the Ig-domain. Comparison of the three structures reveals small inherent flexibility between the two Rossmann-like domains of the GT-B fold.
Keywords: α2–6-sialyltransferase, protein crystal structure, sialic acid, Photobacterium damselae, CMP-3F(a)Neu5Ac
1. Introduction
Sialic acids are α-keto acids usually found as the terminal carbohydrate moieties attached to cell surface glycoconjugates of higher eukaryotes [1]. The terminal position allows the sialic acids to function as key recognition sites that can be used for multi-purposes by cells such as modulating cellular interactions or masking molecules for invasion of host systems [2, 3]. The important roles of sialic acids in numerous pathological and physiological processes, including tumor metastasis and bacterial and viral infections, make sialyltransferases, the key enzymes involved in the biosynthesis of sialic acid-linked glycans, potential therapeutic targets [4].
Sialyltransferases are enzymes that catalyze the transfer of sialic acid from its activated sugar nucleotide form, cytidine 5’-monophosphate sialic acid (CMP-sialic acid or CMP-Sia), to the terminal positions of glycans on glycoproteins and glycolipids [2, 5]. Sialyltransferases can link sialic acids either through an α2–3- or an α2–6-bond to galactose (Gal), an α2–6-bond to Nacetylgalactosamine (GalNAc) or N-acetylglucosamine (GlcNAc), or an α2–8/9-bond to another sialic acid to form polysialic acid. Some bacterial sialyltransferases also display multifunctionality. For example, Pasteurella multocida sialyltransferase 1 (PmST1) has optimal activities as an α2–3-sialyltransferase at pH 7.5–9.0, an α2–6-sialyltransferase at pH 4.5–6.0, an α2-3-sialidase at pH 5.0–5.5 and an α2–3-trans-sialidase at pH 5.5–6.5 [6]. Multifunctionalities including trans-sialidase and sialidase activities in addition to sialyltransferase activities have also been found for Campylobacter jejuni sialyltransferase CstII [7] and Photobacterium damselae α2–6-sialyltransferase Pd2,6ST [8]. Investigation of the structures of sialyltransferases will hence contribute to further understanding of the detailed mechanism and various functions of this important class of enzymes.
Among the sialyltransferases known to date, crystal structures of representative members of Carbohydrate Activated enZyme (CAZy, http://www.cazy.org) [9, 10] sialyltransferases from families GT29, GT42, GT52, and GT80, have been reported. Based on their protein sequence similarity, all mammalian sialyltransferases are grouped into CAZy GT29 family. The crystal structures of two members of this mammalian sialyltransferase family, including porcine ST3Gal I [11] and rat ST6Gal I [12], have been reported. Among bacterial sialyltransferases, the structures of a multifunctional Campylobacter jejuni α2–3/8- sialyltransferase Cst-II [13] and Campylobacter jejuni α2–3-sialyltransferase Cst-I [14] belonging to CAZy GT42 family and Neisseria meningitidis lipopolysaccharide α2–3-sialyltransferase [15] belonging to CAZy GT52 family have been solved by X-ray crystallography. In addition, the protein crystal structures of several GT80 family bacterial sialyltransferases, including PmST1 [2, 16, 17], Vibrionaceae Photobacterium sp. JT-ISH-224 α2–6-sialyltransferase (Psp26ST) [18], and Photobacterium phosphoreum α/β-galactoside α2–3-sialyltransferase (Pp23ST) [19] have been reported. However, structures have not been solved for polysaccharide synthases catalyzing the formation of sialic acid-containing polysaccharides in mammals and bacteria, such as mammalian α2–8-polysialyltransferases belonging to GT29, Neisseria meningitidis serogroups B and C and Escherichia coli K-1 and K-92 α2–8/9-polysiayltransferases belonging to GT38, and capsular polysaccharide synthases of Neisseria meningitidis serogroups W135 and Y belonging to CAZy GT4 family [20].
Among solved sialyltransferase crystal structures, members belonging to GT29, GT42, and GT52 families all have one single Rossmann domain and fall into glycosyltransferase GT-A or GT-A-like structures [21, 22]. In contrast, crystal structures of GT80-family sialyltransferases identified so far belong to glycosyltransferase GT-B type, which contain two Rossmann-like domains.
Here we present the structures of α2–6-sialyltransferase from Photobacterium damselae, Δ15Pd2,6ST(N) [23] in the ligand-free form and complexed with cytidine 5'-monophosphate-3- fluoro(axial)-N-acetylneuraminic acid [CMP-3F(a)Neu5Ac] as well as Δ112Pd2,6ST(N) in the ligand-free form resolved to resolutions of 1.93 Å, 1.70 Å, and 2.30 Å, respectively. Pd2,6ST is an α2–6-sialyltransferase that catalyzes the transfer of sialic acid from the activated sugar nucleotide donor CMP-Sia to the Gal or the GalNAc in an acceptor [24]. It was the first bacterial α2–6-sialyltransferase that was cloned [25]. Pd2,6ST has weaker α2–6-trans-sialidase and α2– 6-sialidase activities [8] and is grouped to the CAZy GT80 family (EC 2.4.99.1). The structures presented here of Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N) can provide valuable information for developing potential drugs and generating mutants for chemoenzymatic synthesis of oligosaccharides [26, 27].
2. Materials and methods
2.1. Expression and Purification of Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N)
Cloning, expression, and purification were performed as previously reported [23]. For expression, E. coli BL21 (DE3) cells containing recombinant plasmid in pET15b vector were cultured in LB-rich medium (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) supplemented with 100 μg/mL ampicillin. The cells were grown to OD600 nm = 0.8–1.0, induced with 0.1 mM of isopropyl-1-thio-β-D-galactopyranoside (IPTG) for over-expression of the target protein, and incubated at 20 °C with shaking at 250 rpm in a C25KC incubator shaker (New Brunswick Scientific, Edison, NJ) for 24 h. The cells were harvested by centrifugation at 4 °C in a Sorvall Legend RT centrifuge with a swinging bucket rotor at 4,000 g for 2 h. The cell pellet was resuspended in 20 mL/L cell culture in lysis buffer (pH 8.0, 100 mM Tris-HCl containing 0.1% Triton X-100) supplemented with lysozyme (1 mg/L culture) and DNaseI (50 μg/L culture) and incubated at 37 °C for 50 min with shaking (125 rpm). The cell lysate was collected by centrifugation (Sorvall RC-5B centrifuge with a S5-34 rotor) at 12,000 g for 30 min and the lysate was applied to a Ni2+-NTA affinity column to purify the target protein. After loading, the Ni2+ column was washed with 10 column volumes of binding buffer (5 mM imidazole, 0.5 M NaCl, and 20 mM Tris-HCl, pH 7.5), 15 column volumes of washing buffer (20–40 mM imidazole, 0.5 M NaCl, and 20 mM Tris-HCl, pH 7.5), followed by 8 volumes of elute buffer (200 mM imidazole, 0.5 NaCl, 20 mM Tris/HCl, pH 7.5). The fractions containing the purified enzyme were collected. The combined sample was dialyzed against Tris-HCl buffer (20 mM, pH7.5), and stored at 4 °C.
