Abstract
The fragile X mental retardation protein FMRP regulates translation of its bound mRNAs through incompletely defined mechanisms. FMRP has been linked to the microRNA pathway and we show here that it associates with the RNA helicase MOV10, also associated with the microRNA pathway. FMRP associates with MOV10 directly and in an RNA-dependent manner and facilitates MOV10-association with RNAs in brain and cells suggesting a cooperative interaction. We identified the RNAs recognized by MOV10 using RNA-IP and iCLIP. Examination of the fate of MOV10 on RNAs revealed a dual function for MOV10 in regulating translation: it facilitates microRNA-mediated translation of some RNAs but also increases expression of other RNAs by preventing AGO2 function. The latter subset was also bound by FMRP in close proximity to the MOV10 binding site, suggesting that FMRP prevents MOV10-mediated microRNA suppression. We have identified a new mechanism for FMRP-mediated translational regulation through its association with MOV10.
Introduction
Fragile X syndrome (FXS) is a disease of aberrant protein production (Bolduc et al., 2008; Kelleher and Bear, 2008; Liu-Yesucevitz et al., 2011). As a result, FXS patients are cognitively impaired and have behavioral abnormalities that include autistic-like features (Hagerman et al., 2009). The fragile X mental retardation protein FMRP is absent in FXS, establishing that FMRP is required for normal cognition. FMRP is an RNA binding protein that binds ~4% of brain mRNAs and regulates their expression— either enhancing or suppressing translation—by an unknown mechanism (Brown et al., 2001, Miyashiro et al., 2003). FMRP is implicated in microRNA (miRNA)-mediated translational suppression (Caudy et al., 2002; Edbauer et al., 2010; Ishizuka et al., 2002; Jin et al., 2004; Muddashetty et al., 2011), although the molecular basis for its role is unknown.
In contrast, much is known about the molecular mechanism of small RNA-mediated silencing (Wilson and Doudna, 2013). Upon engaging a target mRNA, the nucleic acid-binding cleft of Argonaute (AGO) opens to accommodate both the guide and target strands (Pratt and MacRae, 2009). The guide strand, held in a helical conformation by AGO, can increase the affinity of the target up to ~300 fold by decreasing the entropic cost associated with ordering the guide (Wilson and Doudna, 2013). Accordingly, kinetic analyses show that human RISC (minimally the AGO-guide strand complex) increases the ability of the guide to find and cleave its target RNA at a rate 10 times faster than the same small guide and target RNAs can anneal in free solution (Ameres et al., 2007). Importantly, this minimal RISC cannot unfold structured RNA, thus, creating a need for a protein to expose miRNA recognition elements (MRE) located within highly structured RNAs.
MOV10 is a helicase that was initially identified in a screen of mouse embryos intentionally infected with the Moloney leukemia virus (MOV) (Jaenisch et al., 1981; Mooslehner et al., 1991). Like FMRP, MOV10 has been implicated in miRNA-mediated translational suppression (Banerjee et al., 2009; Meister et al., 2005; Sievers et al., 2012). We show here that MOV10 has an effect on the fate of its bound RNAs: usually facilitating translation suppression, reflecting its role in the miRNA pathway; however, for a subset of its RNAs, MOV10 increases their expression by blocking AGO2 binding. These RNAs are also bound by FMRP. We show that FMRP binding at or near the MOV10 binding site blocks the usual role of MOV10 in the miRNA pathway. This is a new role for FMRP in the 3’UTR.
Results
FMRP associates with MOV10
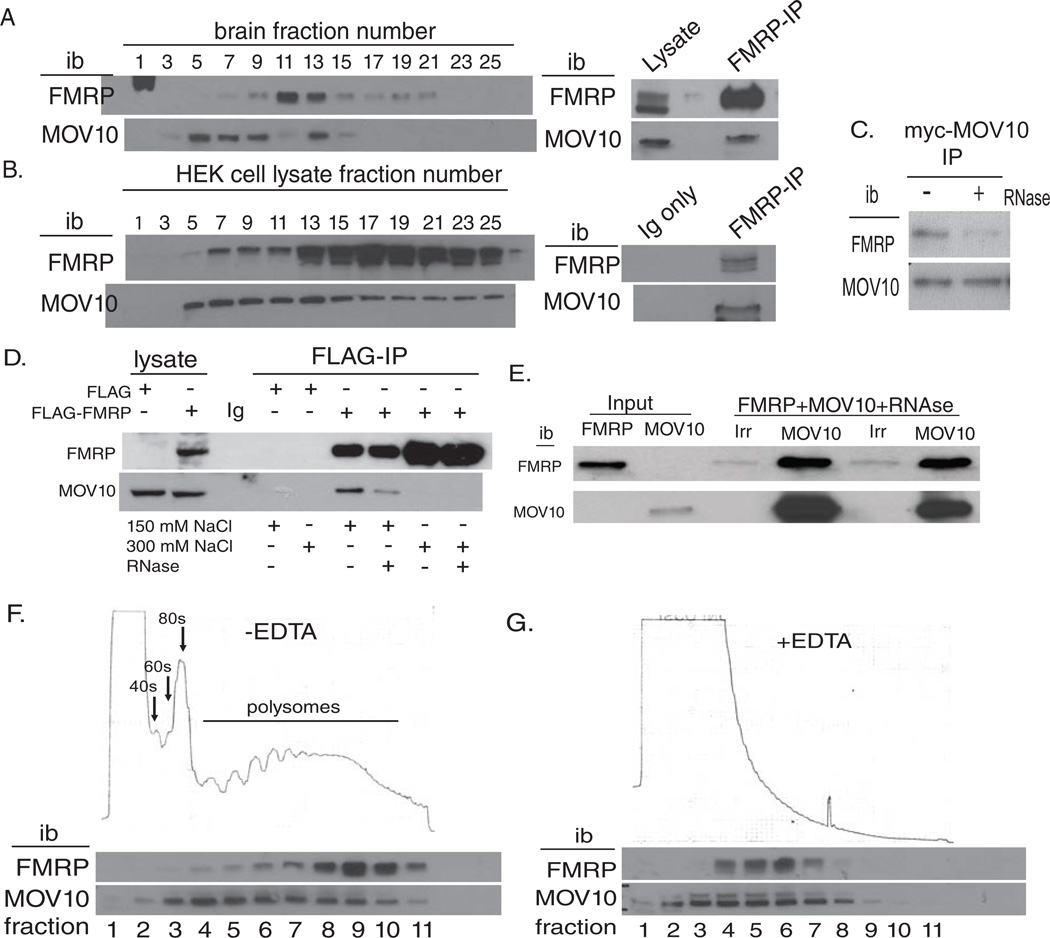
In addition to being implicated in the miRNA pathway, FMRP and MOV10 are both expressed in brain and co-localize in dendritic foci in cultured neurons, demonstrated by immunostaining (Banerjee et al., 2009; Liu-Yesucevitz et al., 2011; Wulczyn et al., 2007). To examine their physical association biochemically, we prepared an RNA sedimentation gradient on brain and HEK293T cells as described (Kanai et al., 2004). Both FMRP and MOV10 were present in fractions 7–15 in brain and 7–25 in HEK293T cells (Fig.1A and B). To show that FMRP and MOV10 are in the same complex in brain, we immunoprecipitated (IP’ed) FMRP from brain lysate and found it associated with MOV10 (Fig 1A, right). To demonstrate that FMRP and MOV10 directly associate in fractions from the RNA sediment gradient (versus being present individually in similarly sized populations), we pooled the MOV10- and FMRP-containing fractions, IP’ed FMRP and showed that MOV10 was associated with FMRP (Fig.1B, right).
Fig. 1. FMRP associates with MOV10 in brain and cell lines.
A, B) Brain and HEK293 extracts were analyzed as in (Kanai et al., 2004) and immunoblotted (ib) for FMRP and MOV10. Fractions with both FMRP and MOV10 were pooled and IP’ed for FMRP and MOV10 (right); C) myc-MOV10 was IP’ed, treated with RNaseA and ib for endogenous FMRP (Devys et al., 1993) and MOV10. D) FLAG-FMRP was IP’ed from L-M(TK-) cells (Ceman et al., 1999) in high EDTA to disrupt polysomes, treated with RNaseA (+) or not (−) and 150 mM or 300 mM NaCl and ib for MOV10 or FLAG. E) Duplicate co-IP of 2uM recombinant FMRP and MOV10 treated with RNAseA with irrelevant (Irr) or MOV10 antibody (MOV10) and ib for FMRP or MOV10. Input- 5ng of FMRP and 10 ng of MOV10. F) Polysome analysis of HEK293: 40S, 60S subunits, 80S ribosomes and mRNAs with multiple ribosomes (polysomes) indicated. G) EDTA treatment disrupts polysomes. The spike between fractions 7 and 8 is a technical artifact. See also Fig.S1.
