Abstract
The ability to obtain gene expression profiles from human disease specimens provides an opportunity to identify relevant gene pathways, but is limited by the absence of data sets spanning a broad range of conditions. Here, we analyzed publicly available microarray data from 16 diverse skin conditions in order to gain insight into disease pathogenesis. Unsupervised hierarchical clustering separated samples by disease and common cellular and molecular pathways. Disease specific signatures were leveraged to build a multi-disease classifier which predicted the diagnosis of publicly and prospectively collected expression profiles with 93% accuracy. In one sample, the molecular classifier differed from the initial clinical diagnosis and correctly predicted the eventual diagnosis as the clinical presentation evolved. Finally, integration of interferon (IFN) regulated gene programs with the skin database revealed a significant inverse correlation between IFN–β and IFN–γ programs across all conditions. Our study provides an integrative approach to the study of gene signatures from multiple skin conditions, elucidating mechanisms of disease pathogenesis. Additionally, these studies provide a framework for developing tools for personalized medicine towards the precise prediction, prevention, and treatment of disease on an individual level.
Introduction
Gene expression profiling technology, such as microarrays, provides the opportunity to identify disease specific genes and pathways. The NCBI Gene Expression Omnibus (GEO) is a community resource of publicly available experimental data sets including those involving human health and disease and that incorporate less common diseases studied in specific laboratories (Edgar et al., 2002). This searchable database contains annotated gene expression profile data (both in summarized and raw formats), primarily from published work. The ability to mine data from GEO provides a tremendous opportunity to compare gene expression profiles across multiple diseases.
Previously, disease profiles have been compared across multiple data sets by normalizing to controls within each set; however, this practice limits the use of available data to those containing equivalent control profiles (Chaussabel et al., 2008; Wong et al., 2012). To overcome this limitation, data integration from multiple sources, particularly those from experiments containing a single disease, can be achieved with Frozen Robust Multi-array Average (fRMA), which normalizes samples to a standard reference set of microarrays, eliminating the need for control samples in each data set (McCall et al., 2010). Here we present a study that uses this approach on gene expression profiles derived from skin biopsy specimens. We assembled a database of publicly available skin microarray samples representing 16 inflammatory, infectious, and neoplastic conditions. This database was used to construct a classifier, perform functional analyses to identify representative pathways, and establish a spectrum of differentially expressed Type I vs. Type II interferon gene programs across these diseases (Figure S1).
Results
Data normalization with Frozen RMA
We searched GEO for microarray experiments performed on human skin samples associated with a dermatological disorder (www.ncbi.nlm.nih.gov/geo) (Edgar et al., 2002). Data for microarrays 311 skin biopsy samples were downloaded, representing 16 conditions from 15 experiments and 14 laboratories (Table S1). All samples used the Affymetrix HG U133 Plus 2.0 platform. To analyze this data as a single set, Frozen RMA (fRMA) was used to normalize samples such that they showed comparable probe set intensity distributions (Figure S2) (McCall et al., 2010).
After unsupervised gene clustering, samples segregate by disease and into groups with related pathogenesis
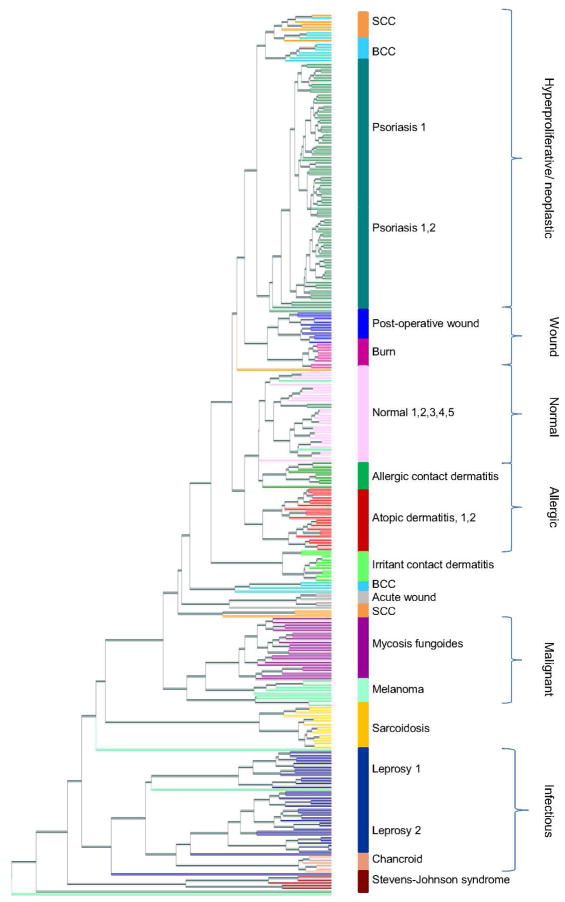
In order to determine whether the batch effects within a given disease were smaller than the differences between diseases, trees of filtered gene expression profiles were constructed for both samples and diseases (Figures 1, S3). Remarkably, we found that in diseases in which there were multiple batches of microarrays from different sources, including psoriasis, atopic dermatitis and leprosy, batches from the same disease nearly always clustered together, despite coming from independent data sets (Figure 1). Furthermore, five batches of normal skin, each obtained from healthy control subjects from different laboratories, clustered together with little differentiation by batch. However, batch effects were not completely eliminated as samples from specific diseases often separated by lab or experiment. There were isolated cases of individual samples clustering with the incorrect disease. However, squamous cell carcinoma (SCC) and basal cell carcinoma (BCC) samples from a single lab were split into two groups. Overall, these results suggest that the use of fRMA allows us to minimize the effect of batch and allow true disease signatures to predominate.