2.2. Crystallization of Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N)
Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N) were concentrated to 10 mg/mL using centrifugal filter units (EMD Millipore, Billerica, MA, USA) and crystallized by hanging drop vapor diffusion in a 1:1 ratio of protein and reservoir solution at 21 °C. The reservoir solution for Δ15Pd2,6ST(N) contained PEG6000 (20%, w/v), NaCl (0.2 M), and HEPES buffer (0.1 M, pH 7.0). The reservoir solution for Δ112Pd2,6ST(N) contained PEG1000 (20%, w/v), Ca(OAc)2 (0.2 M), and imidazole (0.1 M) at pH 8.0. Δ15Pd2,6ST(N) binary structure with CMP-3F(a)Neu5Ac (‘a’ stands for the axial position of the fluorine at carbon 3) was obtained by soaking with 1.25 mM of CMP-3F(a)Neu5Ac (final concentration) for 30 min prior to cryocooling and data collection. The CMP-3F(a)Neu5Ac analogue was synthesized as previously described [16, 28]. The crystals were placed in a reservoir solution containing 20% ethylene glycol (for both crystals of Δ15Pd2,6ST(N)) or Paratone-N oil (for crystal of Δ112Pd2,6ST(N)) and flash cooled in liquid nitrogen prior to data collection.
2.3. Data collection, structure determination, model building, and refinement
X-Ray diffraction data for the crystals were collected at Stanford Synchrotron Radiation Lightsource (SSRL) beam line 7-1 at 100 K. The SSRL data were indexed and integrated with MOSFLM [29], and then scaled with SCALA [30]. A complete data set for the ligand-free Δ15Pd2,6ST(N) structure was collected to a resolution of 1.93 Å (Table 1). Soaking the crystal with the non-hydrolyzable analog of CMP-3F(a)Neu5Ac actually improved the resolution of the crystal. A complete data set was collected to 1.70 Å resolution (Table 1). Finally, the Δ112Pd2,6ST(N) crystal only diffracted to 2.30 Å resolution for collecting a complete data set (Table 1).
Table 1.
Data collection and Refinement Statistics
| Δ15Pd2,6ST(N) Ligand-free | Δ15Pd2,6ST(N) Soaked w/ Neu5Ac-3F(a)-Neu5Ac | Δ112Pd2,6ST(N) Ligand-free | |
|---|---|---|---|
| X-ray Source | SSRL BL 7-1 | SSRL BL 7-1 | SSRL BL 7-1 |
| Wavelength (Å) | 0.97946 | 0.97946 | 0.97607 |
| Space Group | P1 | P1 | C2 |
| Cell parameters | a = 64.7 Å b= 81.3 Å c = 111.8 Å α = 98.60° β = 92.76° γ = 100.85° |
a = 64.3 Å b= 81.4 Å c = 111.2 Å α = 99.02° β = 91.59° γ = 101.20° |
a = 95.4 Å b= 56.5 Å c = 87.4 Å α = 90° β = 119.95° γ = 90° |
| Monomers/ASU | 4 | 4 | 1 |
| VM (Å3/Da) | 2.53 | 2.50 | 2.30 |
| Percent Solvent | 51.4% | 50.9% | 46.5% |
| Resolution (Å) | 1.93 (1.98 – 1.93) | 1.70 (1.79 – 1.70) | 2.30 (2.36 – 2.30) |
| No of reflections | 311,576 (23,059) | 455,871 (61,700) | 63,335 (4,671) |
| No. of unique reflections | 156,324 (11,570) | 229,782 (31,361) | 17,873 (1,287) |
| Completeness (%) | 94.2 (94.0) | 95.7 (89.3) | 99.1 (99.2) |
| Rmergea (%) | 2.9 (39.7) | 4.2 (31.5) | 6.7 (32.0) |
| I/σ | 18.4 (2.16) | 8.8 (2.2) | 16.9 (3.6) |
| Refinement Statistics | |||
| Resolution (Å) | 1.93 | 1.70 | 2.30 |
| No. of reflections (F>0) | 156,284 | 229,738 | 17,868 |
| Rfb (%) | 18.3 | 17.1 | 18.4 |
| Rfreec (%) | 22.4 | 20.0 | 24.8 |
| RMS bond length (Å) | 0.008 | 0.007 | 0.008 |
| RMS bond angle (°) | 1.123 | 1.091 | 1.075 |
| Wilson B-factor (Å2) | 31.9 | 23.8 | 38.3 |
| Refined Average B-factor (Å2) | 45.0 | 33.1 | 44.3 |
| Ramachandran Plot Statistics | |||
| Most Favorable (%) | 1851 (98.1%) | 1850 (98.0%) | 372 (96.9%) |
| Allowed (%) | 31 (1.6%) | 34 (1.8%) | 12 (3.1%) |
| Disallowed (%) | 5 (0.3%) | 4 (0.2%) | 0 (0.0%) |
| Asymmetric Unit Content | |||
| Non-hydrogen protein atoms | 15,261 | 15,274 | 3099 |
| Calcium ions | 4 | 4 | 1 |
| CMP-3F(a)Neu5Ac atoms | 0 | 168 (4 molecules) | 0 |
| Waters | 1,395 | 2,441 | 126 |
| PDB ID | 4R83 | 4R84 | 4R9V |
Rmerge = [ΣhΣi|Ih – Ihi|/ΣhΣiIhi] where Ih is the mean of Ihi observations of reflection h.
Numbers in parenthesis represent highest resolution shell.
R and
Rfree = Σ||Fobs| - |Fcalc|| / Σ|Fobs| x 100 for 95% of recorded data (R) or 5% data (Rfree)
The Δ112Pd2,6ST(N) ligand-free structure was obtained by molecular replacement using a previously solved sialyltransferase structure from Vibrionaceae Photobacterium sp. JTISH- 224 (Δ16Psp26ST) (PDBID: 2Z4T) [18]. The program PHASER [31] as a part of the PHENIX suite [32] was used for molecular replacement by splitting the GT-B domain of Δ16Psp26ST structure into two separate Rossmann-like search domains. The Δ15Pd2,6ST(N) structure was also solved by molecular replacement using the two Rossmann-like domains of the Δ112Pd2,6ST(N) structure and the N-terminal Ig-like domain from Δ16Psp26ST (PDBID: 2Z4T). The atomic model building was carried out with the molecular graphics program COOT [33]. The structures were refined using the program PHENIX [34] using 95% of the measured data as a target function. Noncrystallographic restraints (for Δ15Pd2,6ST(N)) and TLS parameters [35] were included during refinement. The final R and R-free for all three structures, along with the quality of the model based on PROCHECK [36] are listed in Table 1.
2.4. Effects of ethylene glycol tetraacetic acid (EGTA) and Ca2+ on the α2–6-sialyltransferase activity of Δ15Pd2,6ST(N)
EGTA (10 mM), CaCl2 (10 mM), or the mixture of EGTA (15 mM) and CaCl2 (5 mM) were used in Tris-HCl buffer (100 mM, pH 8.5) containing cytidine 5'-monophosphate-Nacetylneuraminic acid (CMP-Neu5Ac) (1 mM) and LacβMU (1 mM) to analyze their effects on the α2–6-sialyltransferase activity of Δ15Pd2,6ST(N) (16.5 ng/μL). A reaction without EGTA or CaCl2 was used as a control. Reactions were allowed to proceed for 15 min at 37 °C followed by quenching with ice-cold acetonitrile (12%, v/v) and tested by high performance liquid chromatography (HPLC) [23].
