Abstract
Our previous studies have shown that the 3' end of metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is involved in colorectal cancer (CRC) cell proliferation and migration/invasion in vitro. The role and mechanism of MALAT1 in CRC metastasis in vivo, however, remain largely unknown. In the present study, we found that MALAT1 was up-regulated in human primary CRC tissues with lymph node metastasis. Overexpression of MALAT1 via RNA activation promoted CRC cell proliferation, invasion and migration in vitro, and stimulated tumor growth and metastasis in mice in vivo. Conversely, knockdown of MALAT1 inhibited CRC tumor growth and metastasis. MALAT1 regulated at least 243 genes in CRC cells in a genome-wide expression profiling. Among these genes, PRKA kinase anchor protein 9 (AKAP-9) was significantly up-regulated at both mRNA and protein levels. AKAP-9 was highly expressed in CRC cells with metastatic potential and human primary CRC tissues with lymph node metastasis, but not in normal cells or tissues. Importantly, knockdown of AKAP-9 blocked MALAT1-mediated CRC cell proliferation, migration and invasion. These data indicate that MALAT1 may promote CRC tumor development via its target protein AKAP-9.
Keywords: Metastasis associated lung adenocarcinoma transcript 1, PRKA kinase anchor protein 9, Colorectal cancer, Metastasis, Long Non-coding RNA
1. Introduction
Long non-coding RNAs (lncRNA) are non-protein coding transcripts longer than 200 nucleotides[1–3]. Emerging evidences show that lncRNAs are associated with a spectrum of biological processes such as gene regulation on transcriptional and post-transcriptional levels[4, 5], chromatin modification and epigenetics, alternative splicing, protein activity modulation, and protein localization, etc. Dysregulation of lncRNAs is also an important feature of many complex human diseases, including Alzheimer’s disease[6] ischemic diseases[7], heart disease[8] and cancer[9, 10]. In fact, lncRNAs have been recognized as a hallmark of the onset and development of various tumors or tumor suppressor pathways[11, 12].
The metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is a 8.1 kb long noncoding RNA transcribed from the nuclear-enriched transcript 2 (NEAT2). As a highly abundant nucleus-restricted RNA, MALAT1 can modulate the activity of serine/arginine (SR) splicing factor including its pre-mRNA splicing and mRNA export[13, 14]. MALAT1 is found to enhance the proliferation and invasion of nasopharyngeal carcinoma cell line CNE-1[15]. Numerous in vivo studies have shown that transient overexpression of MALAT1 enhances tumor formation of gastric cancer[16], gallbladder cancer[17], and lung cancer[18] in nude mice while depletion of MALAT1 in tumor cells reduces tumorigenicity[19]. In fact, MALAT1 is considered as an independent prognostic marker and a potential target for antimetastatic therapy in lung cancer[19]. Although these studies strongly indicate that MALAT1 plays an important role in carcinogenesis, the mechanisms whereby MALAT1 induces colorectal carcinoma growth or metastasis remains largely unknown.
Colorectal Cancer (CRC) is the third leading cause of cancer death worldwide. Metastasis is the major cause of mortality in patients with colorectal tumor[20]. However, little is known about the key mechanisms and factors underlying the complex process of CRC tumor invasion and metastasis. Our previous studies show that a MALAT1 fragment at 3' end of the LncRNA plays a pivotal role in the proliferation, migration and invasion of CRC cells in vitro[14, 21]. However, the role and mechanism of MALAT1 in CRC tumor growth and metastasis in vivo remain to be determined. In the present study, we found that MALAT1 is closely associated with the metastasis of human CRC. By manipulating MALAT1 expression in CRC cells or tumor cubes that were implanted in animals, we have demonstrated the unambiguous role of MALAT1 in tumorigenesis and metastasis in vivo. Most importantly, we have identified anchor protein 9 (AKAP-9) as a downstream target that is responsible for MALAT1 function in CRC cell proliferation and migration/invasion.
2. Materials and methods
2.1 Cell culture and tissue specimens
Human colorectal cancer cell lines HCT116, SW480, SW620, LoVo, CaCo-2, HT-29, Lst174-t, and COLO205 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The M5 cells with enhanced metastatic capability was isolated by an in vivo selection of SW480 cells. The stably-transduced cell lines SW480-RNAi-MALAT1 (RNA interference), SW480-RNAa-MALAT1 (RNA activation), and SW480-control (scramble control) were established by lentiviral vector (pGCSIL-GFP, GeneChem, ShangHai, China) transduction of SW480 cells. All CRC cells were cultured in RPMI 1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) (Hyclone, USA) and 100 U/ml penicillin/streptomycin (Life Technologies, USA) and incubated in a humidified chamber with 5% CO2 at 37°C. The tumor samples were obtained from 27 patients, paired with normal tissues (10 cm away from the colorectal tumor). Nine of them had metastatic lymph-nodes. Patient’s consent and approval from the Ethics Committee of Southern Medical University were obtained before use of these clinical materials for research, and the clinical information about the patients is listed in Supplemental Table S1. In each selected case, pathological diagnosis was performed in the Department of Pathology of Nanfang Hospital, and all patients had undergone elective surgery for CRC in Nanfang Hospital during March to April in 2009.