Figure 1. Unsupervised hierarchical clustering tree of 311 skin samples.
Normalized, filtered gene expression profiles were clustered using gene expression distance (Pearson correlation) and displayed in a tree. Each terminal leaf in the tree represents a biopsy sample and is colored according to disease, with colored bars to the right representing the majority disease diagnosis. Samples that clustered apart from other samples of the same diagnosis can be seen a leaf that differs in color from its neighbors. Numbers following disease name labels denote batches of the same disease, and lists of numbers following a disease name denote multiple batches clustering to the same tree branch with little or no differentiation by batch. Brackets to the far right delineate biological groups of neighboring diseases.
When the higher-level structure of the tree was examined, we found that branches of the tree could be annotated as disease groups with related pathogenesis. These distinct groups were categorized according to the following descriptions: i) keratinocyte proliferation and neoplastic growth (psoriasis, and approximately half the SCC and BCC samples), ii) wound (post-operative wound, burn), iii) normal, iv) allergic (allergic contact dermatitis, atopic dermatitis), v) malignant (mycosis fungoides, melanoma); and, vi) infectious (leprosy, chancroid). These relationships are consistent with those seen in an unrooted disease tree, which was built with one leaf per disease by averaging distances between all pairs of samples (Figure S3).
Proportional median metric for identifying disease specific gene signatures
Gene signatures were identified for each disease in order to build a disease classifier and perform downstream functional analysis. We developed the “proportional median” (PM) metric to identify highly expressed gene probe sets for each disease. The PM of a microarray probe set X in disease Y represents how highly expressed X was in Y, compared to all other diseases. Probe sets were ranked by PM for each disease. Because lowly ranked genes often have low intensity and tend to be noisier than those with higher ranks, subsequent analysis utilized genes with high PM values, reflecting over-expression in a particular disease (Mutch et al., 2002; Tu et al., 2002). The top 25 genes in the psoriasis PM list were confirmed by comparison to RNA-seq data derived from the same samples (Figure S4) (Li et al., 2014).
Random forest classifier accurately predicts disease diagnosis
We built a random forest multi-classifier using our disease expression profiles to predict disease status based on the expression of a limited number of genes (Breiman, 2001). Briefly, the random forest algorithm selects subsets of samples and genes to iteratively build multiple, parallel decision trees. This random subsampling and iteration reduces the effect of noise and outliers on classifier training. In addition, cross-validation is built into the classifier training process by testing each decision tree with samples not used to build that tree.
We used PM to select the most informative probe sets as input to our classifier, which yielded an overall error rate of 4.5% (Figure S5). Performance for each disease was assessed by sensitivity, specificity, precision, and F1 score, a measure of accuracy equivalent to a weighted average of sensitivity and precision that has values between 0 (poor accuracy) and 1 (perfect accuracy). Sensitivity values ranged from 0.60 and 1.00 (mean 0.92) and specificity values ranged from 0.99 to 1.00, which corresponds to an average of 92% of disease samples being accurately classified and 99% of negative classifications being correct. The range of F1 scores was from 0.75 to 1.00 (mean 0.94) (Table S3). We also performed three-fold cross validation, which yielded aggregate F1 scores from 0.75 to 1.00 (mean 0.96) (Table S4).
To assess the classifier’s ability to generalize, two-fold cross validation with separation by batch was carried out. We separated data into two approximately equal groups: in multi-batch conditions (leprosy, psoriasis, atopic dermatitis, and normal), data was separated according to batch; otherwise, data was randomly partitioned. Two independent classifiers were built, each trained on one partitioned set and tested on the other. PM values used in feature selection were computed from only one group of data to ensure that test data did not bias classification. Diseases separated by batch had F1 scores between 0.90 and 0.95 (mean 0.92), and diseases randomly separated had F1 scores between 0.71 and 1.00 (mean 0.92), indicating little loss of accuracy with batch separated cross validation (Table 1).
Table 1.
Two-fold cross validation, with separation by batch where applicable.