3. Results and discussion
3.1. Overall native structure
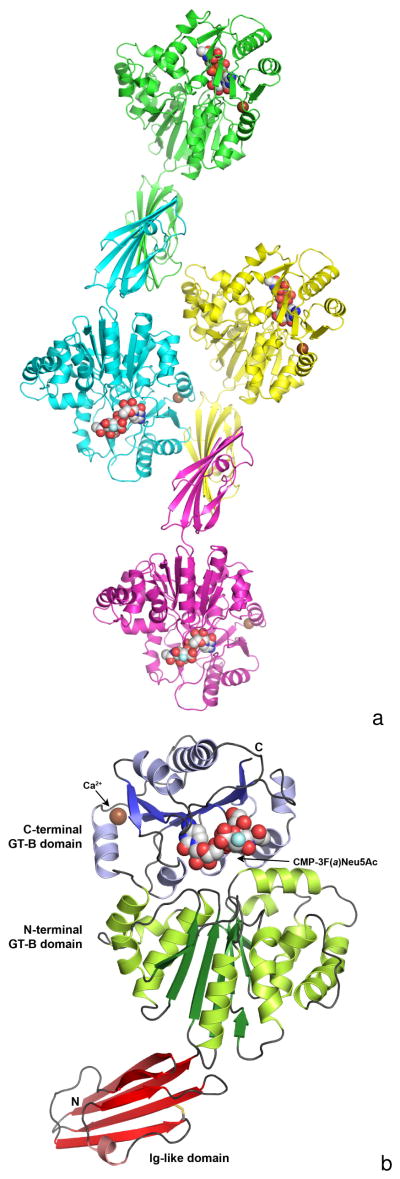
Two soluble Pd2,6ST constructs, Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N) deleting the first 15 and 112 residues respectively, were expressed and crystallized. Residues 1–15 contain a putative membrane anchor, and residues 24–111 contain an Ig-like domain of unknown function. Both constructs lack the C-terminal 178 residues, which constitute a PhoU domain. The PhoU domain is typically involved in phosphate regulation, specifically in regulating phosphate uptake [37]. The role or function of this domain at the C-terminal end of the sialyltransferase in Photobacterium damselae is unknown. Both constructs also include 21 additional residues at the N-terminus corresponding to a His6-Tag, which was not cleaved prior to crystallization. Because the Δ15Pd2,6ST(N) construct was more complete, diffracted to higher resolution, and substrate could be soaked into the active site, much of the discussion will be focused on the structures of this construct.
Crystals of Δ112Pd2,6ST(N) were grown in a couple of days at room temperature. They belong to the monoclinic space group C2 and contain one monomer in the asymmetric unit. Residues 112–497 of this crystal structure are well ordered in the electron density map (residue 112 is from the pET15b vector). Crystals of the larger Δ15Pd2,6ST(N) construct were also grown in a few days, but were crystallized in the triclinic space group P1. The Δ15Pd2,6ST(N) crystals contain four monomers per asymmetric unit, with each monomer consisting of ordered residues 24–497 organized into three separate domains (Figure 1A). In each monomer, the first domain (residues 24–111, Ig-like domain) is the smallest domain and adopts the Ig-like beta-sandwich fold that has been found in some members of the glycosyl hydrolase family (Figure 1B) [38]. The Ig-like domain was also found in Δ16Psp26ST, where it might play a regulatory role because deleting the Ig-like domain increased the specific activity of the sialyltransferase by two-fold [18]. In contrast, deleting the Ig-like domain in the Δ112Pd2,6ST(N) actually decreased kcat/KM about two-fold, mostly by decreasing the kcat value [23].
Figure 1.
Figure 1A. The composition of a crystallographic asymmetric unit. The structure of a Δ15Pd2,6ST(N) crystal soaked with CMP-3F(a)Neu5Ac is shown here in triclinic space group P1 with four monomers per asymmetric unit. Each monomer is colored differently, the calcium ions are illustrated as a brown-colored spheres and the bound CMP-3F(a)Neu5Ac molecules are shown as space filling spheres with white-colored carbon atoms. The fluorine atoms are colored light blue.
Figure 1B. A ribbon representation of the Δ15Pd2,6ST(N) monomer. The N-terminal Ig-like domain is at the bottom and is colored red, while the two-domain GT-B fold is at the top. The N-terminal GT-B domain is colored green (dark green for β-strands and light green for α-helices), while the C-terminal domain is colored blue (dark blue for β-strands and light blue for α-helices). The bound CMP-3F(a)Neu5Ac is represented by space filling spheres between the two Rossmann domains of the GT-B fold. The calcium ion is shown as a brown-colored sphere and the disulfide bond in the Ig-like domain is illustrated as sticks colored yellow.
The Ig-like beta-sandwich domain contains a four-stranded anti-parallel β-sheet on one side (topology β1-β6-β5-β3), covered by a two-stranded anti-parallel β-sheet (β2-β4) and a single short α-helix (between β3-β4) on the other side of the beta-sandwich (Figure 1B). A disulfide bond is formed between Cys60 and Cys89 on β3 and β5 respectively, which is also observed in the Ig-domain of the Δ16Psp26ST structure.
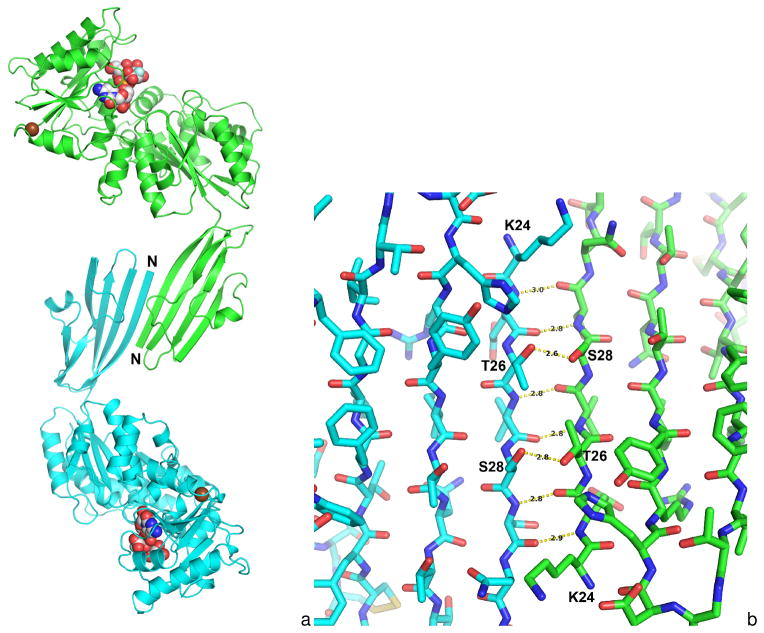
In the Δ15Pd2,6ST(N) structure, the Ig-like domains from two adjacent monomers form a tight crystal contact across a non-crystallographic 2-fold axis. The β1 strands from two monomers adopt a dimeric anti-parallel β-sheet interaction resulting in a continuous eightstranded β-sheet across the two Ig-like domains (Figure 2A). This contact is observed between both the A–B monomers and the C–D monomers within the crystallographic asymmetric unit (Figure 1A). The β1 strand from each monomer is situated at the non-crystallographic 2-fold axis where residues 24–29 formed six main-chain hydrogen bonds together with two side-chain hydrogen bonds between Thr26 and Ser28 (Figure 2B). This Ig-domain dimerization is not observed for the Δ16Psp26ST structure, so the biological relevance of this interaction remains to be determined for Photobacterium damselae.
Figure 2.
Figure 2A. Dimeric contacts between two Δ15Pd2,6ST(N) monomers. A- and B-chain Δ15Pd2,6ST(N) monomers in the asymmetric unit are shown with the Ig-like domain mediating a dimeric contact. The first strand of the Ig-like domain forms β-strand interaction across a noncrystallographic 2-fold resulting in a continuous eight-stranded β-sheet dimer interaction. A similar association is observed for the C- and D-chains in the asymmetric unit.
Figure 2B. A close-up view of the non-crystallographic 2-fold illustrating the dimer contacts between the Ig-like domains of the A and B-subunits in the asymmetric unit. Yellow dashed lines show hydrogen bonds with distances in Ångstroms shown in black texts. Six main-chain hydrogen bonds are formed between the side chains of Thr26 and Ser28 across the dimers.