2.2 RNA isolation and MALAT1 expression analysis
Total RNA was extracted with TRIzol Reagent (Invitrogen). First strand cDNA was synthesized with the PrimeScript™ RT Kit (Takara Biotechnology Co, Japan). MALAT1 expression was detected by both semi-quantitative polymerase chain reaction (PCR) and quantitative qPCR using PrimeScript™ PCR Master Mix (Takara Biotechnology Co) and an ABI 7500 Real-Time PCR system. GAPDH was used as an internal control that is comparable with cyclophilin control. The assay was run in triplicate for each sample.
2.3 Plasmid and lentivirus preparation
MALAT1 was knocked down with RNA interference (RNAi) or overexpressed by RNA activation (RNAa) targeting on mRNA or promoter region of MALAT1 gene. Stealth RNAi™ negative control with medium GC content was purchased from Invitrogen. The promoter of human MALAT1 was analyzed for promoter motifs and high GC domains by using Promoter Scan Searcher and CpG Island Searcher software. RNAi cDNA and the promoter-dsDNA sequence was cloned into the pGCSIL-GFP lentiviral expression vector according to the manufacture’s instruction.
2.4 Cell proliferation assay and cell cycle analysis
Cells were seeded in 96-well plates at 0.8~1 × 103 per well. Cell proliferation was evaluated using Cell Counting Kit-8 (CCK-8, Dojindo, USA) according to manufacturer's instructions. Briefly, 10 µl of CCK-8 solution was added to culture medium, and incubated for 2 h. The absorbance at 450 nm wavelength was determined with a reference wavelength of 570 nm. For cell-cycle analysis, cells were plated in 6-well plates at 5×105 per well. The cell-cycle distribution was analyzed by propidium iodide (Sigma-Aldrich) staining and flow cytometry. All experiments were performed in triplicates.
2.5 Colony formation assay
Cells were plated in 6-well plates at 1–2× 102 per well and maintained in RPMI1640 containing 10% FBS. After 12–14 days, the cells were washed twice with PBS, fixed with methanol and stained with Giemsa solution. The number of colonies containing ≥ 50 cells was counted under a microscope. All these experiments were performed in triplicates.
2.6 Wound healing assay
Cells were cultured in standard conditions until 80–90% confluence and treated with mitomycin C (10 µg/ml) during the wound healing assay. The cell migration was assessed by measuring the movement of cells into the acellular area created by a sterile insert. The wound closure was observed after 48 h.
2.7 Invasion Assay
For invasion assays, matrigel-coated chambers (BD Biosciences, USA) containing 8 µm pores were used for the assays. Briefly, 2×105 cells were seeded into the upper chambers (coated in Matrigel) in serum-free medium. The lower chamber of the transwell was filled with culture media containing 10% FBS as a chemo-attractant. After the chambers were incubated at 37 °C for 48 h, non-invaded cells on the top of the transwell were scraped off with a cotton swab. Cells successfully translocated were fixed with 10% formalin, stained with 0.1% crystal violet, and counted under a light microscope.
2.8 Microarray analysis
Total RNA from SW480-RNAa-MALAT1, SW480-RNAi-MALAT1, and control SW480 cells was extracted using a Qiagen RNeasy kit (Qiagen, USA). The Roche Human Genome NimbleGene Array GeneChips (Roche, USA) were used for microarray analysis. Hybridization and scanning of the genechips were performed by the KangCheng Biotechnology Company (ShangHai, China) according to the standard Roche protocol. The data have been deposited in the public Gene Expression Omnibus Repository (Accession number XXH100360/XXH1010128). Significance analysis of microarrays (SAM) were performed to identify differentially expressed genes between SW480-RNAa-MALAT1 and SW480-control, between SW480-RNAi-MALAT1 and SW480-control, or between SW480-RNAa-MALAT1 and SW480-RNAi-MALAT1. Common differential genes were obtained by cross-comparing the three data sets.
2.9 Functional analysis and gene annotation
The Molecule Annotation System (MAS, KangCheng, ShangHai, China) was used to analyze GO and Pathways. The Database for Annotation, Visualization, and Integrated Discovery was used to discover the enriched function-related gene groups by gene-annotation enrichment analysis (http://david.abcc.ncifcrf.gov/). The genes in the top ten clusters (gene enrichment score >1) from the gene-annotation enrichment analysis were selected.
2.10 In vivo functions assays
2×106 of SW480-RNAa-MALAT1, SW480-RNAi-MALAT1, or control SW480 cells were injected subcutaneously in left and right flank of 4- to 6-week-old Balb/C- nu/nu athymic nude mice (n=6 per group) obtained from Animal Center of Southern Medical University. Tumor growth was visualized and imaged using a whole-body GFP imaging system (Lighttools, Encinitas, CA). Tumor size was measured by a slide caliper, and tumor volume was calculated as follows: volume = (D×d2)/2, where D meant the longest diameter and d meant the shortest diameter. For testing the in vivo metastasis, the subcutaneous tumors were diced into 1 mm3 cubes, and implanted into the mesentery at the caecum terminus of the nude mice. Animal were kept until the end of the experiment (8 weeks). The organs were fixed with 10% neutral buffered formalin and paraffin-embedded. Subsequently, consecutive tissue sections were made for each block and stained with haematoxylin-eosin (H&E) to observe the metastatic nodules in organs under microscope. The experiments were performed according to institutional guidelines and approved by the Institution Animal Care and Use Committee of Southern Medical University.