| Condition | Sensitivity | Specificity | Precision | F1 |
|---|---|---|---|---|
| Leprosy* | 0.86 | 1.00 | 0.97 | 0.91 |
| Psoriasis* | 0.96 | 0.97 | 0.94 | 0.95 |
| Chancroid | 1.00 | 1.00 | 1.00 | 1.00 |
| Allergic contact dermatitis | 0.78 | 1.00 | 1.00 | 0.88 |
| Irritant contact dermatitis | 1.00 | 1.00 | 1.00 | 1.00 |
| Atopic dermatitis* | 0.95 | 0.99 | 0.88 | 0.91 |
| Burn | 1.00 | 1.00 | 1.00 | 1.00 |
| Acute wound | 0.83 | 1.00 | 1.00 | 0.91 |
| Post-operative wound | 1.00 | 1.00 | 1.00 | 1.00 |
| Mycosis fungoides | 1.00 | 1.00 | 1.00 | 1.00 |
| BCC | 0.87 | 1.00 | 0.93 | 0.90 |
| Melanoma | 0.86 | 0.98 | 0.67 | 0.75 |
| SCC | 0.60 | 1.00 | 0.86 | 0.71 |
| Sarcoidosis | 1.00 | 1.00 | 1.00 | 1.00 |
| Stevens Johnson syndrome | 0.80 | 1.00 | 1.00 | 0.89 |
| Normal skin* | 0.96 | 0.98 | 0.84 | 0.90 |
Denotes diseases separated by batch
The classifier’s performance was next tested on data not used for any of the previous analyses, including the feature selection and training steps of this classifier. We generated 26 de-identified gene expression profiles derived from biopsy specimens of leprosy, psoriasis, atopic dermatitis, and normal skin. We also found public data for 168 gene expression profiles derived from biopsy specimens of psoriasis, atopic dermatitis, and melanoma (Table S5). The classifier was used to assess these 194 validation samples, yielding an overall sensitivity of 0.93, specificity of 0.99, and F1 statistics between 0.76 and 1.0 (Table S6).
Upon follow-up of the patients from whom we had collected samples, we discovered that one that was clinically diagnosed with atopic dermatitis, which we classified as psoriasis, had an unusual presentation. This patient had a history of an atopic diathesis including hay fever, increased total IgE levels as well as elevated levels of the eosinophilic cationic protein. The patient presented clinically with chronic dermatitis on the palms of the hands, as well as on the plantar side of the feet. Furthermore, inflammatory skin lesions on the arms and other locations were clinically diagnosed as atopic dermatitis, consistent with the atopic diathesis, and a sample was obtained for the present study. However, later the patient developed inflammatory plaques on the lower back, which were clinically diagnosed as psoriasis. Both atopic dermatitis and psoriasis were considered as a diagnosis for this patient at various stages; however, the co-occurrence of atopic dermatitis and psoriasis is rare, perhaps due to the opposing immunopathogenic mechanisms for the two diseases (Beer et al., 1992; Eyerich et al., 2011; Henseler and Christophers, 1995). Although we cannot be certain of the initial diagnosis, our molecular classifier correctly predicted the diagnosis as the clinical course evolved.
Functional annotation of related disease signatures using cell-type specific deconvolution and k-means clustering shows shared and unique mechanisms of disease
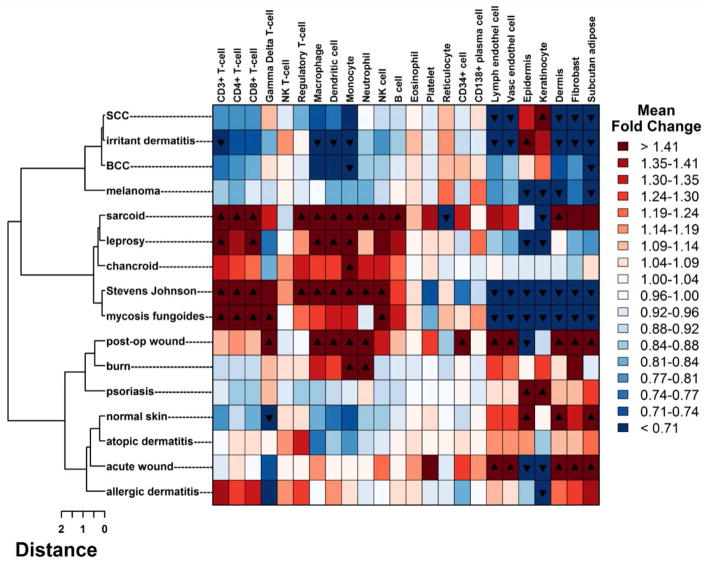
We investigated whether diseases that grouped together on the hierarchical clustering tree shared cell types in their associated lesions. Using cell-type specific gene signatures developed in previous work by Swindell, et al., the relative enrichment of each cell type signature was assessed in each disease, and subjected to hierarchical clustering using Euclidian distance (Figure 2) (Swindell et al., 2013; Swindell et al., 2012).
Figure 2. Cell-type specific signature enrichment.
For each of 24 cell-type specific signatures, log fold changes were calculated using average gene expression for each condition. Each fold change represents the enrichment for a particular cell-type in that condition relative to the other 15 conditions. Enrichment profiles for each condition were clustered using Euclidean distance and displayed in a heatmap, where rows correspond to conditions and columns correspond to cell types. Black triangles denote FDR < 0.05 and directionality of fold change.
In some instances, the clustering by cell-type signatures was consistent with the hierarchical clustering and unrooted trees generated from the entire filtered gene profiles (Figures 1, S3). For example, atopic dermatitis, allergic contact dermatitis and normal skin clustered together in both trees, and deconvolution analysis revealed a T regulatory cell gene signature consistent with previous studies of atopic dermatitis expression profiles (Figure 2) (Clark and Kupper, 2007; Hanafusa et al., 2013). The notable absence of a general T cell specific signature in psoriasis and atopic dermatitis likely reflects that expression of T cell specific genes is much greater in conditions such as mycosis fungoides and Stevens Johnson syndrome, which are characterized by marked T cell activation and proliferation (Chung et al., 2008; Nassif et al., 2004; Yamashita et al., 2012). In other instances, the clustering differed by cell type versus hierarchical clustering, suggesting that other factors must be involved in the observed groupings.