The interaction between MOV10 and FMRP was characterized by IP of myc-MOV10 in the presence or absence of RNAse: we found that FMRP co-IP’ed in a partially RNA-dependent manner (Fig.1C). Further, anti-FLAG IPs of murine fibroblast L-M(TK-) cells stably expressing either empty FLAG vector (VC) or FLAG-FMRP (Ceman et al., 1999) showed that MOV10 associated with FMRP in a complex that was disrupted in 300 mM NaCl and was partially disrupted by treatment with RNaseA (Fig.1D). These data suggest that association of FMRP with MOV10 not only occurs in an RNA-dependent manner, but also has some protein-protein association. Accordingly, purified recombinant FMRP incubated with similarly prepared MOV10 in the presence of RNAseA showed a direct association, demonstrating a protein-protein interaction between FMRP and MOV10 (Fig.1E).
FMRP associates with translating ribosomes (polysomes) (Feng et al., 1997; Khandjian et al., 1996). We hypothesized that MOV10 interacts with FMRP to regulate translation. Examination of the distribution of FMRP and MOV10 on a sucrose gradient showed both proteins in the same fractions as actively translating polysomes (Fig.1F). Treatment with EDTA disrupts polysomes and removes MOV10 and FMRP from the heavier fractions, as described previously for FMRP (Feng et al., 1997; Khandjian et al., 1996) (Fig.1G). Thus, FMRP associates with MOV10 in an RNA- and protein-dependent manner and on polysomes, suggesting a role in translational regulation.
Identification of cellular RNAs bound by endogenous MOV10
The RNAs associated with FMRP in brain and cell lines have been extensively characterized (Ascano et al., 2012; Brown et al., 2001; Darnell et al., 2011; Miyashiro et al., 2003). In addition, there are two studies of RNAs associated with ectopically-expressed MOV10 (Gregersen et al. 2014; Sievers et al 2012), which can result in altered intracellular localization (Messaoudi-Aubert et al., 2010). Because FMRP and MOV10 associate in a partially RNA-dependent manner (Fig.1C,D), we used two independent approaches to identify mRNAs associated with endogenous MOV10 (Fig.S1). First, we IP’ed MOV10 and prepared libraries from the associated RNAs (RNA-IP) (Brown et al., 2001; Tenenbaum et al., 2000). Second, we used the approach of individual-nucleotide resolution UV cross-linking and IP (iCLIP) (Konig et al., 2010, 2011), which allowed determination of the MOV10 binding regions on the identified RNAs. As a control for non-specific RNA binding, an irrelevant rabbit antibody (Winograd et al., 2008) (ir) was used in parallel to the MOV10-specific IP (Fig. S1A).
RNAs that co-IP’ed with MOV10 in the RNA-IP were sequenced and compared to the transcriptome of HEK293 cells. Approximately 80% of the reads were aligned with sequences from exonic regions of the genome representing 18,831 protein-coding genes and 4757 non-coding genes. We used EdgeR to compare the RNA-IP samples with the total transcriptome and identified 2117 genes (2039 protein coding genes and 78 noncoding) that were significantly enriched (Table S1).
For the iCLIP experiments, random barcodes were used in two independent library preparations and similar amounts of RNA from each sample were sequenced. A total of 1.4M mapped reads were obtained from both MOV10 iCLIP experiments (698,430 and 686,898 mapped reads, respectively), hereafter referred to as C5 and C7 libraries (Fig.S1B). In contrast, only 0.2M mapped reads were obtained from the combined irrelevant IPs (ir1 and ir2, Fig.S1B). Among the genome-mapped reads from C5 and C7, 67% of the reads were mapped to regions within genes, of which 33% aligned within 3’UTRs (Fig.S1C). Normalization of the reads (RPKM) indicated that the 3’ UTR had the highest depth of coverage (Fig.S1D).
In C5 and C7, 32,331 clusters were identified within 8986 genes, with 15,475 of these clusters found in 3’UTRs. We followed standard iCLIP protocols (Konig et al., 2010, 2011) to determine the cross-link sites, obtain reads and their subsequent clusters, which were used to identify MOV10-associated genes. Within the 32,331 clusters, 78% had multiple cross-link sites and 5% had 10 or more cross-link sites, indicating that the MOV10 clusters were of high quality. Within these clusters, 62% of the cross-link sites were within one nucleotide of each other and 88% were within 5 nucleotides of each other (Fig.S1E). Only 1687 clusters were observed in the irrelevant IP samples and were filtered for downstream analysis.
We compared the RNA-IP data to the iCLIP data (Tables S1 and S2, respectively) and found that most of the RNAs identified in the iCLIP data (1850 genes) were also significantly enriched (2117 genes) in the RNA-IP data (Fig.S1F), indicating consistency between the two experimental approaches. Although some of the iCLIP targets were not enriched in the RNA-IP (Fig.S1G, orange boxes), in general, the iCLIP targets were well represented in the RNA-IP at different expression levels.
FMRP and MOV10 bind a subset of RNAs and FMRP facilitates association of MOV10 with target mRNAs
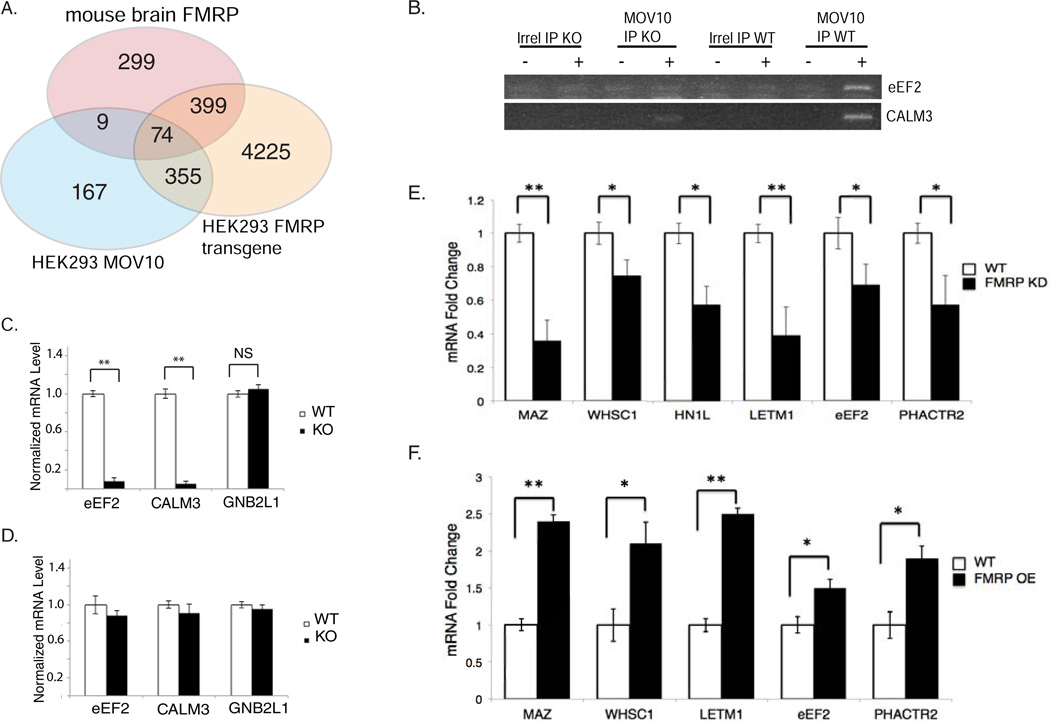
Because we found MOV10 and FMRP associated in a partially RNA-dependent manner in Figure 1, we identified the commonly bound RNAs by comparing the CLIP lists of FMRP isolated from HEK293 (Ascano et al., 2012) and from brain polysomes (Darnell et al., 2011) with the MOV10 iCLIP sites found at least once in both C5 and C7 libraries. Using the permutation approach described in the Experimental Approach, we found statistically significant overlap in a number of shared target mRNAs (Fig.2A).
Fig. 2. MOV10 recognizes a subset of RNAs bound by FMRP.