For the second and third domains of Δ15Pd2,6ST(N) (residues 112–334 named as Nterminal GT-B domain and 335–497 named as C-terminal GT-B domain, respectively, equivalent to the Δ112Pd2,6ST(N) construct), each contains an α/β/α fold resembling the Rossmann nucleotide-binding domain. Together, the domains form a structure found in the glycosyltransferase B (GT-B) superfamily, which is composed of two Rossmann domains separated by a deep cleft that forms a substrate-binding site (Figure 1B) [39, 40]. In the Δ15Pd2,6ST(N) and Δ112Pd2,6ST(N) structures, the N-terminal GT-B domain (residues 112– 334) contains a central seven-stranded parallel β-sheet (dark green, Topology β9-β8-β7-β10-β11-β12-β13) flanked by seven α-helices on one side and four helices (light green) on the other side of the central β-sheet. The C-terminal GT-B domain (residues 335–497) is a smaller Rossmann fold composed of a central six-stranded parallel β-sheet (dark blue, Topology β16-β15-β14-β17-β18-β19) flanked by two α-helices on one side and five α-helices (light blue) on the other side. The CMP-3F(a)Neu5Ac substrate analogue interacts more with this C-terminal domain (see below).
Overall, the structure of Δ15Pd2,6ST(N) has a similar architecture to sialyltransferases from Photobacterium sp. JT-ISH-224 [18] and Photobacterium leiognathi [41, 42], which contain a membrane anchor, an N-terminal Ig-like domain, followed by a GT-B sialyltransferase domain. It is interesting to point out that the Photobacterium damselae sialyltransferase also contains a C-terminal PhoU domain that shares amino acid sequence similarity to phosphate regulatory proteins [43]. The Δ112Pd2,6ST(N) structure lacks the Ig-like domain and simply contains the GT-B fold. Nevertheless, its structure and conformation are similar to those found in the Δ15Pd2,6ST(N) structure (see below).
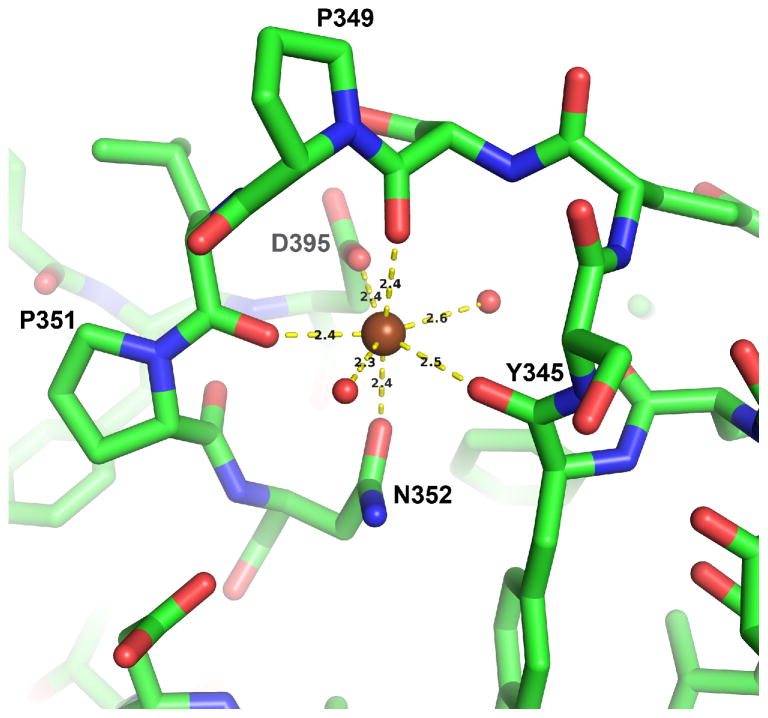
3.2. Metal binding site
Strong electron density is observed at the loop between the first α-helix of the C-terminal domain and β14 in all four monomers within the asymmetric unit of the Δ15Pd2,6ST(N) structures and in the Δ112Pd2,6ST(N) structure. This density is consistent with a divalent metal ion, which we modeled as a calcium ion based on results from the metal identification feature in PHENIX [44], and the CheckMyMetal web server [45]. The Ca2+ ion is heptavalent and displays a pentagonal bipyramidal geometry. The ion is coordinated by main-chain oxygen atoms of Tyr345, Ser348, and Leu350, along with side chain oxygens of Asn352 and Asp395, and two water molecules (Figure 3). All but one of the residues that coordinate the Ca2+ ion come from the loop preceding strand β14. Asp395 is the first residue of neighboring strand β15.
Figure 3.
The calcium binding site. A strong electron density is modeled as a calcium ion in the loop between the first helix and the β-strand of the C-terminal Rossmann domain of the GT-B fold. Yellow dashed lines show coordination interactions with distances in Ångstroms shown in black text. Water molecules are shown as small red-colored spheres. The geometry of coordination is pentagonal bipyramidal. The calcium ion (shown as a brown sphere) is observed in all four monomers in the asymmetric unit for both the Δ15Pd2,6ST(N) ligand-free and the CMP-3F(a)Neu5Ac-bound structures as well as in the Δ112Pd2,6ST(N) structure.
As no divalent metal ions were included in the crystallization of Δ15Pd2,6ST(N), the Ca2+ ion may be retained through protein purification process. A similar divalent metal ion was also observed in the Δ16Psp26ST structure, which was modeled as a Mg2+ ion [18]. The Ca2+ ion in Δ15Pd2,6ST(N) is not near the active site and may play a structural role. Nevertheless, the enzyme still displayed activity in the presence of ethylenediaminetetraacetic acid (EDTA) [23]. Enzymatic activity assays in the presence of 10 mM of ehtylene glycol tetraacetic acid (EGTA), which is more selective than EDTA for calcium ions, revealed that Δ15Pd2,6ST(N) still retained more than 92% activity (data not shown), suggesting the ion is not crucial for sialyltransferase activity.
3.3 Δ15Pd2,6ST(N)—CMP-3F(a)Neu5Ac binary structure
Previous co-crystallization studies of sialyltransferases with their sugar nucleotide donor substrate, CMP-Neu5Ac, only led to the observation of CMP bound in the active site, due to the cleavage of Neu5Ac during crystallization [2, 13]. To prevent the hydrolysis of the donor sugar to identify amino acid residues that may interact with the Neu5Ac moiety, inert analogues of CMP-Neu5Ac, such as CMP-3F(a)Neu5Ac, have been synthesized and used in crystallization. An electronegative fluorine substitution at the C-3 of Neu5Ac in CMP-Neu5Ac destabilizes the oxocarbenium ion-like transition state for the reaction and hence slows down the enzyme turnover rate. Previously, sialyltransferases from Pasteurella multocida and Campylobacter jejuni have been co-crystallized with CMP-3FNeu5Ac to identify residues involved in binding of the nucleotide and the sugar components of the sialyltransferase donor [14, 16]. Here, crystals of Photobacterium damselae Δ15Pd2,6ST(N) were soaked with CMP-3F(a)Neu5Ac and a data set was collected (Table 1).