2.11 2D electrophoresis proteomics analysis
SW620-scramble (SW620-Ctrl) and SW620-RNAa-MALAT1 cells were collected and lysed. Protein concentrations were determined using a modified Bradford assay (Ramagli 1999). The first-dimensional isoelectric focusing was carried out at 18°C with a maximum current setting of 50mA/strip using conditions programmed in an IP Gphor IEF unit (Amersham Biosciences). Second-dimension isoelectric focusing was carried out at 10°C with a maximum current setting of 400mA in Ettan DIGE unit (Amersham Biosciences). The resulting gel was then visualized by SYPRO Ruby staining. The image was scanned at an excitation wavelength of 400 nM and an emission wavelength of 630 nM. All images were captured on a Typhoon 9400 (Amersham Biosciences). Quantification of the protein expression and statistical analysis were carried out using the Decyder-DIA software program (Amersham Biosciences).
2.12 Protein MALDI-TOF/TOF Analysis
Proteins were extracted from gel plugs and were purified using Ziptip C18 (Millipore, USA). The eluted proteins were mixed with MATRIX solution and spotted on MALDI plate using dry droplet method and analyzed using AbSciexTof/Tof instruments. Protein identification using peptide mass fingerprinting (PMF) was performed by the MASCOT search engine (http://www.matrixscience.com/) (MatrixSicence Ltd., UK) against the MSDB protein database. Proteins matching more than four peptides with a MASCOT score higher than 63 were considered significant (P<0.05). Carboamidomethylation of cysteine was selected as the static modification and oxidation of methionine as the differential modification. Protein identification was filtered with peak Erazor software.
2.13 Western blot analysis
Cells were washed twice with cold PBS and lysed in RIPA buffer (1× PBS, 1% NP40, 0.1% SDS, 5 mM EDTA, 0.5% sodium deoxycholate, and 1 mM sodium orthovanadate) containing protease inhibitors. Whole protein extracts were resolved on a 10% SDS polyacrylamide gel and electrotransferred to polyvinylidene fluoride membranes (Millipore, USA), which were then blocked in 5% non-fat dry milk in Tris-buffered saline (pH 7.5; 100 mM NaCl, 50 mM Tris, and 0.1% Tween-20) and immunoblotted with AKAP9 (Santa Cruz, USA) at 4°C overnight followed by HRP (horseradish peroxidase)-labeled secondary antibody (Santa Cruz) and detected by chemiluminescence. α-Tubulin expression was used as a protein-loading control. The intensity of protein bands was quantified with the Quantity One software (4.5.0 basic, Bio-Rad).
2.14 Immunofluorescence
Cells plated on poly-L-lysine-coated glass coverslips were fixed with 4% paraformaldehyde, and washed with PBS. Cells were then permeabilized with 0.1% Triton X-100/PBS for 10 min and subsequently incubated with primary antibodies followed by incubation with fluorescein isothiocyanate-tagged secondary antibodies. 4′,6-Diamidino-2-phenylindole (DAPI) was used for the nuclear counterstain. The fluorescence was recorded using an inverted fluorescence microscope (Leica, Germany).
2.15 Statistical analysis
The SPSS statistical software (Abbott Laboratories, USA) was used for conducting all the statistical analyses. All results were confirmed by statisticians in the Department of Health Statistics, Southern Medical University. The comparison between the two groups was analyzed by Mann-Whitney U test. RT-PCR gray values, RT-qPCR assay, flow cytometry assay, plate clone formation assay, wound healing assay, and tumor cell invasion assay were considered two-sided T tests. In vitro cell growth assay was tested using factorial design ANOVA. For differential expression of matched gels, protein spots whose intensities were either increased or decreased two-folds or more were marked and then confirmed by manual inspection of all relevant 2-D gel to ensure consistency. In all samples, P<0.05 was considered to be statistically significant.