A) Venn diagram of HEK293 MOV10 iCLIP targets, brain FMRP CLIP RNAs (Darnell et al., 2011) and FMRP isoform1 CLIP RNAs from transfected HEK293 (Ascano et al., 2012). See Methods for details and statistical analysis and Table S4. B) RT-PCR of eEF2 and CALM3 RNAs synthesized with (+) or without (−) RT, Irrelevant or MOV10 IP from FMR1 KO and WT brains on a 2% ethidium bromide gel. C) qPCR of MOV10-associated RNAs (eEF2, CALM3 and GNB2L1) IP’ed from WT and FMR1 KO brains (n=3) and normalized to GAPDH. D) qPCR of RNAs from WT and FMR1 KO brains (n=3), E) qPCR of MOV10 RNAs IP’ed from WT and FMR1 KD HEK293 cells (n=3). F) qPCR of MOV10 RNAs IP’ed from WT and FMR1-OE HEK293 cells (n=3). See also Fig.S2
The functional relationship of mRNAs bound by both FMRP and MOV10 was examined using targets CALM3 and eEF2, which were identified in both the RNA-IP and iCLIP and were also present in the FMRP brain CLIP list (Darnell et al., 2011). IP with an irrelevant antibody from WT and FMR1 knockout (KO) brains showed background levels of both RNAs (Fig.2B). In contrast, IP of MOV10 from WT brains showed significantly more eEF2 and CALM3 mRNA than from FMR1 KO brains (Fig.2B,C), suggesting that FMRP facilitates association of MOV10 with commonly bound RNAs. As a control, we examined the amount of MOV10-associated GNB2L1 RNA, which is bound by MOV10 (Table S1 and S2) but not FMRP in brain (Darnell et al., 2011). MOV10-associated GNB2L1 was the same in both the WT and FMRP KO mice (Fig. 2C). The total level of all assessed RNAs was the same in both WT and KO brains (Fig. 2D).
To further explore the effect of FMRP on MOV10 association with RNA, we IP’ed MOV10 from cells in which FMRP was knocked down (KD) or overexpressed (OE). We found that MOV10-associated RNAs decreased in the absence of FMRP (Fig.2E) while those same mRNAs increased in association with MOV10 when FMRP was OE (Fig.2F). Our results suggest a cooperative interaction between FMRP and MOV10, wherein FMRP binding to RNAs facilitates association with MOV10. Total mRNA levels of target RNAs were unchanged by FMRP KD or OE (Fig.S2), agreeing with previous work (Ascano et al., 2012).
MOV10 and AGO bind near predicted MREs and recognize G-rich sequences
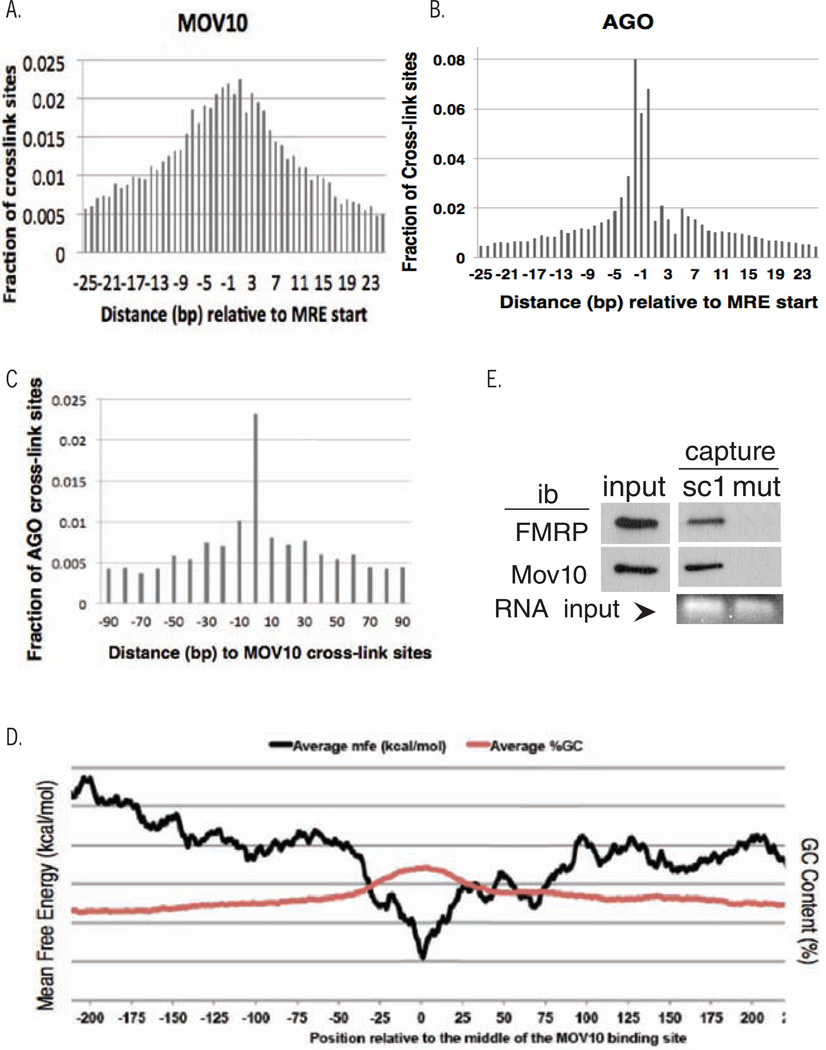
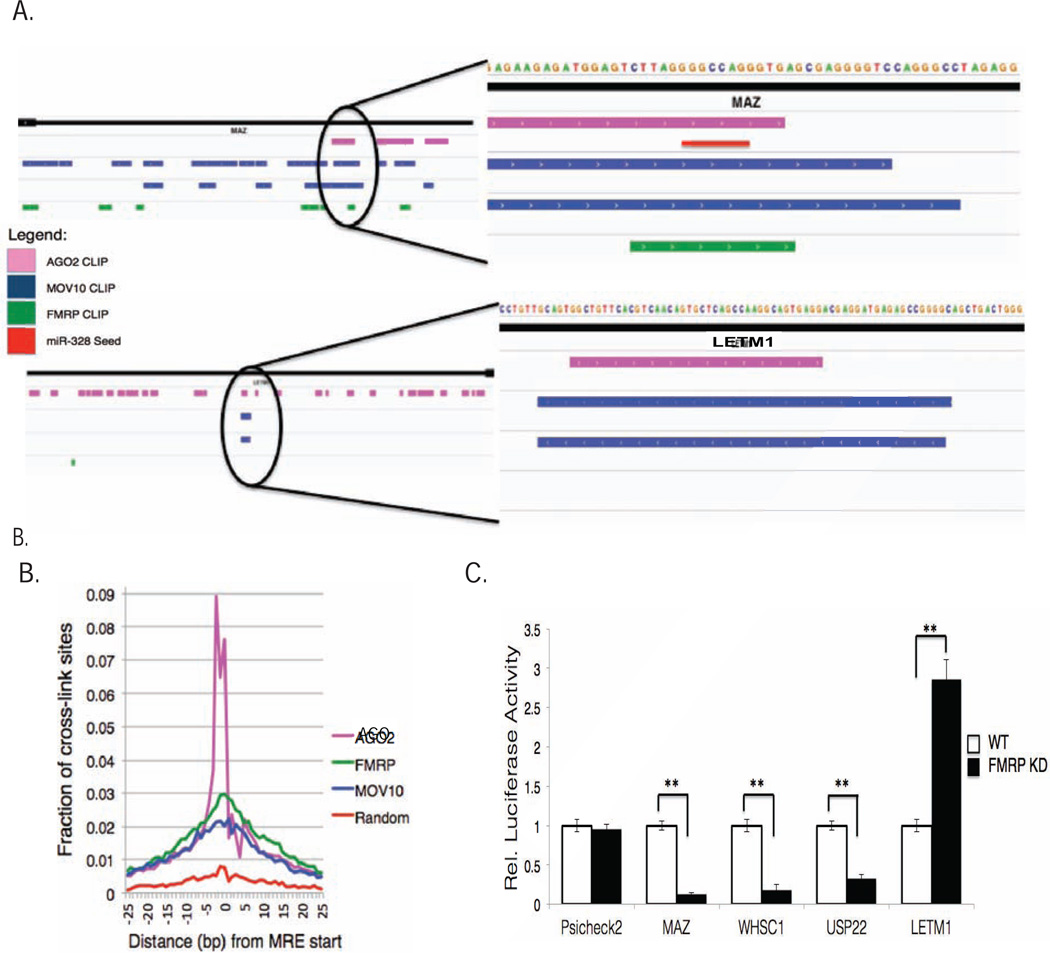
MOV10 has a physical and functional association with AGO (Meister et al., 2005; Sievers et al., 2012). The high abundance of MOV10 mapped sites in the 3’ UTRs of target mRNAs is consistent with a role for MOV10 in post-translational regulation through the miRNA pathway. Thus, we analyzed the relationship between MOV10 binding sites and MREs. Based on the 6-mer (2–7 nt at the 5` end of the miRNA) rules of miRNA binding, we predicted the MREs for the top 100 highest expressed miRNAs in HEK293 cells (Hafner et al., 2010) and calculated the distance between the cross-link sites and its closest MREs. The cross-link sites with the most reads within each cluster were selected for analysis. Interestingly, 22.3% of the MOV10 cross-link sites were located within the 5 nucleotides (nts) flanking the MREs, 62% of the MOV10 cross-link sites in the clusters were identified within 25 nts of their closest MREs and 75.5% within 100 nts of the closest MRE (Fig.3A). We also studied the distributions of the AGO binding sites (Hafner, et al., 2010) relative to the MOV10 binding sites and found significant overlap among the mRNAs targeted by both MOV10 and AGO, confirming the results of others (Sievers et al., 2012). A specific example is shown with the RNA target GLE1 (Fig.S3A). Despite differences between the samples and CLIP protocols, there were still 74% (4777 out of 6436 AGO targets) of genes targeted by both AGO and MOV10. Thus, MOV10 and AGO bind many of the same mRNAs; however, we did not find significant precise overlaps of their respective cross-link sites compared with what we have observed between their cross-link sites and MREs (Figs.3A,B). Only 918 of the AGO cross-link sites were within 25 bp from MOV10 target sites, although the cross-link sites that did overlap increased with decreasing distance (Fig.3C). In conclusion, MOV10 and AGO bind the same mRNAs in proximity to MREs.