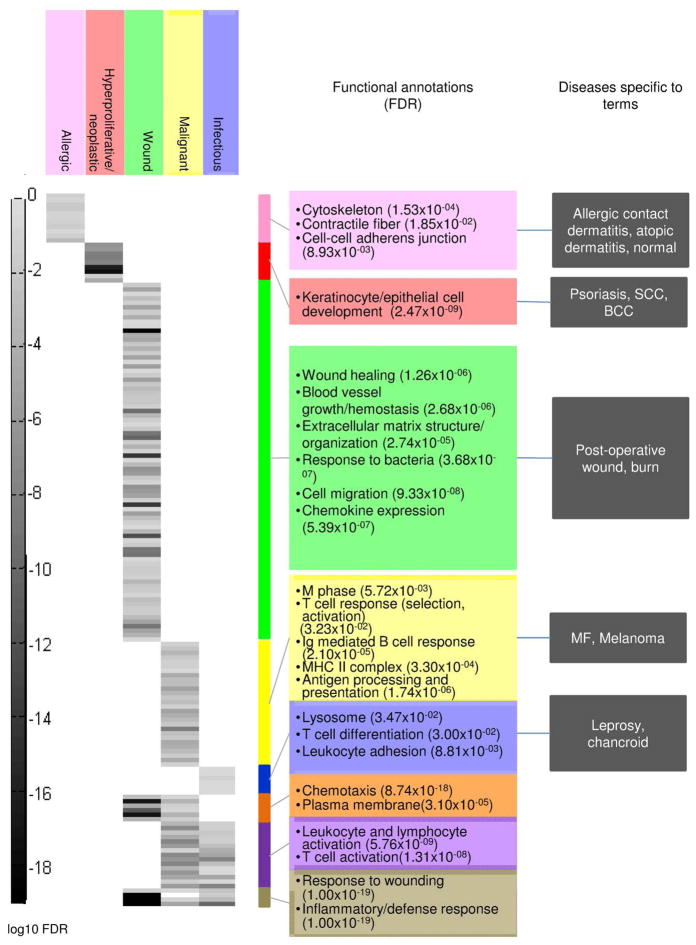
We used pathway analysis of gene signatures to further investigate common pathways within each disease group. For each group of diseases indicated by hierarchical clustering, combined gene signatures were constructed and evaluated for enriched functional terms. K-means clustering was performed on the p-values associated with each term (Figure 3). Significantly enriched terms support previous findings in the literature: allergic diseases were enriched for “cell-cell adherens junction” (p-value=8.93×10−03), and hyperproliferative/neoplastic diseases were enriched for “keratinocyte and epithelial cell development” (p-value=2.47×10−09) (Cai et al., 2012; O’Regan et al., 2008). Wound, malignant, and infectious groups share overlapping enriched GO terms, associated with response to wounding.
Figure 3. Functional annotation and k-means clustering of group signatures.
PM signatures for each biological group from unsupervised hierarchical clustering (Figure 1) were calculated and annotated with enriched Gene Ontology (GO) terms. Corresponding false discovery rates (FDR) for each GO term were clustered using k-means clustering (k=8) and visualized in a heatmap, where rows correspond to GO terms and columns correspond to disease groups. Each gray bar represents the log10 FDR for a particular GO term in a particular disease group. Colored bars to the right demarcate clusters of GO terms, and a summary of terms and p-values are provided for each cluster.
Functional analysis of PM signatures shows enrichment for genes and pathways corresponding to single diseases
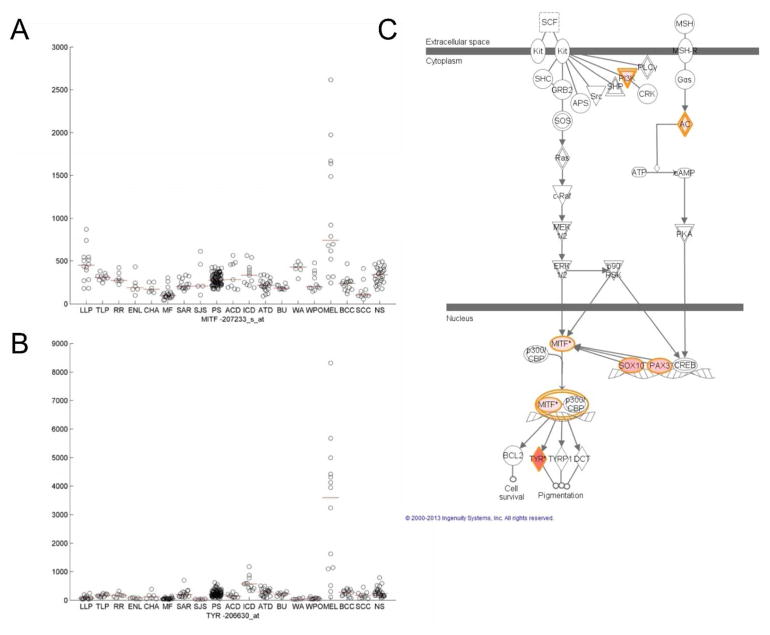
In order to assess the relevance of individual disease PM signatures, additional pathway analysis was performed on the 250 probe sets with the highest PM values for each disease. DAVID and Ingenuity Pathways functional analyses of individual disease signatures often showed a correspondence to the disease of origin (Tables S7, S8). For example, Ingenuity analysis of the melanoma PM signature revealed an enrichment of “biologic functions” relevant to melanocyte development and disorders (“differentiation of melanocytes,” p-value=4.4×10−09), as well as a significant enrichment of the “Melanocyte Development and Pigmentation Signaling canonical pathway” (p-value=9.2×10−05) (Figure 4C). We developed a web-based visualization tool (http://pathways-pellegrini.mcdb.ucla.edu/goTeles/dot_plot.html) that plots the expression of a gene within our database; shown are two melanocyte development genes (Figure 4A,B).