3. Results
3.1 MALAT1 was upregulated in metastatic CRC cells and human primary CRC tissues with lymph node metastasis
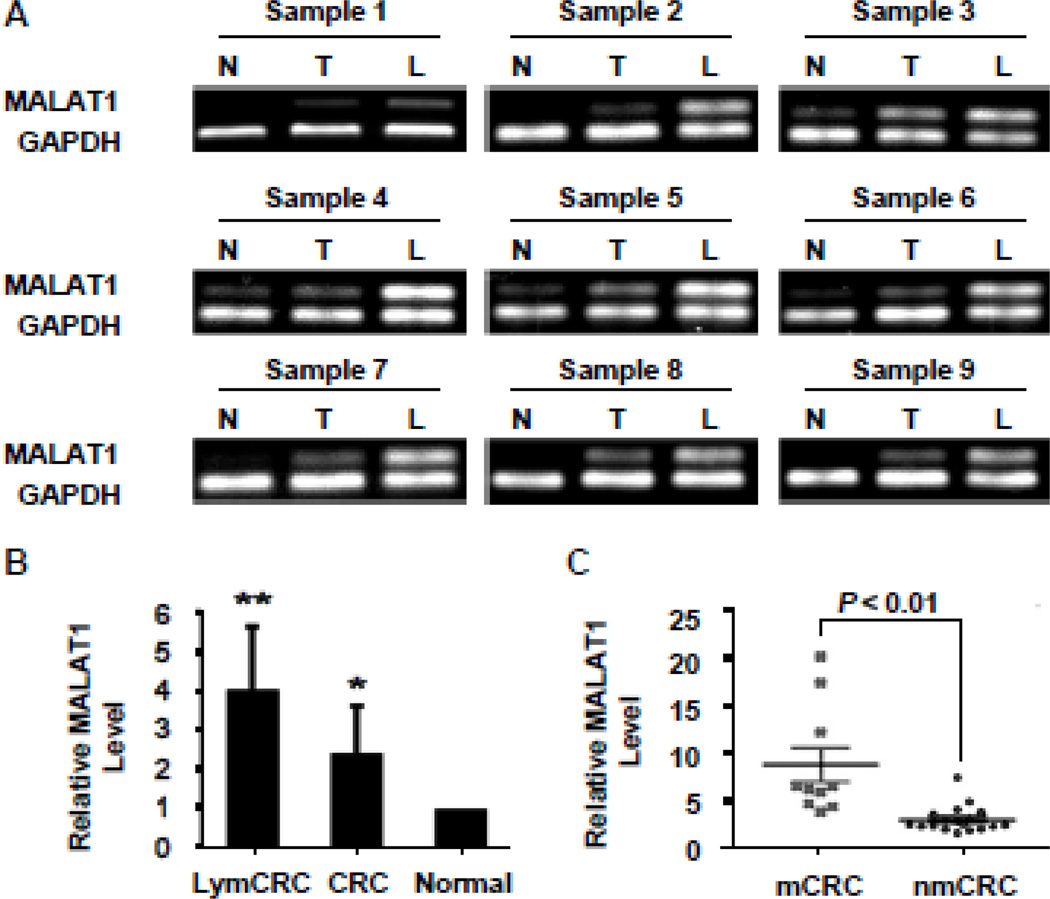
The expression of MALAT1 was examined in nine CRC cell lines, 18 human CRC tissues paired with adjacent non-neoplastic normal tissues and additional nine carcinoma tissues with lymphoma metastasis. We found that MALAT1 expression was higher in COLO-205, Lovo, SW620, M5 cells that originate from metastatic CRC compared with cells derived from primary CRC including HT29, HCT116, SW480, LS174t, and CaCo-2 (Supplemental Figure S1A–B). Importantly, MALAT1 expression level in tumors was significantly higher than those in the corresponding normal tissues (Supplemental Figure S1C–D). Moreover MALAT1 expression was consistently increased even more in CRC tissues with lymph node metastases (Figure 1), suggesting that MALAT1 may be involved in colorectal cancer metastasis.
Figure 1. MALAT1 was up-regulated in human CRC tissues with metastasis.
(A) Semi-quantitative analyses of MALAT1 levels in human CRC tissues. MALAT1 expression was higher in CRCs (T) than adjacent normal tissues (N). The highest level of MALAT1 was observed in CRCs with lymph metastasis (L) in all groups. (B) MALAT1 expression in human CRC tissues was detected by real-time PCR and normalized to GAPDH. MALAT1 levels in all 9 groups (CRC, CRC with lymph metastasis (LymCRC), and adjacent normal tissues) were consistent with the semi-quantitative analyses shown in A. *P<0.05 compared to normal tissue group; **P<0.05 compared to CRC and normal tissue group. (C) MALAT1 expression in human CRCs with and without metastasis relative to match-normal tissues. The level of MALAT1 in human CRCs with metastasis (mCRC, n=9) was significantly higher than that in CRCs without metastasis (nmCRC, n=18).
3.2 RNA activation (RNAa)-mediated overexpression of MALAT1 promoted CRC cell proliferation and migration/invasion
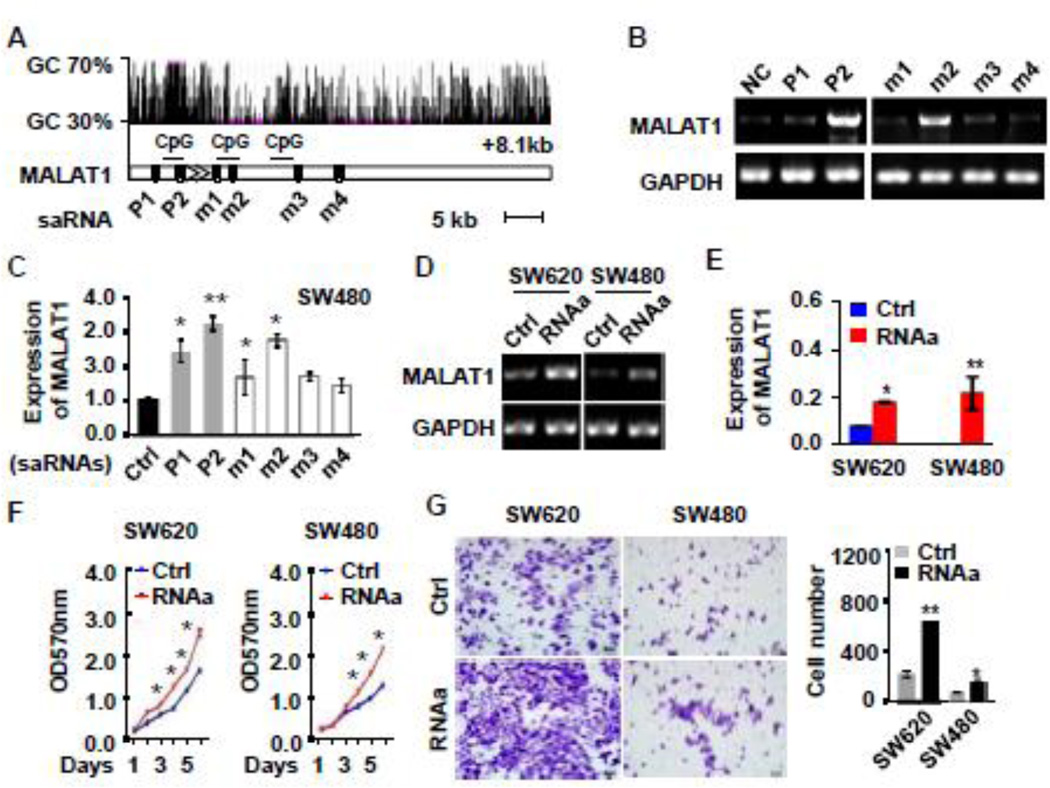
RNAa is a newly identified mechanism for gene induction and is triggered by small activating RNA (saRNAs). saRNAs interaction with promoter and GC rich region of mRNAs triggers expression of endogenous genes. We designed six specific saRNAs targeting either the promoter or GC rich regions of MALAT1 gene (Figure 2A). The saRNAs or negative control RNA were individually transfected into SW480 cells, and the MALAT1 expression was detected by semi-quantitative PCR and qPCR. Compared to the negative control, most of the saRNAs upregulated MALAT1 levels by 1.5 to 3.5 fold (Figure 2B and 2C). saRNA-348bp (P2) appeared to be an optimal activator of MALAT1 expression (3.5-fold increase). Therefore, this activator was selected for subsequent experiments.