Fig. 3. Characterization of MOV10 binding sites in the 3’UTR.
A, B) Fraction of MOV10 or AGO UV-cross-link sites plotted against distance in bps to predicted MRE start site (Hafner et al., 2010);. C) Relative distance between AGO CLIP sites and MOV10 cross-link sites. D) Mean free energy plot of MOV10 3’UTR iCLIP sites. ΔGfolding was calculated across 55 nt windows +/− 275 nt from center of iCLIP site (black). % GC content plotted in red. E) RNA capture assay with GQ sc1 and sc1 mutant of in vitro synthesized FMRP and recombinant MOV10. Bottom: RNA input-ethidium gel. See also Fig.S3.
MOV10 recognizes G-rich and GQ-containing motifs
To identify the recognition motif of MOV10, we determined the strandedness of the 3’UTR CLIP sites identified at least once in both C5 and C7 libraries in Integrated Genomics Viewer (IGV2.3), entered these sites into MEME (Bailey and Elkan, 1994) and identified three sequences that were collectively found in 153 of the 454 genes analyzed (Fig.S3B). Because not all of the CLIP sites contained one of these three linear motifs, we analyzed the structural features of the 3’UTR CLIP sites by calculating the mean free energy using a modified version of ViennaRNA Package (Hofacker et al., 1994). The decrease in ΔGfolding at the CLIP sites along with a correlative increase in GC content suggests that MOV10 recognizes and binds GC-rich secondary structures (Fig.3D).
The motifs recognized by FMRP have been extensively characterized (Ascano et al., 2012; Brown et al., 2001; Chen et al., 2003; Darnell et al., 2001) and include both G-rich sequences and G-quadruplexes (GQs) (Muddashetty et al., 2011; Phan et al., 2011; Westmark and Malter, 2007). GQs are stable nucleic acid structures that can be substrates for helicases, as is the case for the primarily nuclear helicases G4R1/RHAU and DHX9 (Chakraborty and Grosse, 2011; Creacy et al., 2008).
To examine the MOV10 iCLIP sites for putative GQs, we used the GQ prediction program QGRS Mapper (Kikin et al., 2006; Menendez et al., 2012) and found that 27.2% of the MOV10 3’UTR CLIP sites contained predicted GQs--nearly twice that predicted in a large-scale screen of 3’UTRs (Beaudoin and Perreault, 2013). To ask directly whether MOV10 bound GQs, we tested its ability to bind the RNA sc1, a model GQ that binds FMRP with nanomolar affinity (Darnell et al., 2001). Like FMRP, MOV10 specifically bound sc1 and was unable to bind the nucleotide-substituted sc1-mutant, in which formation of the GQ is disrupted (Fig.3E). Thus, both FMRP and MOV10 are able to bind GQs.
MOV10 regulates expression through the 3’UTR and modulates AGO2 function
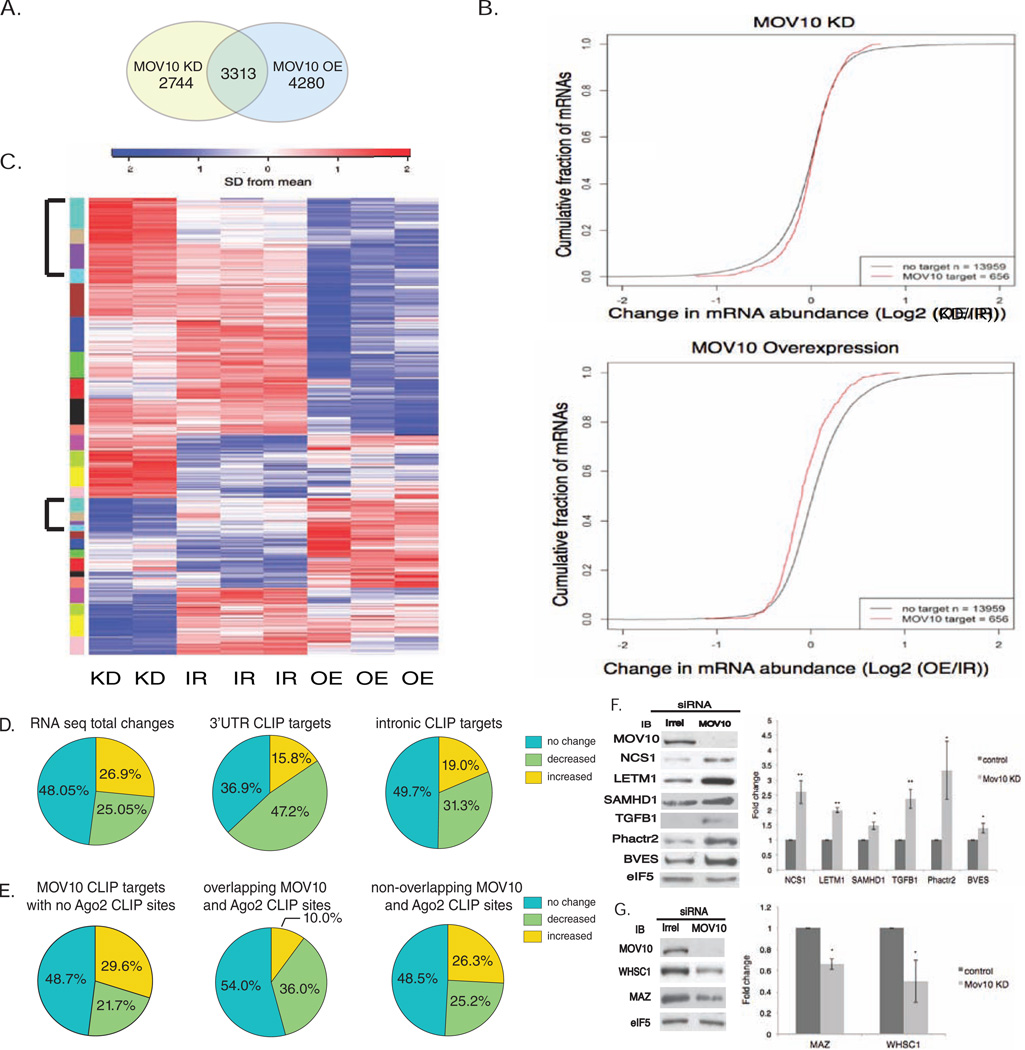
Because MOV10 binds in close proximity to MREs and AGO binding sites (Fig. 3), we hypothesized that MOV10 functions in the miRNA pathway. As a consequence of miRNA-mediated translational suppression, it would be expected that a larger percentage of MOV10 target mRNAs would decrease (Baek et al., Hendrickson; Guo et al., 2010; Lim et al., 2005). Accordingly, KD of MOV10 should lead to an increase in mRNA levels. To examine the effect of MOV10 on total mRNA levels, we treated cells with either MOV10 siRNAs for KD, irrelevant siRNAs as a control (IR), or overexpressed a MOV10 transgene (OE) and evaluated mRNA levels by RNA-Seq (Table S3). We identified 14,679 RNAs in the total RNA pool and found that 6057 RNAs changed significantly in the KD while 7593 RNAs changed in the OE (Fig.4A). The changes in RNA levels in both the KD and OE were significant (p<0.05, False Discovery Rate [FDR]) compared to the control treatment. 3313 genes significantly changed expression in both treatment conditions. Of these, 1216 RNAs changed in opposite directions in the KD or OE: specifically, in the absence of MOV10, 604 RNAs increased while 612 decreased. In the OE, those same RNAs changed in the opposite direction.