Figure 4. Visualization of melanoma proportional median (PM) signature.
A,B. Visualization of normalized intensities for representative probe sets of MITF and TYR, two genes in the melanocyte development pathway. Each black circle represents an intensity value on a microarray for the specific probe set indicated. Red lines show median intensity values for each disease. Disease abbreviations are as follows: lepromatous leprosy (LLP), tuberculoid leprosy (TLP), reversal reaction leprosy (RR), erythema nodosum leprosum (ENL), chancroid (CH), mycosis fungoides (MF), sarcoid (SAR), Stevens Johnson Syndrome (SJS), psoriasis (PS), allergic contact dermatitis (ACD), irritant contact dermatitis (ICD), atopic dermatitis (ATD), burn (BU), acute wound (WA), post-operative wound (WPO), melanoma (MEL), basal cell carcinoma (BCC), squamous cell carcinoma (SCC), normal skin (NS). C. Visualization of melanocyte development pathway in Ingenuity Pathways Analysis. The 250 probe sets with highest PM for melanoma were annotated for enriched functional pathways using Ingenuity Pathways Analaysis and one representative pathway is shown. Red genes indicate presence in a 250 probe set PM melanoma signature, with darker coloring denoting higher PM values. The melanocyte pathway was significantly enriched in the top 250 probe sets for melanoma (p-value=9.19×10−05).
Network analysis of the PM signatures was carried out using Ingenuity Pathways Analysis to visualize connected genes and pathways in each disease and further evaluate the functional significance of our signatures. Notably, a psoriasis network showed connections between TCN1, OASL, and SPRR3 (Figure S6). This network appears to provide a cellular nexus connecting key pathogenesis elements, including differentiation-associated pathways, IFN-directed responses, and infiltrating inflammatory cells such as neutrophils. Furthermore, cell-type specific deconvolution of the psoriasis PM signature demonstrated expression patterns consistent with the presence of neutrophils (Figure S7) (Fujimoto et al., 1997; Suárez-Fariñas et al., 2012; Suárez-Fariñas et al., 2011; Swindell et al., 2013; Yao et al., 2008).
Type I vs. Type II interferon gene programs have a negative inverse correlation across a spectrum of skin diseases
Type I and Type II interferons (IFN) have opposing immunoregulatory roles in human disease, and previous work has shown different diseases or subtypes of disease to exhibit a range of IFN responses. Recent work in leprosy has demonstrated a distinct pattern of Th1 versus Th2 cytokines, which we have used as a model for interrogating our skin disease database in order to identify similar patterns across a range of conditions (Teles et al., 2013; Wong et al., 2012; Yamamura et al., 1991). IFN–γ (Type II IFN) is involved in macrophage activation to fight bacterial infection, and is opposed by IFN–α/β (Type I IFN), which combats viral infection. Because IFNs are weakly detected on microarrays, we used Type I and Type II-specific induced transcriptional profiles of human peripheral blood mononuclear cells to infer the expression of IFN signatures (Teles et al., 2013; Waddell et al., 2010).
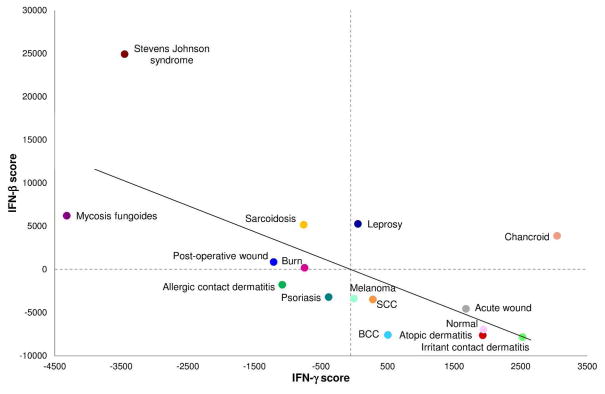
Integration of the IFN gene expression profiles with our data set containing 16 different skin conditions demonstrated a significant, inverse correlation between IFN–β and IFN–γ regulated genes across all skin diseases studied (r=−0.66, p-value=0.006), underscoring the opposing roles of IFN–β and IFN–γ in skin disease (Figure 5). The Stevens Johnson syndrome samples, which were obtained from blister fluid rather than full thickness biopsies, had the most extreme IFN–β IFN–γ profiles. Nevertheless, even if these samples were omitted, the anti-correlation between IFN–β vs. IFN–γ profiles was still significant (r=−0.53, p-value=0.04).