Figure 2. RNA activation-mediated overexpression of MALAT1 promoted CRC cell proliferation and invasion.
(A) Schematic locations of small activation RNA (saRNA) target sites relative to the transcription starting site of MALAT1 gene including CpG island, Ctrl (scramble control), P1 (saRNA targeted in MALAT1 promoter region −592 bp), P2 (saRNA targeted in MALAT1 promoter region −348 bp), M1 (saRNA targeted in MALAT1 mRNA +258 bp), M2 (saRNA targeted in MALAT1 mRNA +380 bp), M3 (saRNA targeted in MALAT1 mRNA +1474 bp), M4 (saRNA targeted in MALAT1 mRNA +2584 bp). (B–C) saRNA-induced MALAT1 expression. SW480 cells were transfected with scramble (Ctrl) or different saRNAs (P1, P2, m1–m4). MALAT1 expression were assessed by semi-quantitative analysis (B) and real-time PCR (C). GAPDH was used as an internal control. (D, E) SW620 and SW480 cells expressed an elevated level of MALAT1 due to RNA activation (RNAa) as detected by semi-quantitative (D) and real-time PCR (E). (F) RNAa-activated MALAT1 promoted CRC cell proliferation as measured by CCK-8 assay. (G) RNAa-activated MALAT1 promoted CRC cell invasion as determined by matrigel invasion chamber analysis. *P < 0.05, **P <0.01 compared to corresponding control cells (Ctrl) (n=3).
To determine the role of MALAT1 in CRC tumourigenesis and progression, SW480 and SW620 cells were transfected with saRNA-348bp (MALAT1-RNAa or RNAa) and control RNA, respectively. As shown in Figure 2D and 2E, MALAT1 transcript was robustly induced in both SW480 and SW620 cells, which increased the proliferation of both cancer cells (Figure 2F). The role of MALAT1 in CRC proliferation was confirmed by using RNAi-mediated blockage of MALAT1 expression (Supplemental Figure S2). The ability of MALAT1-RNAa cells to form colonies was also much higher than those transfected with control RNA (Supplemental Figure S3A). Cell cycle analysis with flow cytometry revealed that MALAT1 caused a significant decrease of cells in G1/G0 phase with an increase of cells in S phase (Supplemental Figure S3B).
To determine if RNAa-mediated expression of MALAT1 affects CRC migration and invasion, we performed wound-healing and matrigel invasion assay using SW480 and SW620 cells. Over-expression of MALAT1 caused a significant increase in CRC cell migration (Supplemental Figure S4A) and invasion (Figure 2G). On the contrary, migration and invasive capacity of the CRC cells were markedly decreased when MALAT1 was blocked by siRNA (Supplemental Figure S4B–C). These data demonstrate that endogenous activation of MALAT1 promoted CRC cell proliferation and invasion.