Fig. 4. MOV10 affects mRNA and protein levels of iCLIP targets.
A) Venn diagram of significantly changed RNAs (FDR p<0.05) in MOV10 KD (6057 RNAs) and OE experiments (7593 RNAs). B) iCLIP targets in MOV10 KD and OE were significantly changed compared to non-CLIP RNA (top, p=0.0068, Chi-square test; bottom, p=1.83E-26, Chi-square test). C) Heatmap of significantly changed MOV10 iCLIP RNAs in MOV10 KD, irrelevant siRNA (IR), and MOV10 OE created using Weighted Gene Correlation Network Analysis (WGCNA). Colored bars indicate discrete modules of RNAs. Individual experiments indicated at bottom. Black brackets indicate anti-correlated groups: top-MOV10 iCLIP targets increased upon KD and decreased upon OE (100 genes); bottom- MOV10 iCLIP targets decreased upon KD and increased upon OE (39 genes). D) Distribution of RNAs in MOV10 OE that did not change (blue), significantly decreased (green) or significantly increased (yellow) for total RNA (left), 3’UTR CLIP targets (center) or intronic CLIP targets (right). See methods for RNA-seq statistics. E) Distribution of RNAs in MOV10 KD with no 3’UTR AGO2 CLIP sites (left, 115 RNAs), overlapping MOV10 and AGO2 CLIP sites in 3’ UTR (center, 50 RNAs) and non-overlapping MOV10 and AGO2 CLIP sites in 3’UTR (left, 167 RNAs) Decreased: 21.7% vs. 36% is significant (X2= 2.9752, df = 1, p-value = 0.04228) (1-tailed). Increased: 10% vs. 26.3% = X2 = 4.9842, df = 1, p-value = 0.01279 (1-tailed); Increased: 10% vs. 29.6% = X2 = 6.3465, df = 1, p-value = 0.005881 (1-tailed). F, G) irrelevant (Irrel) or MOV10 siRNA treated HEK293 (MOV10 KD was >80%); loading control-eIF5. Right: fold change in protein levels from three independent experiments. Error bars were plotted using Std. deviation and p-values using Student’s t-test, * p <0.05, ** p<0.01). See also Table S3 and Fig.S4.
We compared the fate of the MOV10 iCLIP targets to the non-CLIP targets. As expected, direct MOV10 binding had a significant impact on the mRNA levels, with an overall increase in mRNA expression in MOV10 KD and an overall decrease in expression in MOV10 OE (Fig.4B top, shift to right; bottom, shift to left). MOV10 iCLIP targets changed significantly in the KD (p=0.0068) and the OE (p=1.833E-26) when compared to the non-CLIP targets (Fig.4B and Table S3). The larger effect observed in the OE experiment likely reflects OE of the MOV10 transgene (>30-fold, Table S3). Thus, RNAs that are directly bound by MOV10 are more likely to have significantly altered expression than the RNAs that are not CLIP targets under conditions of MOV10 KD or OE.
We created a heat map to visualize the fate of the MOV10 iCLIP targets after MOV10 KD or OE. 312 genes had FDR p < 0.05 in KD vs IR (mock treatment) (172 up/140 down) and 412 had FDR p < 0.05 in OE vs IR (123 up/289 down). Combining these lists yielded 541 genes in which 139 genes were anti-correlated: 100 genes increased in KD and decreased in OE (indicated toward the top by a black bar) as would be expected if MOV10 participates in miRNA-mediated silencing. Importantly, there were also clusters of RNAs that demonstrated the opposite expression pattern (39 genes decreased in KD and increased in OE, bottom black bar) (Fig.4C). These two expression patterns suggested that MOV10 binding has two distinct fates: MOV10 binding decreases the RNA levels of some iCLIP targets but increases the levels of others. Because of the hypothesized role of MOV10 in the miRNA pathway and the greater efficacy of MREs in the 3’UTR (Bartel, 2009; Eulalio et al., 2008; Hausser et al., 2013), we were particularly interested in the fate of mRNAs in which MOV10 bound in the 3’UTR. Examination of the effect of MOV10 OE on 3’UTR CLIP targets revealed that 47.2% were decreased (Fig.4D, center) compared to the fate of non-CLIP RNAs (~ 25%) and intronic CLIP targets (~30%) (Fig.4D), consistent with MOV10 having a role in miRNA-mediated degradation.
We next analyzed the 3’UTR CLIP targets for the presence of AGO2 CLIP sites (Xue et al., 2013), hypothesizing that MOV10 modulates miRNA-mediated translation suppression by either facilitating or blocking AGO2 association. MOV10 and AGO2 bind in proximity to one another (Fig.3). We now examined the fate of MOV10 iCLIP mRNAs upon MOV10 KD and observed three categories of RNAs with 3’UTR MOV10 iCLIP sites: 1) RNAs that contained no AGO2 CLIP sites in their 3’UTR (Fig.4E, left); 2) RNAs with overlapping MOV10 and AGO2 CLIP sites (Fig.4E, center); 3) RNAs with MOV10 and AGO2 CLIP sites that did not overlap (Fig.4E, right). When there was overlap between the MOV10 and AGO2 CLIP sites, the percentage of RNAs that decreased upon MOV10 KD was significantly larger than the percentage of RNAs that decreased when there were no AGO2 CLIP sites (36% compared to 21.7%, p=0.042). This observation suggested a protective role for MOV10 on those RNAs where the MOV10 and AGO2 CLIP sites overlapped such that loss of MOV10 led to decreased RNAs. Accordingly, the percentage of RNAs that increased upon MOV10 KD when the MOV10 and AGO2 sites overlapped was significantly reduced compared to the percentages of increased RNAs in the other two categories (10% compared to 26.3% and 29.6%, p<0.05). Thus, when the MOV10 and AGO2 CLIP sites overlap, MOV10 binding appeared to antagonize AGO2-mediated transcript reduction.
To evaluate the fate of MOV10 on steady-state protein levels encoded by individual CLIP targets, we examined the 3’UTR MOV10 iCLIP targets whose RNAs are regulated by miRNAs in HEK293 cells (Schmitter et al., 2006). MOV10 KD significantly increased the expression of the endogenous proteins (Fig.4F), as would be expected if MOV10 participated in miRNA-mediated suppression. In contrast, endogenous protein levels of MAZ and WHSC1 were significantly decreased upon MOV10 KD (Fig.4G). Both proteins have overlapping MOV10 and AGO2 CLIP sites in their RNAs, suggesting that MOV10 blocks miRNA-mediated suppression by inhibiting AGO2 binding—either through steric hindrance or through an inability of MOV10 to unwind and expose MREs. To both verify the effect of MOV10 on the 3’UTR and to test iCLIP targets for which antibodies were not available, luciferase reporters of 3’UTRs were examined (Fig.S4A). Three other MOV10 targets with overlapping MOV10 and AGO2 CLIP sites, namely eIF4B, HN1L and USP22, were also decreased in the absence of MOV10 (Fig.S4A, right), supporting the idea that MOV10 binding blocks AGO2 function when bound to the AGO2 site. However, realizing that overlap of MOV10 and AGO2 on mRNA did not always lead to increased mRNA or protein levels in the absence of MOV10 (Fig.4e center), we hypothesized that another co-factor might be playing a role in blocking translation suppression by AGO2.
FMRP modulates MOV10’s role as a translational suppressor of co-bound mRNAs
Knowing that MOV10 binding is facilitated by FMRP on co-bound mRNAs (Fig. 2), we wondered if there was an additional role for FMRP in translation regulation. We examined binding patterns within the 3’UTR (which often contained multiple FMRP binding sites) that correlated with increased or decreased mRNA expression in the MOV10 KD. In the mRNAs in which expression levels decreased upon MOV10 KD, we observed clear overlap in AGO2, MOV10, and FMRP CLIP sites, of which MAZ, WHSC1, USP22, HN1L, and eIF4B were included. We hypothesized that FMRP binding at the site of MOV10/AGO2 overlap blocked AGO2-mediated suppression. 3’UTRs from two representative RNAs with opposite fates (and opposite FMRP binding profiles) are shown in Figure 5A. LETM1 has no FMRP CLIP sites overlapping the MOV10 and AGO2 CLIP sites and increases upon MOV10 KD (Fig.4F,S4). In contrast, MOV10, FMRP and AGO2 CLIP sites overlap in MAZ, suggesting that proximal binding of FMRP and MOV10 protects from AGO2 binding and suppression (Fig.4G,5A,S4).