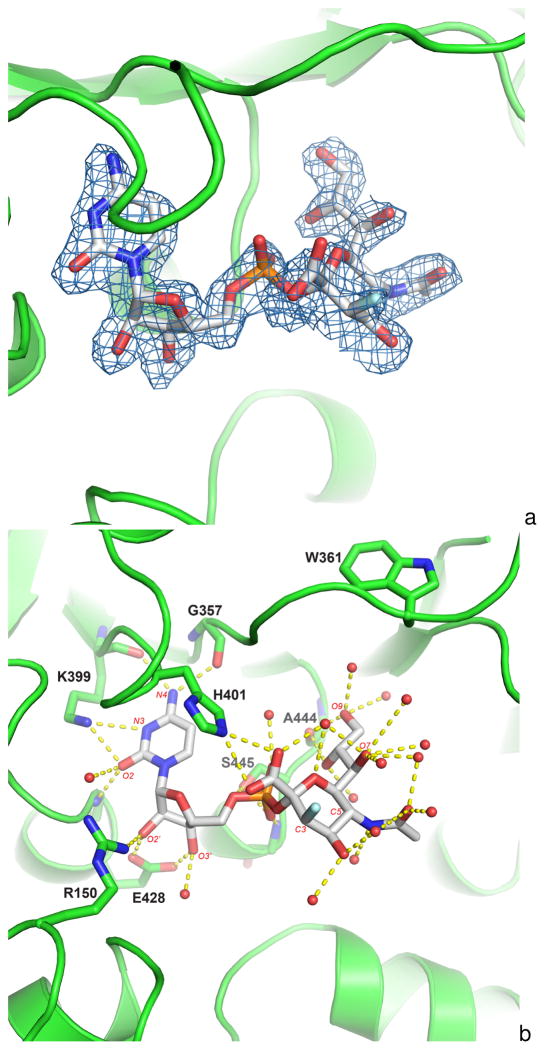
The resulting CMP-3F(a)Neu5Ac-soaked structure (Figure 4A) reveals clearly defined electron density of CMP-3F(a)Neu5Ac in a catalytic site between the two nucleotide binding domains of all four monomers within the crystallographic asymmetric unit. The structure (Figure 4B) identifies similar interactions between the enzyme and the substrate as seen in previous reported structures complexed with a CMP moiety [18]. The CMP moiety interacts almost exclusively with the C-terminal nucleotide-binding domain through a total of ten hydrogen bonds. The cytosine base interacts through N4 with two hydrogen bonds to the main-chain carbonyl oxygens of residues Gly357 and Lys399. The side chain amine group of Lys399 hydrogen bonds to O2 of the cytosine ring. These hydrogen-bonding interactions with the base have also been observed in the previous CMP-bound structures belonging to the GT-B group [2, 18, 19]. Additionally, as seen in other GT-B sialyltransferase structures with CMP bound, a conserved Glu side chain makes a bidentate interaction with both the 2’ and 3’ hydroxyls of the ribose ring (Figure 4B). The phosphate group hydrogen bonds to main chain nitrogen of Ser446 and side chain of Ser445.
Figure 4.
Figure 4A. The original electron density map around the donor analogue after soaking Δ15Pd2,6ST(N) crystals with CMP-3F(a)Neu5Ac prior to data collection. The electron density is calculated from the original molecular replacement solution using the three ligand-free structure domains as search models. The electron density is contoured at 1σ displayed with the final refined structure. Similar density is seen for all four active sites in the asymmetric unit. CMP- 3F(a)Neu5Ac is shown with white-colored carbon atoms. Fluorine and phosphorous atoms are colored cyan and orange respectively.
Figure 4B. The substrate analogue CMP-3F(a)Neu5Ac binding site of Δ15Pd2,6ST(N). The protein is illustrated as a cartoon. The side chain and the main chain atoms of important residues are drawn as sticks. CMP-3F(a)Neu5Ac is drawn as sticks with white-colored carbon atoms with some atoms labeled in red text. Yellow dashes show potential hydrogen bonds between the substrate analogue and the protein or ordered water molecules (red spheres). The fluorine atom in the sialic acid is colored cyan. Trp361 is labeled. It has been observed to swing down and interact with the sialic acid in other sialyltransferases.
Surprisingly, conserved His401 that normally interacts with the CMP phosphate group in other sialyltransferases [16], is seen to form an ion-pair with the carboxylate group of the sialic acid moiety. Also unexpectedly, conserved Arg150 of the N-terminal domain, which typically ionpairs with the sialic acid carboxylate group [16], interacts with the 2’-OH of the ribose ring here in the Δ15Pd2,6ST(N) structure, and represents the only interaction between CMP- 3F(a)Neu5Ac and the N-terminal Rossmann domain.
For the sialic acid moiety of CMP-3F(a)Neu5Ac in the Δ15Pd2,6ST(N) structure, the majority of contacts are with ordered water molecules (Figure 4B). Only O8 and O9 of the sialic acid are in hydrogen bonding distance to carbonyl oxygen of Ala444 of the C-terminal domain, and no interactions are observed to the N-terminal Rossmann domain. These unique interactions are likely due to the binding of CMP-3F(a)Neu5Ac to the sialyltransferase in an open conformation.
Because CMP-3F(a)Neu5Ac was soaked into ligand-free Δ15Pd2,6ST(N) crystals to obtain this binary structure, crystal packing and numerous lattice contacts, especially between the C-terminal domain of the GT-B fold, impedes the substrate-induced large conformational changes seen in other sialyltransferases that were obtained by co-crystallization with substrate analogues [16, 18]. We have previously shown large conformational changes in PmST1 upon binding to CMP and CMP-3F(a)Neu5Ac [2, 16]. In PmST1, distances between loops that close in over the substrate-binding site move from ~30Å separation to ~15Å [2, 16]. The equivalent loop distances in the Δ15Pd2,6ST(N) move only from ~27Å apart in the ligand-free structure to ~25Å in the CMP-3F(a)Neu5Ac-binary structure. Additionally, in the previously reported structure of the homologous Δ16Psp26ST [18] which contains an N-terminal Ig-like domain as Δ15Pd2,6ST(N), CMP binding in the active site resulted in a closed conformation of the enzyme where the distance between equivalent loops are ~14 Å apart [18]. Thus, the Δ15Pd2,6ST(N) binary structure with CMP-3F(a)Neu5Ac bound presented here provides a unique glimpse of sugar nucleotide donor binding prior to a conformational change. This also suggests that the conformational change (closure of the two Rossmann domains) is not required for donor substrate binding.
The previously determined sialyltransferase structures with nucleotide bound in the closed conformation also reveal a big movement of a conserved tryptophan (Trp) residue of the C-terminal domain. In the ligand-free structure, this Trp residue (Trp270 of PmST1) is buried in the hydrophobic core of the C-terminal Rossmann domain. Upon binding to nucleotide, Trp270 residue pops out of the core and swings down where the indole nitrogen hydrogen bonds with sialic acid, which helps to shape the acceptor sugar binding site [16]. In the Δ15Pd2,6ST(N) structure here, Trp361 (equivalent to Trp270 of PmST1) remains buried in the C-terminal hydrophobic core (Figure 4B), suggesting CMP-3F(a)Neu5Ac binding alone is not enough to release Trp361. In the Δ16Psp26ST structure, which is in the closed conformation with CMP and lactose bound, homologous Trp365 is released from the C-terminal core and interacts with lactose [18].
The conformational closure upon binding to donor substrate does not only help to shape the acceptor binding site, but also position catalytically important residues in close proximity to the acceptor substrate to catalyze the sialic acid transfer. Specifically, a conserved aspartic acid in the N-terminal domain serves as a catalytic base to deprotonate the acceptor hydroxyl to create a better nucleophile to attack the anomeric carbon of CMP-Neu5Ac [16]. This aspartate residue hydrogen bonds with the O3 of lactose in the PmST1 ternary structure, leading to the formation of an α2-3-linkage with sialic acid catalyzed by the enzyme. It also hydrogen bonds to the O6 of lactose in the Δ16Psp26ST ternary structure, allowing the formation of the α2–6-sialyl linkage catalyzed by the enzyme. In the Δ15Pd2,6ST(N) structure here, Asp229 is over 10 Å away from the sialic acid anomeric carbon because the conformation of Δ15Pd2,6ST(N) is in the open state. Therefore, the Δ15Pd2,6ST(N) binary structure with CMP-3F(a)Neu5Ac bound reported here likely represents a conformation of a pre-Michaelis complex structure when the donor substrate binds prior to a conformational change.