3.3 MALAT1 promoted CRC tumor growth and metastasis in vivo
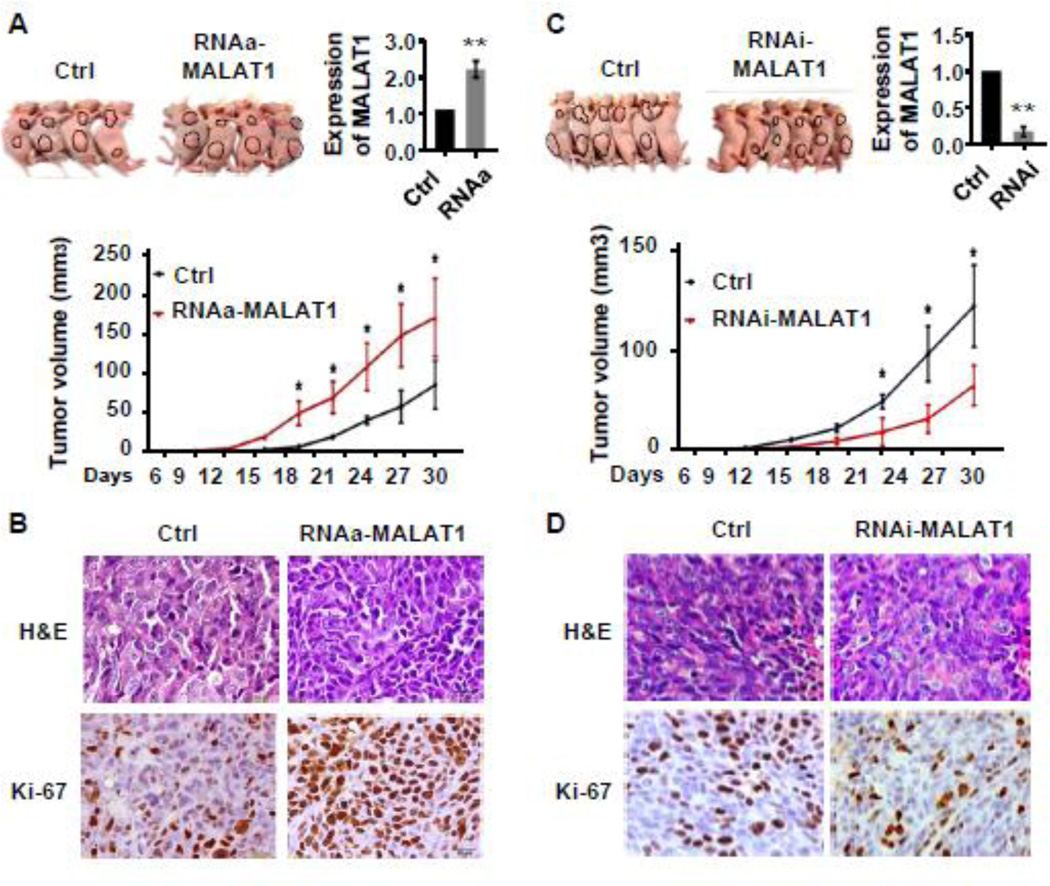
To assess the effect of MALAT1 on CRC tumor growth in vivo, we injected MALAT1-activated SW480 cells, MALAT1-depleting SW480 cells, and their control SW480 cells (with scramble saRNA or control shRNA) subcutaneously into nude mice respectively, and monitored the growth of the resultant primary tumors. As shown in Figure 3A, the xenograft tumors developed at the injection site after 15 days. During the 4 weeks of observation period, the tumor growth in the MALAT1-RNAa group was significantly faster than that in the control groups. Consequently, the tumor volumes of the MALAT1-RNAa group were significantly larger than those of control groups. Immunohistochemistry staining demonstrated that the tumors of MALAT1-activated group displayed a much higher Ki-67 index than the control group (Figure 3B). On the other hand, tumor growth, tumor volume and Ki67 expression in the MALAT1-blocked group was significantly attenuated compared to the controls (Figure 3C–3D). These data demonstrate that MALAT1 plays a crucial role in CRC growth in vivo.
Figure 3. MALAT1 promoted CRC tumor growth in vivo.
SW480 cells with scramble RNA (Ctrl), stable overexpression of MALAT1 (RNAa-MALAT), or MALAT1 knockdown (RNAi-MALAT1) were injected subcutaneously into nude mice. Tumors were allowed to grow for 30 days. (A) External whole-body images, MALAT1 expression in 30 day-old tumors, and quantification of the tumor sizes. Tumors derived from RNAa-MALAT1 cells grew significantly faster than that from control cells. *P<0.05 compared to the control groups in each time point. **P<0.01 compared to the control group at 30 day after injection (n=6). (B) Representative H&E images and immunohistochemical Ki-67 staining of tumor tissues derived from control (Ctrl) or MALAT1- overexpressed cells (RNAa-MALAT1). (C) Tumor of the RNAi-MALAT1 cells grew much slower than that of control cells. *P<0.05 compared to MALAT1 knockdown group (RNAi-MALAT1). **P<0.01 compared to the control group at 30 day after injection (n=6). (D) Representative H&E images and immunohistochemical Ki-67 staining of tumor tissues derived from control (Ctrl) or MALAT1-blocked (RNAi-MALAT1) cells. RNAa-mediated MALAT1 overexpression stimulated while MALAT1 knockdown inhibited CRC cell proliferation in vivo.