Fig. 5. FMRP modulates MOV10’s role in translational regulation.
A) IGV screen capture of MAZ and LETM1 MOV10 iCLIP sites from C5 and C7 libraries, AGO2 CLIP sites (Xue et al., 2013) and FMRP CLIP sites (Ascano et al., 2012). B) Overlay of MOV10 (blue), AGO (pink) and FMRP (green) cross-link sites plotted against distance in bps to predicted MRE start site. Random sites in random genes were selected as a control (red). C) Effect of FMRP KD on luciferase expression of 3’UTRs of MOV10 iCLIP targets. Error bars represent standard deviation, p-values obtained by Student’s t-test, * p <0.05, ** p <0.01. See also Fig.S5.
To determine if FMRP has a role in regulating miRNA-mediated translation, we analyzed the distance between predicted MREs and the FMRP crosslink sites obtained from FMRP CLIP data (Ascano et al., 2012). Similar to what we found for MOV10, the majority of the FMRP 3’UTR CLIP sites (78%) were within 25 nucleotides of a predicted MRE (Fig.5B, green line). The strong correlation in the proximity of iCLIP sites to known MRE sites for AGO, FMRP and MOV10 suggests an interactive role of these proteins in miRNA-mediated regulation (Fig.5B).
To establish a role for FMRP in RISC function, we knocked down FMRP and examined luciferase expression of the MAZ, WHSC1, and USP22 3’UTR reporters (Fig.5C). In the FMRP KD, we observed the same result as in the MOV10 KD, which was a significant decrease in luciferase expression, suggesting that FMRP protects those mRNAs from suppression. In contrast, LETM1, which has an FMRP CLIP site at the 3’ end of the 3’UTR but does not overlap with the MOV10 iCLIP sites, showed an increase in luciferase expression in the absence of FMRP, reflecting typical miRNA-mediated translation suppression. The same effect of FMRP KD was observed on the endogenous MAZ and LETM1 proteins (Fig.S5).
MOV10 modulates association with AGO
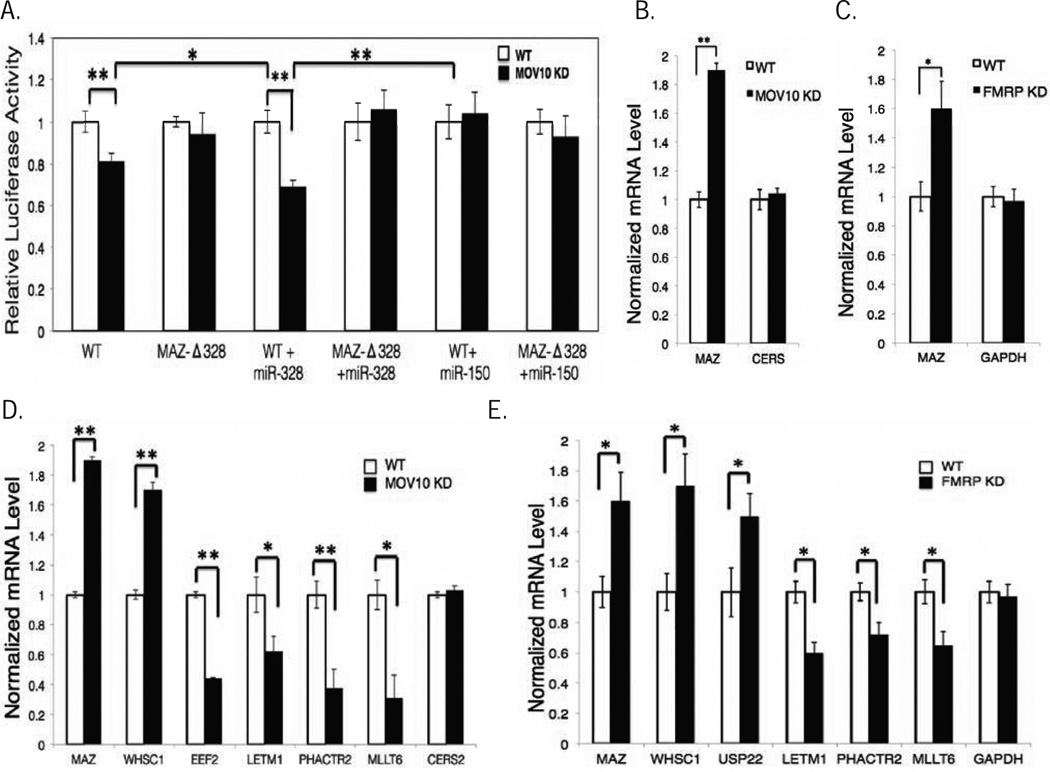
We used MAZ to demonstrate that MOV10 binding blocked miRNA-mediated silencing by identifying candidate miRNAs using TargetScan that were tested for their ability to suppress MAZ expression upon MOV10 KD (Fig.S6A). miR-328 was identified as the primary miRNA associated with MOV10-mediated suppression of the MAZ reporter (Fig.5A red bar, S6A). We found that ectopic expression of miR-328 significantly decreased MAZ 3’UTR reporter expression after MOV10 KD (Fig.6A, compare 3rd column set to 1st column set), suggesting MOV10 blocks the effect of miR-328. To demonstrate specificity, mutation of two nucleotides within the MRE of miR-328 in the MAZ 3’UTR reporter eliminated the effect of MOV10 KD on expression, as well as the ability of exogenously introduced miR-328 to suppress the reporter (Fig.6A, 3rd column set, 4th column set, respectively). miR-150 was used as a negative control. We conclude that MOV10 blocks miR-328 mediated suppression of MAZ.
Fig. 6. FMRP modulates effect of MOV10 on AGO function.
A) Effect of MOV10 KD on MAZ 3’UTR (WT) expression and miR-328 site deletion (Δ-328) co-transfected with miRNAs indicated. Error bars represent standard deviation, p-values obtained by student t-test, * p <0.05, ** p <0.01 See Fig. S6. B) Effect of MOV10 KD on AGO-associated RNAs (X-axis) by Q-PCR. C) Effect of FMRP KD on AGO-associated RNAs (X-axis) by Q-PCR. D, E) Q-PCR of additional AGO-associated RNAs in the presence or absence of MOV10 or FMRP. See also Fig.S6.
To determine whether MOV10 modulated AGO association with the MAZ mRNA, we IP’ed AGO and quantified associated MAZ RNA in the presence or absence of MOV10. Significantly more MAZ mRNA associated with AGO in the MOV10 KD than in the presence of MOV10 (Fig.6B), suggesting that MOV10 blocked AGO binding to MAZ. As a control, the CERS2 mRNA, which is bound and regulated by AGO (Chi et al.,2009) but not by MOV10 (Table S4), was bound equally well under both conditions.
FMRP affects AGO binding to mRNA
Our hypothesis is that MOV10 generally facilitates AGO-mediated suppression but its interaction with FMRP blocks suppression of a subset of mRNAs including MAZ. To test this hypothesis, we IP’ed AGO and determined whether FMRP modulated AGO association with the MAZ mRNA by quantifying AGO-associated RNAs in the presence or absence of FMRP. AGO association with MAZ, was increased upon FMRP KD (Fig.6C), suggesting that like MOV10, FMRP blocks association of AGO with MAZ.
MOV10 can modulate mRNA expression through its association with FMRP
Based on the results with MAZ, we examined mRNAs that contained similar overlapping CLIP sites of FMRP, MOV10, and AGO compared to mRNAs with non-overlapped sites. We then determined whether AGO association with these target RNAs changed in the presence or absence of MOV10 and FMRP. In the MOV10 KD, we observed that WHSC1, which contained overlapped binding sites and was reduced upon MOV10 KD (Fig.4G), showed an increase in AGO association, similar to what we observed with MAZ (Fig.6D). In contrast, mRNA targets that did not contain overlapped sites like LETM1 (Fig.5A) and Phactr2 and had increased expression upon MOV10 KD (Fig.4F) resulted in less AGO association upon MOV10 KD (Fig 6D).