Figure 5. Type I and II interferon program cross-regulation.
IFN–β and IFN–γ scores were calculated by ranking genes specifically regulated by IFN-β or IFN-γ relative to total gene expression and centering relative to the mean rank of each gene across all conditions. Individual up-or-down-regulated scores for each type of IFN were obtained by summing centered ranks, and an overall IFN score was obtained by subtracting the up-regulated sum of ranks by the down-regulated sum of ranks. Intuitively, high scores for each type of IFN represent high expression of IFN-stimulated genes, low expression of IFN-repressed genes, or both, such that placement on each axis shows the magnitude of expression of IFN–β or IFN–γ gene programs. The plot shows a significant negative inverse correlation (r=−0.66, p-value=0.006). Removing the outlier Stevens Johnson syndrome, the correlation remains significant (r=−0.53, p-value=0.04).
Discussion
Insights into disease pathogenesis obtained by comparison of gene expression profiles are often limited because these comparisons are performed between either two different diseases or one disease versus healthy controls, and therefore cannot identify distinct and common mechanisms of pathogenesis. Here, we performed a cross-disease analysis of molecular profiles from multiple skin diseases. Using fRMA, it was possible to assemble a database of gene expression profiles from 311 samples spanning 16 conditions and visualize disease relationships on a hierarchical clustering tree. Remarkably we found that samples of a particular disease that were taken from different batches colocalized to the same branch. This was particularly striking in the case of normal skin, where five batches of samples taken from healthy control subjects not only clustered on the same branch, but were arranged with little differentiation by batch.
Our approach demonstrates that a multi-disease classifier can be built from disparate public data sources comprising over a dozen different conditions in a single tissue. We built this classifier from disease specific gene signatures, and found that it was accurate and robust to batch effect. The potential utility of molecular classification over the classic clinical criteria was demonstrated by the correct classification of an ambiguous case of psoriasis. A multi-disease classifier using epithelial cells from patients with psoriasis, atopic dermatitis, allergic contact dermatitis, and irritant contact dermatitis was previously constructed, although this was limited to diseases which had epidermal involvement (Kamsteeg et al., 2010). Our work expands this principle to a wide range of both inflammatory and neoplastic diseases, and demonstrates the potential value of this approach in comparing diverse conditions. This approach can be expanded to include a more diverse spectrum of diseases, as more data is publicly available, allowing for the comparative study of diseases for which skin biopsy specimens may not be widely available.
We also analyzed this data by three supervised approaches: analysis of cell-type signatures, Gene Ontology pathways enrichment, and interferon response signatures. Together, these bioinformatic analyses provided insight into the distinct and related pathogenesis of the diseases. For example, hierarchical clustering of gene expression profiles revealed that leprosy and chancroid were located on the same branch which we termed “infectious” based upon their known etiologies. In the deconvolution analysis of cell type signatures, both diseases were characterized by the enrichment of similar lymphoid (CD3+, CD4+, CD8+, regulatory T cells, B cells) and myeloid (monocytes, macrophages, dendritic cells, neutrophils) expression profiles. Furthermore, Gene Ontology enrichment analysis identified the terms “lysosome”, “T cell differentiation” and “leukocyte adhesion”. Additionally, we found an anti-correlation of Type I and Type II IFN responses across a wide range of skin diseases of different etiologies, consistent with our previous studies using leprosy as a model (Teles et al., 2013). An earlier multi-disease comparison found that the magnitude of an IFN gene signature distinguished different inflammatory skin diseases, but could not distinguish between the Type I vs. Type II IFN patterns (Wong et al., 2012).
The present findings provide a rationale for further investigations to determine how these different IFN programs contribute to the pathogenesis of these diseases and identify treatment targets. The spectrum of IFN–β vs. IFN–γ gene program expression in the skin diseases studied here is consistent with current practices in treatment of skin disease. Typically, Type I IFN exhibits anti-proliferative effects, and is used to treat neoplasms, such as melanoma and BCC, which have a negative Type I IFN score. It should be noted that IFN–γ has been used to treat acute atopic dermatitis, even though chronic atopic dermatitis lesions, as studied here, express IFN–γ consistent with our findings (Alazemi and Campos, 2006; Grassegger and Höpfl, 2004; Grewe et al., 1994; Grewe et al., 1995; Hamid et al., 1994). Anti-IFN–γ has also been shown in preliminary studies to have a positive effect on Th1-mediated autoimmune skin diseases, including psoriasis, which we found had a high Type II IFN score (Skurkovich and Skurkovich, 2006).
High throughput analysis of gene expression profiles are a step towards the development of tools for personalized medicine. These studies have led to the discovery of individual genes and pathways that underlie disease pathology, which can be leveraged as biomarkers to diagnose and predict the course of disease, as well as identify targets for therapeutic intervention. For example, human leukocyte antigen (HLA) allele type has been linked to adverse responses to commonly used drugs such as the antiretroviral abacavir and the antiepileptic carbamazepine (Illing et al., 2013). Subsequent diagnostic tests would be based on such individual genes or sets of genes, making them more economical for clinical use. The integrative molecular classification and functional analysis of multiple skin conditions reported here demonstrates the value of such a comparison to gain insight into the pathogenesis of human disease.