3.4. Structural comparisons
Superposition of the four monomer structures found in the triclinic asymmetric unit shows very similar structures with the largest differences coming from the relative orientation of the N-terminal Ig-like domain with respect to the GT-B sialyltransferase domain (Figure 5A). Overall, the root-mean-square deviations (rmsd) between the four CMP-3F(a)Neu5Ac-bound structures within the asymmetric unit range from 0.35–1.07 Å for all 474 α-carbons. Aligning only the GT-B domains of each monomer reveals that the GT-B domains are more closely aligned with rmsd range 0.21–0.32 Å for 374 equivalent α-carbons.
Figure 5.
Figure 5A. A superposition of all four monomers of the CMP-3F(a)Neu5Ac-bound Δ15Pd2,6ST(N) structures seen in the triclinic P1 asymmetric unit. Each monomer is colored differently and only the GT-B folds are aligned, illustrating a small shift in Ig-like domain orientations. The subunits GT-B domains are superimposed with rmsd ranges 0.21–0.32 Å for 374 equivalent α-carbons.
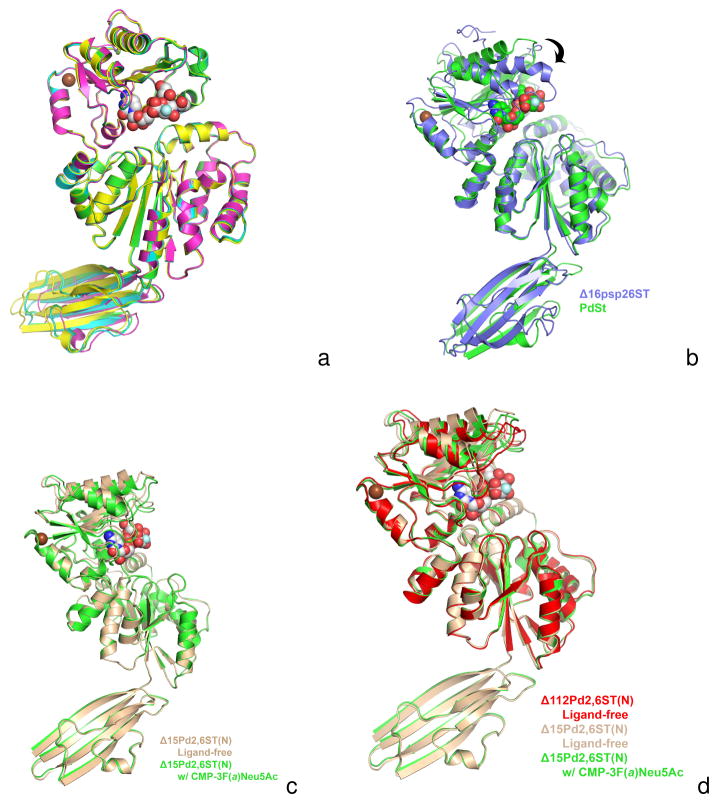
Figure 5B. A superposition of the Δ15Pd2,6ST(N) structure (green) onto V. Photobacterium sp. JT-ISH-224 sialyltransferase (Δ16Psp26ST) (blue). The donor substrate analogue CMP- 3F(a)Neu5Ac bound in Δ15Pd2,6ST(N) is shown in space filling spheres with green-colored carbon atoms. The CMP of the Δ16Psp26ST structure is not shown for clarity, but lies very close to the CMP moiety of CMP-3F(a)Neu5Ac bound in Δ15Pd2,6ST(N). Only the N-terminal Rossmann domains of the GT-B fold are aligned (rmsd 0.95 Å) illustrating the differences in closure around the active site (C-terminal Rossmann domain swings down as shown by arrow) upon binding activated donor. The Δ15Pd2,6ST(N) structure with CMP-3F(a)Neu5Ac bound resembles, more closely, other GT-B sialyltransferases in the ligand-free state (not shown). The alignment also illustrates the different orientations of the Ig-like domains of two structures.
Figure 5C. A superposition between the ligand-free (tan) and CMP-3F(a)Neu5Ac-bound (green) structures of Δ15Pd2,6ST(N). The alignment shows a small closure between the two domains of the GT-B fold upon soaking in CMP-3F(a)Neu5Ac (white-colored carbon spheres). Crystal packing and lattice contacts may prevent the larger closure seen in other GT-B sialyltransferases [2, 18].
Figure 5D. A superposition of the Δ112Pd2,6ST(N) construct lacking the Ig-like domain and substrates (red) and the ligand-free (tan) and CMP-3F(a)Neu5Ac-bound (green) structures of Δ15Pd2,6ST(N). The alignment shows the most closed structure occurs in the ligand free Δ112Pd2,6ST(N). This is likely due to crystal contacts illustrating the inherent flexibility between the two Rossmann-like domains. The CMP-3F(a)Neu5Ac seen in the Δ15Pd2,6ST(N) binary structure is drawn as white-colored carbon spheres.
Comparing the binary Δ15Pd2,6ST(N) structure to the ternary Δ16Psp26ST structure cocrystallized with CMP and lactose (also containing an N-terminal Ig-like domain) clearly shows the structural conformational change that occurs upon binding substrates to generate a productive complex. As shown in Figure 5B, superposing the N-terminal Rossmann domains reveals a large difference in orientation between the two C-terminal Rossmann domains. Each individual domain superimposes with rmsd of 0.95 Å and 1.37 Å for the N-terminal and C-terminal Rossmann domains respectively, and an rmsd of 1.95 Å for the overall GT-B fold between the Δ15Pd2,6ST(N) and the Δ16Psp26ST. If all three domains are included, the rmsd is 2.49 Å, which also includes the effect due to the difference in orientation between the N-terminal Ig-like domain and the overall GT-B domain (Figure 5B).
In spite of crystal packing contacts, which prevent large conformational changes upon substrate binding during the crystal soaking, a slight closure between the two Rossmann domains upon substrate binding is identified when comparing the binary complex to the ligand-free structure (Figure 5C). The loop containing Trp361 closes in about 1.8 Å on the substrate, which compares to a 7.4 Å α-carbon movement observed in PmST1 upon substrate binding [16]. The rmsd between the GT-B domains alone of the binary complex and the ligand-free structure is 0.64 Å for 374 equivalent α-carbons, and the superposition reveals a slight movement in orientation between the Ig-like domain and the GT-B fold (Figure 5C).
Comparing the Δ15Pd2,6ST(N) to the Δ112Pd2,6ST(N) lacking the Ig-like domain reveals greater conformational flexibility between the two Rossmann-like domains. The ligand-free Δ112Pd2,6ST(N) structure is actually in a slightly more closed conformation compared to the substrate-bound Δ15Pd2,6ST(N) structure (Figure 5D). This is likely because the truncated Δ112Pd2,6ST(N) crystallizes in a different space group thus creating different lattice contacts. While this structure is slightly more closed, it still does not resemble the completely closed structure seen in other GT-B folded sialyltransferases co-crystallized with CMP or CMP-Neu5Ac analogues. However, comparing all three Pd2,6ST structures suggests there is some inherent flexibility between the two Rossmann-like domains of the GT-B sialyltransferases even in the absence of substrates.