We previously showed that FMRP facilitates MOV10 binding to co-bound mRNAs (Fig.2). Therefore, we expected to observe an overall decrease in AGO associated mRNAs in FMRP KD as a result of reduced MOV10 recruitment, which would have facilitated AGO association by exposing MREs. Accordingly, in the absence of FMRP, there was a decrease in AGO-associated mRNAs in which FMRP binding sites were not proximal to MOV10 binding sites, as seen in LETM1, PHACTR2, and MLLT6 (Fig.6E). However, AGO association with MAZ, WHSC1, and USP22, all of which contain FMRP sites coincident with MOV10 binding sites, was increased upon FMRP KD (Fig.6E), suggesting that FMRP blocks association with AGO. Taken together, this result indicates that multiple FMRP sites within the RNA may facilitate MOV10 binding unless the AGO-FMRP-MOV10 sites are coincident, in which case, FMRP and MOV10 block AGO association.
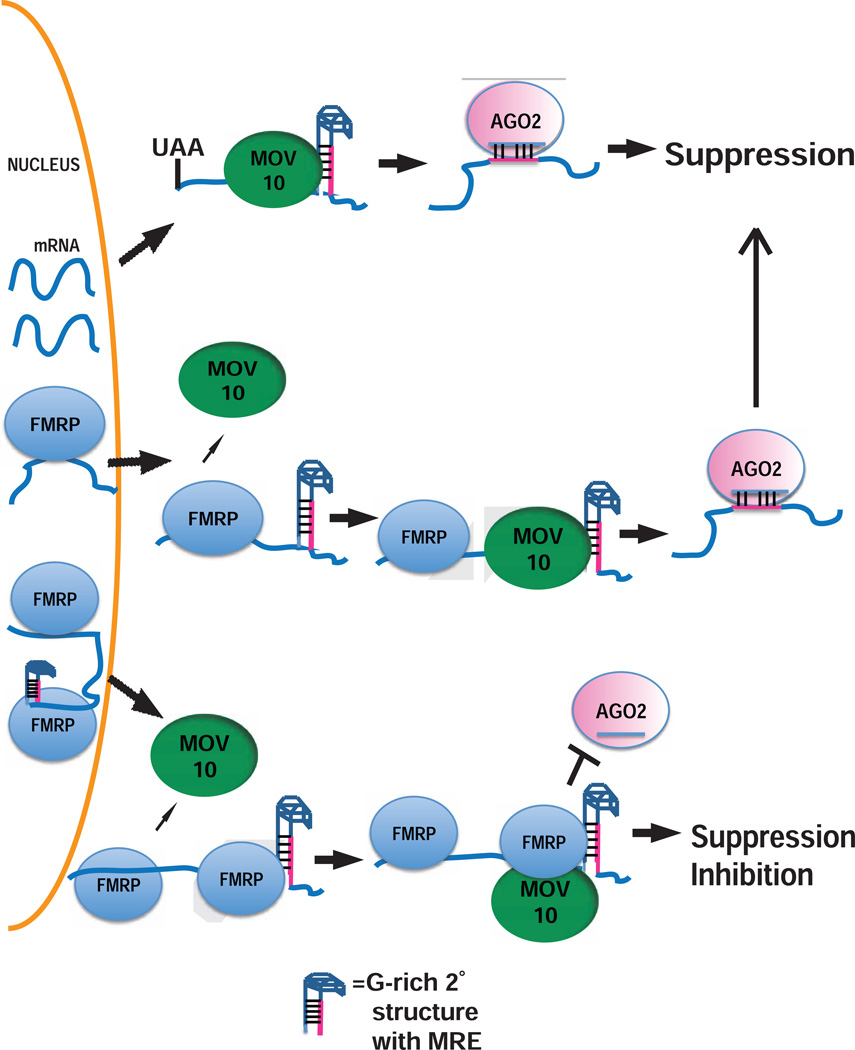
In summary, we propose a model where MOV10 binding to GC-rich structures facilitates RISC association by exposing MREs in the mRNA. In the subset of mRNAs that are co-bound with FMRP, there are two distinct fates determined by where they bind and interact in the 3’UTR. mRNAs that do not contain an overlapped MOV10-FMRP site are translationally suppressed, with FMRP facilitating MOV10 binding to the mRNA. However, if FMRP is bound at or near the MOV10 binding site, then MOV10 is unable to facilitate AGO interaction and the mRNA is protected from AGO-mediated suppression (Fig.7). This is a new, previously undescribed function for FMRP in the 3’UTR.
Fig. 7. Model for MOV10-FMRP association in translation regulation.
A) Fate of RNAs bound by MOV10: MOV10 binds 3’UTR-encoded G-rich structure to reveal MREs for subsequent AGO2 association; B) Fate of RNAs bound by FMRP: FMRP binds RNAs in the nucleus. Upon export, FMRP recruits MOV10, which ultimately unwinds MREs for association with AGO2; C) FMRP recruits MOV10 to RNAs; however, binding of both FMRP and MOV10 in proximity of MRE blocks association with AGO2.
Discussion
We hypothesize that FMRP binds the mRNAs first--perhaps in the nucleus (Kim et al., 2009). Upon export to the cytoplasm, FMRP may participate in granule formation through its low complexity sequence (Kato et al., 2012), recruiting MOV10 and other proteins.
One hypothesis for FMRP facilitating MOV10 binding is that it may stabilize areas of single stranded mRNA, which allows MOV10 to bind and initiate its helicase activity. Recent work by Gregersen and colleagues showed that MOV10 binds single stranded RNA and translocates in a 5’ to 3’ direction within the 3’UTR (Gregersen et al., 2014). A second hypothesis based on the observation that MOV10 and FMRP are found on polyribosomes and evidence that FMRP directly binds ribosomes (Chen et al., 2014) is that the FMRP/MOV10 complex may translocate with the ribosome to the translation termination site. In fact, through the use of non-translocating MOV10 mutants, Gregersen and colleagues found that initial MOV10 binding takes place at the accessible region at the beginning of the 3’UTR ~21 nt downstream of the terminating ribosome (Gregersen et al. 2014). In both hypotheses, MOV10 unwinds GC-rich secondary structure, allowing AGO2 to bind formerly inaccessible MREs, consequently facilitating translational suppression. However, if FMRP also binds at this site, AGO2 association is blocked. At this point, FMRP may be competing with MOV10 for the same G-rich sequence because both proteins bind GQs, or, FMRP may physically associate with MOV10 to prevent it from translocating to unwind and reveal MREs. FMRP binds and stabilizes G-Quadruplexes (Phan et al., 2011). Thus, it is possible that MOV10’s function as a helicase is hindered by FMRP’s ability to stabilize the structure, therefore not allowing unwinding and consequently access of AGO2 to the MRE.
In contrast to our work, Gregersen and colleagues observed no enrichment of MOV10 PAR-CLIP reads around predicted MREs in the 3’UTR. We suspect that this disparity reflects differences in how the data were interpreted and analyzed. We used the actual MOV10 cross-link sites to infer a relationship with MREs (as opposed to using read coverage as they did). Using actual MOV10 cross-link sites is a more reliable method to identify high quality binding sites (Hafner et al., 2010), and is not influenced by transcript abundance (Uren et al., 2012). Using cross-link sites also circumvents issues regarding PCR duplicates, which can confound coverage density. Another significant difference between their study and ours is that they did not observe FMRP association with MOV10. This important relationship was probably not apparent in their data because of the NaCl concentration used in their washes. We show that the association of FMRP and MOV10 is preserved in 150 mM NaCl but completely lost in 300 mM NaCl (Fig.1).
We provide evidence here of a role for FMRP in miRNA-mediated regulation where in concert with MOV10, a subset of mRNAs is protected from AGO2-mediated translation suppression. We also show that FMRP binds in proximity to MREs in the 3’UTR. KD of FMRP did not affect transcript levels in our hands or in others (Ascano et al., 2012), which is at first surprising in light of the genome-wide studies that have proposed that miRNA-mediated repression mainly leads to mRNA decay at steady state (Baek et al., 2008; Guo et al., 2010; Hendrickson et al., 2009; Selbach et al., 2008). However, as described in (Bethune et al., 2012), other studies show that miRNA-mediated suppression can be rapidly reversed in response to different cellular cues (Bhattacharyya et al., 2006; Muddashetty et al., 2011; Schratt et al., 2006). Reversible silencing may be of critical importance in cells such as neurons, where localized translation at synapses occurs in response to stimulation, requiring that target mRNAs are repressed translationally without major mRNA decay (Bhattacharyya et al., 2006; Muddashetty et al., 2011; Schratt et al., 2006).
In demonstrating that MOV10 has a dual role in facilitating and blocking AGO2 activity, this relationship is similar to the novel function described for polypyrimidine tract binding protein PTB, which suppresses or enhances miRNA targeting by competitive binding on target mRNAs or by altering local RNA secondary structure (Xue et al., 2013). Like PTB, MOV10 joins the growing list of RNA binding proteins that have been implicated in modulating miRNA targeting (van Kouwenhove et al., 2011).