Materials and Methods
Microarray acquisition, processing, and clustering
Data was obtained from the NCBI Gene Expression Omnibus (GEO) as described in supplemental methods and Table S1 (http://www.ncbi.nlm.nih.gov/geo/) (Edgar et al., 2002). Additional validation samples were obtained with written informed consent from de-identified biopsy specimens as described in the supplement and Table S5. Data was normalized using the Frozen Robust Multiarray Average method and filtered by mean intensity of at least 15 in any disease. Unsupervised hierarchical clustering was used to group the normalized, filtered expression profiles using Pearson correlation.
Proportional median
Probe sets were ranked using the Proportional Median (PM), which we define as the median intensity of a probe set within one disease divided by the median intensity of the same probe set across all samples. For each disease, probe sets were ranked in descending order by PM.
Random forest classifier
A random forest classifier was built using the Matlab TreeBagger class (see supplemental methods for full description). The classifier feature space was reduced by selecting the 25 probe sets with the highest PM values across the training set for each disease. Classifier validation was performed in three ways: three-fold cross validation, two-fold cross validation with separation by experimental batch where applicable, and external validation using independent samples.
Pathway analysis
The 250 probe sets with the highest PM value for each disease were selected for pathway analysis using Ingenuity Pathways Analysis (http://www.ingenuity.com) and DAVID Functional Annotation Analysis (http://david.abcc.ncifcrf.gov/), using the top 250 probe sets by PM from each disease signature (Huang et al., 2009a, b; www.ingenuity.com).
Cell-type specific signature enrichment
Cell-type specific expression profiles for 24 cell types were calculated as previously described and is described in more detail in the supplemental methods (Swindell et al., 2013).
Group signatures
PM values were calculated for five disease group signatures, based on hierarchical clustering, and Gene Ontology enrichment was performed for each group (see supplement for details).
IFN profile integration
IFN–β and IFN–γ specific scores were calculated by adapting a previously described method (see supplement for details) (Teles et al., 2013).
Supplementary Material
Acknowledgments
We would like to thank Barry Bloom for his thoughtful input on the manuscript, and Steve Horvath for his helpful discussion regarding classifiers and clustering.
Funding: JTE was supported by NIH grant R01 AR054966 and by the Ann Arbor Veterans Affairs Hospital. MSI was supported in part by NIH/NIAMS grant P50 5P50AR063020. RLM is a recipient of the CHANEL-CERIES Research Award.
Footnotes
Conflict of Interest:
No conflicts of interest to report.
Author contributions: MSI, POS, WRS, JTE, RLM, and MP wrote and edited the manuscript. DL built the website. MSI, WRS, JTE, RLM, RMBT, TGG, and MP designed components of the analysis. MG, BH, and SM provided patient samples and microarray data. WRS performed deconvolution analysis. MSI performed all other analyses.
References
- Alazemi S, Campos MA. Interferon-induced sarcoidosis. Int J Clin Pract. 2006;60:201–11. doi: 10.1111/j.1742-1241.2005.00651.x. [DOI] [PubMed] [Google Scholar]
- Beer WE, Smith AE, Kassab JY, et al. Concomitance of psoriasis and atopic dermatitis. Dermatology. 1992;184:265–70. doi: 10.1159/000247564. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- Cai Y, Fleming C, Yan J. New insights of T cells in the pathogenesis of psoriasis. Cell Mol Immunol. 2012;9:302–9. doi: 10.1038/cmi.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussabel D, Quinn C, Shen J, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–64. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WH, Hung SI, Yang JY, et al. Granulysin is a key mediator for disseminated keratinocyte death in Stevens-Johnson syndrome and toxic epidermal necrolysis. Nat Med. 2008;14:1343–50. doi: 10.1038/nm.1884. [DOI] [PubMed] [Google Scholar]
- Clark RA, Kupper TS. IL-15 and dermal fibroblasts induce proliferation of natural regulatory T cells isolated from human skin. Blood. 2007;109:194–202. doi: 10.1182/blood-2006-02-002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyerich S, Onken AT, Weidinger S, et al. Mutual antagonism of T cells causing psoriasis and atopic eczema. N Engl J Med. 2011;365:231–8. doi: 10.1056/NEJMoa1104200. [DOI] [PubMed] [Google Scholar]
- Fujimoto W, Nakanishi G, Arata J, et al. Differential expression of human cornifin alpha and beta in squamous differentiating epithelial tissues and several skin lesions. J Invest Dermatol. 1997;108:200–4. doi: 10.1111/1523-1747.ep12334240. [DOI] [PubMed] [Google Scholar]
- Grassegger A, Höpfl R. Significance of the cytokine interferon gamma in clinical dermatology. Clin Exp Dermatol. 2004;29:584–8. doi: 10.1111/j.1365-2230.2004.01652.x. [DOI] [PubMed] [Google Scholar]
- Grewe M, Gyufko K, Schöpf E, et al. Lesional expression of interferon-gamma in atopic eczema. Lancet. 1994;343:25–6. doi: 10.1016/s0140-6736(94)90879-6. [DOI] [PubMed] [Google Scholar]
- Grewe M, Walther S, Gyufko K, et al. Analysis of the cytokine pattern expressed in situ in inhalant allergen patch test reactions of atopic dermatitis patients. J Invest Dermatol. 1995;105:407–10. doi: 10.1111/1523-1747.ep12321078. [DOI] [PubMed] [Google Scholar]
- Hamid Q, Boguniewicz M, Leung DY. Differential in situ cytokine gene expression in acute versus chronic atopic dermatitis. J Clin Invest. 1994;94:870–6. doi: 10.1172/JCI117408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafusa T, Matsui S, Murota H, et al. Increased frequency of skin-infiltrating FoxP3+ regulatory T cells as a diagnostic indicator of severe atopic dermatitis from cutaneous T cell lymphoma. Clin Exp Immunol. 2013;172:507–12. doi: 10.1111/cei.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henseler T, Christophers E. Disease concomitance in psoriasis. J Am Acad Dermatol. 1995;32:982–6. doi: 10.1016/0190-9622(95)91336-x. [DOI] [PubMed] [Google Scholar]
- Huang dW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Illing PT, Vivian JP, Purcell AW, et al. Human leukocyte antigen-associated drug hypersensitivity. Curr Opin Immunol. 2013;25:81–9. doi: 10.1016/j.coi.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Ingenuity® Systems. www.ingenuity.com.