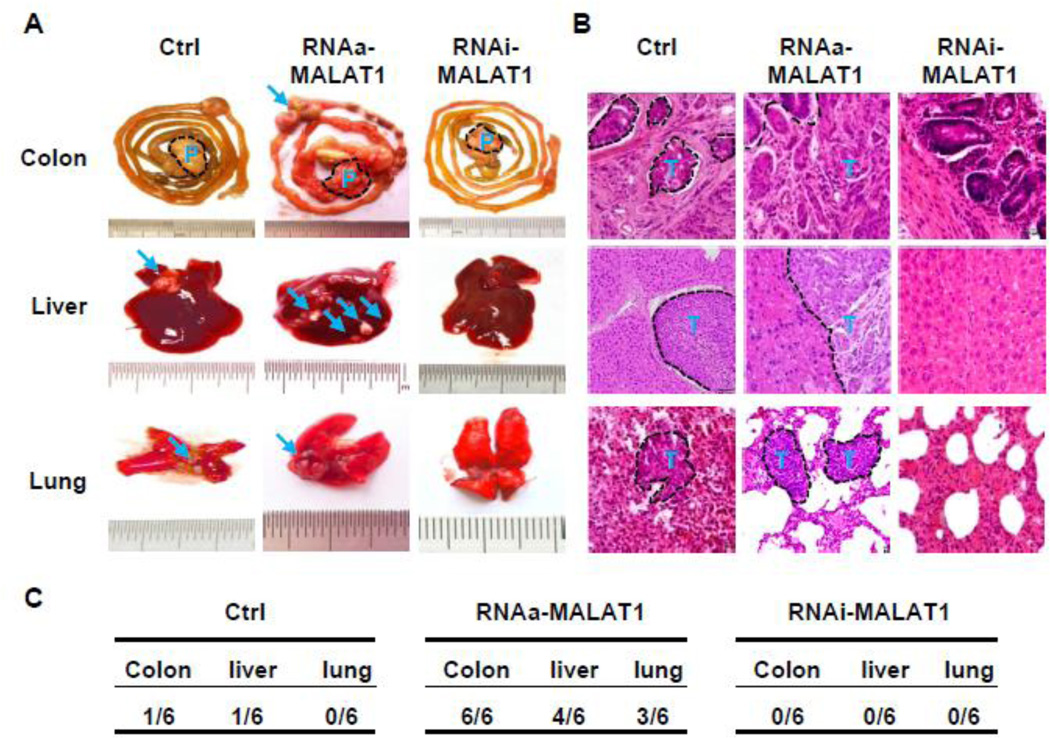
In order to determine the effect of MALAT1 on metastasis of CRC, we performed surgical orthotopic implantation of CRC tumor cubes that were grown from SW480 cells. The tumors from control, MALAT1-activated, or MALAT1-blocked SW480 cells were implanted into mouse caecum terminus. After 8 weeks, MALAT1-RNAa cells exhibited dramatic increase in the metastasis in colon, lung, and liver as compared to the controls (Figure 4A). Conversely, the tumor invasion in intestinal wall and metastasis to liver and lung of the MALAT1-blocked tumor were diminished (Figure 4A). Histological analysis showed an increased invasion of tumor cells in intestinal wall and metastasis in liver and lung with the MALAT1-activated tumor compared to control tumor while no tumor was observed in these tissues with the MALAT1-blocked tumor (Figure 4B). Quantitative analysis revealed that RNAa-MALAT1 significantly promoted while knockdown of MALAT1 blocked the metastasis (Figure 4C).
Figure 4. MALAT1 enhanced tumor invasion and metastasis in vivo.
Small cubes of SW480 tumors grew from cells with stable overexpression of MALAT1 (RNAa-MALAT1), stable knockdown of MALAT1 (RNAi-MALAT1), or left untreated (Ctrl) were implanted into the caecum terminus of nude mice as described in the Methods. P denotes primary tumor. T and arrows indicate the metastatic nodules. (A) Tissue images of metastasis. RNAa-activated MALAT1 expression promoted while MALAT1 knockdown by RNAi inhibited CRC metastasis to lung and liver. (B) Histological images of metastatic nodules in organs. Xenograft tumors with MALAT1 overexpression significantly promoted the invasion of implanted tumor in colon and metastasis in liver and lung. (C) Incidence of metastasis in mice implanted with SW480 cells containing scramble RNA (Ctrl), RNAa-MALAT1, or RNAi-MALAT1 as indicated.
Collectively, these results demonstrated that MALAT1 promotes tumor growth and metastasis in vivo.
3.4 Identification of MALAT1 targets by proteomic and cDNA microarray analyses
To identify the downstream targets mediating MALAT1 function in CRC tumor growth and metastasis, we performed proteomic analysis using 2D gel electrophoresis (Supplemental Figure S5A). Compared with cells without MALAT1 induction (Scramble saRNA), three proteins were significantly up-regulated in MALAT1-activated cells including AKAP-9, Peptidyl-prolyl cis-tRNAs isomerase A, PPLase1), and Glial fibrillary acidic protein (GFAP1). Meanwhile, coiled-coil domain-containing protein 155 (KASH5), Ras-related protein Rab-3A (Rab3A), and GTP-binding nuclear protein RNA (Ran) were down-regulated (Supplemental Figure S5B).