In summary, we have identified a novel functional partner for FMRP that modulates miRNA-mediated translation regulation by AGO, giving new insight into how FMRP regulates translation of a subset of RNAs. In addition to its role as a translation suppressor when it binds in the coding sequence (Darnell et al., 2011), we have now identified a novel role for FMRP when it binds in the 3’UTR.
Experimental Procedures
Cell lines, siRNA and reporter transfection, Q-PCR and molecular procedures including antibodies are described in Supplemental Experimental Procedures.
RNA-IP and polysome analysis were performed as in (Ceman, et al., 2003). iCLIP protocol was described (Konig et al., 2010, 2011) with modifications in the Supplemental Experimental approaches.
Bioinformatic and statistical methods used for RNA analysis and identification and comparisons between data sets are described in the Supplemental Experimental Approaches.
Supplementary Material
Acknowledgements
We thank Tri Cong Nguyen and Sheng Zhong for sharing iCLIP reagents, Auinash Kalsotra for advice on iCLIP and for critically reading this manuscript, along with Albert Himoe; Tod Jebe, Todd Patrick and Kyle Chipman for data analysis, John Martin for the TGFB1 luciferase constructs, Director of DNA Services Alvaro Hernandez, PhD. in the High-Throughput Sequencing and Genotyping Unit, NIH grant R01 MH093661 [to S.C and K.S.K], NIH Cell and Molecular Biology Training Grant [to M.K.], UIUC Research Board Arnold O. Beckman award [to S.C.], Spastic Paralysis Research Foundation of Illinois-Eastern Iowa District of Kiwanis International, estates of Catherine Mary Paulsen and Anita Feller [to S.C.], Dr. Miriam and Sheldon Adelson Medical Research Foundation and the Rainwater Charitable Foundation [to K.S.K.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers All iCLIP data files and RNA-Seq files are available from NCBI Gene Expression Omnibus (http://www.ncbi.nlm.gov/geo/) under GSE51443 (MOV10 iCLIP-SEQ) and GSE50499 (RNA seq)
Author contributions: S.C. and K.S.K. conceived the project and wrote the manuscript. S.C. supervised the project and performed experiments. P.K. and M.K. performed experiments and cowrote the manuscript. H.Z. analyzed the MOV10, AGO and FMRP CLIP data, MOV10 RNA-IP data and performed statistical analyses, G.S. examined protein expression and purified recombinant proteins, R.K. analyzed CLIP data, facilitated access to AGO data and guided genome browser analysis, J.D. performed statistical analyses on iCLIP and RNA-seq, M.L.A. prepared libraries for deep sequencing. There are no conflicts of interest.
References
- Ameres SL, Martinez J, Schroeder Re. Molecular Basis for Target RNA Recognition and Cleavage by Human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS. A Coordinated Local Translational Control Point at the Synapse Involving Relief from Silencing and MOV10 Degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin J-D, Perreault J-P. Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Research. 2013;41:5898–5911. doi: 10.1093/nar/gkt265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-Mediated Translational Repression in Human Cells Subjected to Stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila Fragile X mutants impairs long-term memory. Nat Neurosci. 2008;11:1143. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, Brown V, Warren ST. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol Cell Biol. 1999;19:7925–7932. doi: 10.1128/mcb.19.12.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair. 2011;10:654–665. doi: 10.1016/j.dnarep.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Chen E, Sharma Manjuli R, Shi X, Agrawal Rajendra K, Joseph S. Fragile X Mental Retardation Protein Regulates Translation by Binding Directly to the Ribosome. Molecular Cell. 2014;54:407–417. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yuna SW, Setoa J, Liua W, Toth M. The fragile x mental retardation protein binds and regulates a novel class of mRNAs containing u rich target sequences. Neurosci. 2003;120:1005–1017. doi: 10.1016/s0306-4522(03)00406-8. [DOI] [PubMed] [Google Scholar]
- Creacy SD, Routh ED, Iwamoto F, Nagamine Y, Akman SA, Vaughn JP. G4 Resolvase 1 Binds Both DNA and RNA Tetramolecular Quadruplex with High Affinity and Is the Major Source of Tetramolecular Quadruplex G4-DNA and G4-RNA Resolving Activity in HeLa Cell Lysates. Journal of Biological Chemistry. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van†Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devys D, Lutz Y, Rouyer N, Bellocq J-P, Mandel J-L. The FMR-1 protein is cytoplasmic, most abundant in neurons, and appears normal in carriers of the fragile X premutation. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang C-F, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of Synaptic Structure and Function by FMRP-Associated MicroRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the Root of miRNA-Mediated Gene Silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Gregersen Lea H, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. MOV10 Is a 5′ to 3′ RNA Helicase Contributing to UPF1 mRNA Target Degradation by Translocation along 3′ UTRs. Molecular Cell. 2014;54:573–585. doi: 10.1016/j.molcel.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp A-C, Munschauer M, et al. Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Berry-Kravis E, Kaufmann WE, Ono MY, Tartaglia N, Lachiewicz A, Kronk R, Delahunty C, Hessl D, Visootsak J, et al. Advances in the Treatment of Fragile X Syndrome. Pediatrics. 2009;123:378–390. doi: 10.1542/peds.2008-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser J, Syed AP, Bilen B, Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013;23:604–615. doi: 10.1101/gr.139758.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JS, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, SP F, Bonhoeffer S, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte f Chemie. 1994;125:167–188. [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Jahner D, Nobis P, Simon I, Lohler Jr, Harbers K, Grotkopp D. Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell. 1981;24:519–529. doi: 10.1016/0092-8674(81)90343-3. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kato M, Han Tina W, Xie S, Shi K, Du X, Wu Leeju C, Mirzaei H, Goldsmith Elizabeth J, Longgood J, Pei J, et al. Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, Bear MF., 3rd The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Khandjian EW, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- Kikin O, D’Antonio L, Bagga P. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Research. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Bellini M, Ceman S. Fragile X mental retardation protein FMRP binds mRNAs in the nucleus. Mol Cell Biol. 2009;29:214–228. doi: 10.1128/MCB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP - Transcriptome-wide Mapping of Protein-RNA Interactions with Individual Nucleotide Resolution. J Vis Exp. 2011:e2638. doi: 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA Translation at the Synapse and in Disease. The Journal of Neuroscience. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Menendez C, Frees S, Bagga PS. QGRS-H Predictor: a web server for predicting homologous quadruplex forming G-rich sequence motifs in nucleotide sequences. Nucleic Acids Research. 2012;40:W96–W103. doi: 10.1093/nar/gks422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi-Aubert SE, Nicholls J, Maertens GN, Brookes S, Bernstein E, Peters G. Role for the MOV10 RNA helicase in Polycomb-mediated repression of the INK4a tumor suppressor. Nat Struct Mol Biol. 2010;17:862–868. doi: 10.1038/nsmb.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Mooslehner K, Muller U, Karls U, Hamann L, Harbers K. Structure and expression of a gene encoding a putative GTP-binding protein identified by provirus integration in a transgenic mouse strain. Mol Cell Biol. 1991;11:886–893. doi: 10.1128/mcb.11.2.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible Inhibition of PSD-95 mRNA Translation by miR-125a, FMRP Phosphorylation, and mGluR Signaling. Molecular Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt AJ, MacRae IJ. The RNA-induced Silencing Complex: A Versatile Gene-silencing Machine. Journal of Biological Chemistry. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Research. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sievers C, Schlumpf T, Sawarkar R, Comoglio F, Paro R. Mixture models and wavelet transforms reveal high confidence RNA-protein interaction sites in MOV10 PAR-CLIP data. Nucleic Acids Research. 2012;40:e160. doi: 10.1093/nar/gks697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren PJ, Bahrami-Samani E, Burns SC, Qiao M, Karginov FV, Hodges E, Hannon GJ, Sanford JR, Penalva LOF, Smith AD. Site identification in high-throughput RNA-protein interaction data. Bioinformatics. 2012;28:3013–3020. doi: 10.1093/bioinformatics/bts569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–656. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- Westmark CJ, Malter JS. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol. 2007;5:e52. doi: 10.1371/journal.pbio.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annual Review of Biophysics. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winograd C, Clayton D, Ceman S. Expression of fragile X mental retardation protein within the vocal control system of developing and adult male zebra finches. Neuroscience. 2008;157:132–142. doi: 10.1016/j.neuroscience.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. The FASEB Journal. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. Direct Conversion of Fibroblasts to Neurons by Reprogramming PTB-Regulated MicroRNA Circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.