4. Conclusions
The crystal structures of Δ15Pd2,6ST(N) have provided additional insights into the reaction mechanism and the function of conserved residues important in catalyzing the transfer of sialic acids. Δ15Pd2,6ST(N) is found to contain three separate domains. The first domain at the N-terminus adopts an Ig-like fold. The second and third domains are each a Rossmann fold bearing a deep catalytic binding cleft in between, a characteristic of members in the GT-B structural group. Soaking the donor substrate analogue CMP-3F(a)Neu5Ac into the crystal prior to data collection provided a structure with a potential pre-Michaelis complex of sugar nucleotide bound in the enzyme. This represents a stage before a conformational closure between the two Rossmann domains is induced. It is interesting to note that other glycosyltransferases with GT-B folds, that are not sialyltransferases, do not exhibit large closures between the two Rossmann domains observed for other reported GT-B sialyltransferase structures. Most likely this is due to the presence of the C-terminal “backbone” that traverses both the C-terminal and N-terminal Rossmann domains in non-sialyltransferase glycosyltransferases with GT-B folds, such as peptidoglycan glycosyltransferase MurG [46], T4 phage β-glucosyltransferase [47], and UDPglucosyltransferase GtfB [48]. The significance for the large conformational changes of the GTB sialyltransferases is unknown. However, many of these enzymes display substrate promiscuity and alternate activities such as catalyzing the formation of different (α2–3 and α2– 6) sialyl linkages [6, 7, 49, 50], or having even sialidase or trans-sialidase activities [6–8]. Therefore the large conformational changes may be important for these biological functions. Nevertheless, the hypothesis remains to be tested.
Highlights.
Sialyltransferases couple sialic acid to glycans on glycoproteins and glycolipids
Crystal structure of sialyltransferase from Photobacterium damselae has been determined
Structure contains an N-terminal Ig-like domain with the GT-B sialyltransferase fold
Binary structure with CMP-3F(a)Neu5Ac reveals a non-productive pre-Michaelis complex
Acknowledgments
This work was supported by Camille Dreyfus Teacher-Scholar Fund to X. C., National Institutes of Health grants R01HD065122 and R01GM094523. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NICHD, NIGMS, or NIH.
Abbreviations
- CMP-3F(a)Neu5Ac
Cytidine 5'-monophosphate-3-fluoro(axial)-N-acetylneuraminic acid
- CMP-Neu5Ac
cytidine 5'-monophosphate-N-acetylneuraminic acid
- GalNAc
Gal; Nacetylgalactosamine
- EDTA
ethylenediaminetetraacetic acid
- EGTA
ethylene glycol tetraacetic acid
- GlcNAc
N-acetylglucosamine
- Neu5Ac
N-acetylneuraminic acid
- Gal
galactose
- Ig
immunoglobulin
- Sia
sialic acid
- Δ15Pd2,6ST(N)
Photobacterium damselae α2–6-sialyltransferase lacking the N-terminal 15 residues and the C-terminal 178 residues
- Δ112Pd2,6ST(N)
Photobacterium damselae α2–6-sialyltransferase lacking the N-terminal 112 residues and the C-terminal 178 residues
- Δ16Psp26ST
Vibrionaceae Photobacterium sp. JT-ISH-224 α2–6-sialyltransferase lacking the N-terminal 16 residues
Footnotes
Protein coordinates have been deposited in the Protein Data Bank [IDs: 4R83 (ligand-free Δ15Pd2,6ST(N)), 4R84 (Δ15Pd2,6ST(N) with CMP-3F(a)Neu5Ac) and 4R9V (ligand-free Δ112Pd2,6ST(N)].
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen X, Varki A. Advances in the biology and chemistry of sialic acids. ACS Chem Biol. 2010;5:163–176. doi: 10.1021/cb900266r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ni L, Sun M, Yu H, Chokhawala H, Chen X, Fisher AJ. Cytidine 5’-monophosphate (CMP)-induced structural changes in a multifunctional sialyltransferase from Pasteurella multocida. Biochemistry. 2006;45:2139–2148. doi: 10.1021/bi0524013. [DOI] [PubMed] [Google Scholar]
- 3.Schauer R. Chemistry, metabolism, and biological functions of sialic acids. Adv Carbohydr Chem Biochem. 1982;40:131–234. doi: 10.1016/s0065-2318(08)60109-2. [DOI] [PubMed] [Google Scholar]
- 4.Rillahan CD, Antonopoulos A, Lefort CT, Sonon R, Azadi P, Ley K, Dell A, Haslam SM, Paulson JC. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol. 2012;8:661–668. doi: 10.1038/nchembio.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harduin-Lepers A, Recchi MA, Delannoy P. 1994, the year of sialyltransferases. Glycobiology. 1995;5:741–758. doi: 10.1093/glycob/5.8.741. [DOI] [PubMed] [Google Scholar]
- 6.Yu H, Chokhawala H, Karpel R, Yu H, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. A multifunctional Pasteurella multocida sialyltransferase: A powerful tool for the synthesis of sialoside libraries. J Am Chem Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- 7.Cheng J, Yu H, Lau K, Huang S, Chokhawala HA, Li Y, Tiwari VK, Chen X. Multifunctionality of Campylobacter jejuni sialyltransferase CstII: characterization of GD3/GT3 oligosaccharide synthase, GD3 oligosaccharide sialidase, and trans-sialidase activities. Glycobiology. 2008;18:686–697. doi: 10.1093/glycob/cwn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng J, Huang S, Yu H, Li Y, Lau K, Chen X. Trans-sialidase activity of Photobacterium damsela α2,6-sialyltransferase and its application in the synthesis of sialosides. Glycobiology. 2010;20:260–268. doi: 10.1093/glycob/cwp172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326( Pt 3):929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coutinho P, Deleury E, Davies G, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 11.Rao FV, Rich JR, Rakic B, Buddai S, Schwartz MF, Johnson K, Bowe C, Wakarchuk WW, Defrees S, Withers SG, Strynadka NC. Structural insight into mammalian sialyltransferases. Nat Struct Mol Biol. 2009;16:1186–1188. doi: 10.1038/nsmb.1685. [DOI] [PubMed] [Google Scholar]
- 12.Meng L, Forouhar F, Thieker D, Gao Z, Ramiah A, Moniz H, Xiang Y, Seetharaman J, Milaninia S, Su M, Bridger R, Veillon L, Azadi P, Kornhaber G, Wells L, Montelione GT, Woods RJ, Tong L, Moremen KW. Enzymatic basis for N-glycan sialylation: structure of rat alpha2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. J Biol Chem. 2013;288:34680–34698. doi: 10.1074/jbc.M113.519041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu CP, Watts AG, Lairson LL, Gilbert M, Lim D, Wakarchuk WW, Withers SG, Strynadka NC. Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat Struct Mol Biol. 2004;11:163–170. doi: 10.1038/nsmb720. [DOI] [PubMed] [Google Scholar]
- 14.Chiu CP, Lairson LL, Gilbert M, Wakarchuk WW, Withers SG, Strynadka NC. Structural analysis of the alpha-2,3-sialyltransferase Cst-I from Campylobacter jejuni in apo and substrate-analogue bound forms. Biochemistry. 2007;46:7196–7204. doi: 10.1021/bi602543d. [DOI] [PubMed] [Google Scholar]
- 15.Lin LY, Rakic B, Chiu CP, Lameignere E, Wakarchuk WW, Withers SG, Strynadka NC. Structure and mechanism of the lipooligosaccharide sialyltransferase from Neisseria meningitidis. J Biol Chem. 2011;286:37237–37248. doi: 10.1074/jbc.M111.249920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni L, Chokhawala HA, Cao H, Henning R, Ng L, Huang S, Yu H, Chen X, Fisher AJ. Crystal structures of Pasteurella multocida sialyltransferase complexes with acceptor and donor analogues reveal substrate binding sites and catalytic mechanism. Biochemistry. 2007;46:6288–6298. doi: 10.1021/bi700346w. [DOI] [PubMed] [Google Scholar]