- Kamsteeg M, Jansen PA, van Vlijmen-Willems IM, et al. Molecular diagnostics of psoriasis, atopic dermatitis, allergic contact dermatitis and irritant contact dermatitis. Br J Dermatol. 2010;162:568–78. doi: 10.1111/j.1365-2133.2009.09547.x. [DOI] [PubMed] [Google Scholar]
- Li B, Tsoi LC, Swindell WR, et al. Transcriptome Analysis of Psoriasis in a Large Case-Control Sample: RNA-Seq Provides Insights into Disease Mechanisms. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA) Biostatistics. 2010;11:242–53. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutch DM, Berger A, Mansourian R, et al. The limit fold change model: a practical approach for selecting differentially expressed genes from microarray data. BMC Bioinformatics. 2002;3:17. doi: 10.1186/1471-2105-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif A, Bensussan A, Boumsell L, et al. Toxic epidermal necrolysis: effector cells are drug-specific cytotoxic T cells. J Allergy Clin Immunol. 2004;114:1209–15. doi: 10.1016/j.jaci.2004.07.047. [DOI] [PubMed] [Google Scholar]
- O’Regan GM, Sandilands A, McLean WH, et al. Filaggrin in atopic dermatitis. J Allergy Clin Immunol. 2008;122:689–93. doi: 10.1016/j.jaci.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Skurkovich B, Skurkovich S. Inhibition of IFN-gamma as a method of treatment of various autoimmune diseases, including skin diseases. Ernst Schering Res Found Workshop. 2006:1–27. doi: 10.1007/3-540-37673-9_1. [DOI] [PubMed] [Google Scholar]
- Suárez-Fariñas M, Li K, Fuentes-Duculan J, et al. Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J Invest Dermatol. 2012;132:2552–64. doi: 10.1038/jid.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Fariñas M, Tintle SJ, Shemer A, et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J Allergy Clin Immunol. 2011;127:954–64. e1–4. doi: 10.1016/j.jaci.2010.12.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Johnston A, Voorhees JJ, et al. Dissecting the psoriasis transcriptome: Inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC Genomics. 2013;14:527. doi: 10.1186/1471-2164-14-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Xing X, Stuart PE, et al. Heterogeneity of inflammatory and cytokine networks in chronic plaque psoriasis. PLoS One. 2012;7:e34594. doi: 10.1371/journal.pone.0034594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles RM, Graeber TG, Krutzik SR, et al. Type I Interferon Suppresses Type II Interferon-Triggered Human Anti-Mycobacterial Responses. Science. 2013 doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Stolovitzky G, Klein U. Quantitative noise analysis for gene expression microarray experiments. Proc Natl Acad Sci U S A. 2002;99:14031–6. doi: 10.1073/pnas.222164199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell SJ, Popper SJ, Rubins KH, et al. Dissecting interferon-induced transcriptional programs in human peripheral blood cells. PLoS One. 2010;5:e9753. doi: 10.1371/journal.pone.0009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D, Kea B, Pesich R, et al. Interferon and biologic signatures in dermatomyositis skin: specificity and heterogeneity across diseases. PLoS One. 2012;7:e29161. doi: 10.1371/journal.pone.0029161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura M, Uyemura K, Deans RJ, et al. Defining protective responses to pathogens: cytokine profiles in leprosy lesions. Science. 1991;254:277–9. doi: 10.1126/science.254.5029.277. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Abbade LP, Marques ME, et al. Mycosis fungoides and Sézary syndrome: clinical, histopathological and immunohistochemical review and update. An Bras Dermatol. 2012;87:817–28. doi: 10.1590/S0365-05962012000600001. quiz 29–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Richman L, Morehouse C, et al. Type I interferon: potential therapeutic target for psoriasis? PLoS One. 2008;3:e2737. doi: 10.1371/journal.pone.0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.