- 17.Kim DU, Yoo JH, Lee YJ, Kim KS, Cho HS. Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar. BMB Rep. 2008;41:48–54. doi: 10.5483/bmbrep.2008.41.1.048. [DOI] [PubMed] [Google Scholar]
- 18.Kakuta Y, Okino N, Kajiwara H, Ichikawa M, Takakura Y, Ito M, Yamamoto T. Crystal structure of Vibrionaceae Photobacterium sp. JT-ISH-224 α2,6- sialyltransferase in a ternary complex with donor product CMP and acceptor substrate lactose: Catalytic mechanism and substrate recognition. Glycobiology. 2008;18:66–73. doi: 10.1093/glycob/cwm119. [DOI] [PubMed] [Google Scholar]
- 19.Iwatani T, Okino N, Sakakura M, Kajiwara H, Takakura Y, Kimura M, Ito M, Yamamoto T, Kakuta Y. Crystal structure of alpha/beta-galactoside alpha2,3- sialyltransferase from a luminous marine bacterium, Photobacterium phosphoreum. FEBS Lett. 2009;583:2083–2087. doi: 10.1016/j.febslet.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Chen X. Sialic acid metabolism and sialyltransferases: natural functions and applications. Appl Microbiol Biotechnol. 2012;94:887–905. doi: 10.1007/s00253-012-4040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breton C, Snajdrova L, Jeanneau C, Koca J, Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16:29R–37R. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- 22.Audry M, Jeanneau C, Imberty A, Harduin-Lepers A, Delannoy P, Breton C. Current trends in the structure-activity relationships of sialyltransferases. Glycobiology. 2011;21:716–726. doi: 10.1093/glycob/cwq189. [DOI] [PubMed] [Google Scholar]
- 23.Sun M, Li Y, Chokhawala HA, Henning R, Chen X. N-Terminal 112 amino acid residues are not required for the sialyltransferase activity of Photobacterium damsela alpha2,6-sialyltransferase. Biotechnol Lett. 2008;30:671–676. doi: 10.1007/s10529-007-9588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, Huang S, Chokhawala H, Sun M, Zheng H, Chen X. Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: a P. damsela alpha-2,6-sialyltransferase with extremely flexible donorsubstrate specificity. Angew Chem Int Ed Engl. 2006;45:3938–3944. doi: 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto T, Nakashizuka M, Terada I. Cloning and expression of a marine bacterial beta-galactoside alpha2,6-sialyltransferase gene from Photobacterium damsela JT0160. J Biochem. 1998;123:94–100. doi: 10.1093/oxfordjournals.jbchem.a021921. [DOI] [PubMed] [Google Scholar]
- 26.Sugiarto G, Lau K, Li Y, Khedri Z, Yu H, Le DT, Chen X. Decreasing the sialidase activity of multifunctional Pasteurella multocida alpha2–3-sialyltransferase 1 (PmST1) by site-directed mutagenesis. Mol BioSyst. 2011;7:3021–3027. doi: 10.1039/c1mb05182b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugiarto G, Lau K, Qu J, Li Y, Lim S, Mu S, Ames JB, Fisher AJ, Chen X. A sialyltransferase mutant with decreased donor hydrolysis and reduced sialidase activities for directly sialylating Lewis(x) ACS Chem Biol. 2012;7:1232–1240. doi: 10.1021/cb300125k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chokhawala HA, Cao H, Yu H, Chen X. Enzymatic synthesis of fluorinated mechanistic probes for sialidases and sialyltransferases. J Am Chem Soc. 2007;129:10630–10631. doi: 10.1021/ja072687u. [DOI] [PubMed] [Google Scholar]
- 29.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihoodenhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schomaker V, Trueblood KN. On the rigid-body motion of molecules in crystals. Acta Crystallogr Sec B. 1968;24:63–76. [Google Scholar]
- 36.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 37.Muda M, Rao NN, Torriani A. Role of PhoU in phosphate transport and alkaline phosphatase regulation. J Bacteriol. 1992;174:8057–8064. doi: 10.1128/jb.174.24.8057-8064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain S, Drendel WB, Chen ZW, Mathews FS, Sly WS, Grubb JH. Structure of human beta-glucuronidase reveals candidate lysosomal targeting and activesite motifs. Nat Struct Biol. 1996;3:375–381. doi: 10.1038/nsb0496-375. [DOI] [PubMed] [Google Scholar]
- 39.Chang A, Singh S, Phillips GN, Jr, Thorson JS. Glycosyltransferase structural biology and its role in the design of catalysts for glycosylation. Curr Opin Biotechnol. 2011;22:800–808. doi: 10.1016/j.copbio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unligil UM, Rini JM. Glycosyltransferase structure and mechanism. Curr Opin Struct Biol. 2000;10:510–517. doi: 10.1016/s0959-440x(00)00124-x. [DOI] [PubMed] [Google Scholar]
- 41.Mine T, Katayama S, Kajiwara H, Tsunashima M, Tsukamoto H, Takakura Y, Yamamoto T. An α2,6-sialyltransferase cloned from Photobacterium leiognathi strain JT-SHIZ-119 shows both sialyltransferase and neuraminidase activity. Glycobiology. 2010;20:158–165. doi: 10.1093/glycob/cwp157. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto T, Hamada Y, Ichikawa M, Kajiwara H, Mine T, Tsukamoto H, Takakura Y. A β-galactoside α2,6-sialyltransferase produced by a marine bacterium, Photobacterium leiognathi JT-SHIZ-145, is active at pH 8. Glycobiology. 2007;17:1167–1174. doi: 10.1093/glycob/cwm086. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Lou Y, Yokota H, Adams PD, Kim R, Kim SH. Crystal structure of a PhoU protein homologue: a new class of metalloprotein containing multinuclear iron clusters. J Biol Chem. 2005;280:15960–15966. doi: 10.1074/jbc.M414117200. [DOI] [PubMed] [Google Scholar]
- 44.Echols N, Morshed N, Afonine PV, McCoy AJ, Miller MD, Read RJ, Richardson JS, Terwilliger TC, Adams PD. Automated identification of elemental ions in macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2014;70:1104–1114. doi: 10.1107/S1399004714001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng H, Chordia MD, Cooper DR, Chruszcz M, Muller P, Sheldrick GM, Minor W. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat Protoc. 2014;9:156–170. doi: 10.1038/nprot.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Y, Chen L, Ha S, Gross B, Falcone B, Walker D, Mokhtarzadeh M, Walker S. Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc Natl Acad Sci U S A. 2003;100:845–849. doi: 10.1073/pnas.0235749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morera S, Imberty A, Aschke-Sonnenborn U, Ruger W, Freemont P. T4 phage beta-glucosyltransferase: substrate binding and proposed catalytic mechanism. J Mol Biol. 1999;292:717–730. doi: 10.1006/jmbi.1999.3094. [DOI] [PubMed] [Google Scholar]
- 48.Mulichak AM, Losey HC, Walsh CT, Garavito RM. Structure of the UDPglucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure. 2001;9:547–557. doi: 10.1016/s0969-2126(01)00616-5. [DOI] [PubMed] [Google Scholar]
- 49.Fox K, Cox A, Gilbert M, Wakarchuk W, Li J, Makepeace K, Richards J, Moxon E, Hood D. Identification of a bifunctional lipopolysaccharide sialyltransferase in Haemophilus influenzae: incorporation of disialic acid. J Biol Chem. 2006;281:40024–40032. doi: 10.1074/jbc.M602314200. [DOI] [PubMed] [Google Scholar]
- 50.Freiberger F, Claus H, Gunzel A, Oltmann-Norden I, Vionnet J, Muhlenhoff M, Vogel U, Vann WF, Gerardy-Schahn R, Stummeyer K. Biochemical characterization of a Neisseria meningitidis polysialyltransferase reveals novel functional motifs in bacterial sialyltransferases. Mol Microbiol. 2007;65:1258–1275. doi: 10.1111/j.1365-2958.2007.05862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]