To explore the potential mechanism underlying MALAT1-mediated invasion and metastasis of CRC cells and to further screen MALAT1-dependent genes, we performed cDNA array analysis of SW480 cells with altered MALAT1 expression using Roche HumanGene chips (Supplemental Figure S6A). Compared with control cells, 396 genes were up-regulated while 458 genes down-regulated in RNAa-MALAT1 cells (Figure S6B). In MALAT1-depleted cell (RNAi-MALAT1), 422 genes were up-regulated while 518 genes down-regulated (Figure S6B). Paired-comparison revealed that the expression of 243 genes were depended on MALAT1 in SW480 cells. The molecule annotation system (MAS) and the DAVID gene-annotation enrichment analysis (gene enrichment score >1) revealed that the 243 target genes are involved in cell differentiation, cell cycle, cytoskeleton, and macromolecule metabolic process. By comparing the 243 genes from microarray analysis with the 6 genes identified by proteomic analysis, we found that AKAP-9 was the only gene that was detected by both approaches (Supplemental Figure S6C and Figure S7), suggesting that AKAP-9 may be related to MALAT1-dependent CRC tumorigenesis. To validate the role of MALAT1 in AKAP-9 expression, we activated or knocked down MALAT1 in SW480 cells and detected AKAP-9 protein expression. As shown in Supplemental Figure S8A, AKAP9 protein was up-regulated in MALAT1-activated SW480 cells but attenuated in MALAT1-blocked cells. The protein expression was also confirmed by immunostaining (Figure S8B).
3.5 AKAP-9 was expressed in CRC cells and tumors
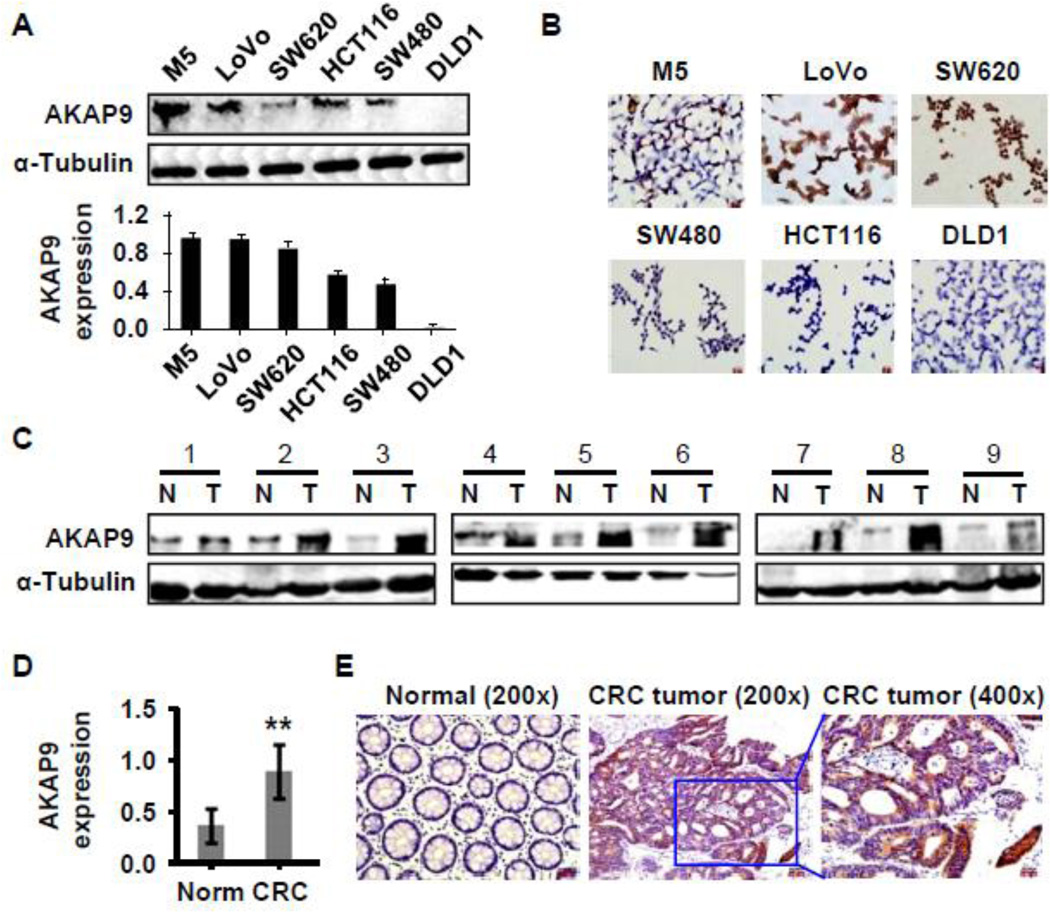
In order to determine if AKAP-9 mediates MALAT1 function in CRC cell proliferation and migration/invasion, we first detected AKAP-9 expression in CRC cells and tumor tissues. As shown in Figure 5A–5B, AKAP-9 expression was higher in M5, Lovo, and SW620 cells originating from metastatic CRC compared with cells derived from primary CRC included SW480, HCT116, and DLD1, consistent with the MALAT1 expression (Supplemental Figure S1A–B). Importantly, AKAP-9 expression in tumors was significantly elevated compared to the corresponding normal tissues in nine human CRC tumors (Figure 5C–5E). These results suggested that AKAP-9 may be involved in the colorectal cancer development.
Figure 5. AKAP-9 expressed in CRC cells and tumor tissues.
(A) AKAP-9 protein expression in six CRC cell lines was detected by western blot and normalized to α-Tubulin expression. (B) AKAP-9 expression in six CRC cell lines was examined by immunocytochemistry staining. (C) AKAP-9 expression was detected in nine human CRC tumor tissues. AKAP-9 levels in CRC tumor (T) was significantly higher than adjacent normal tissues (N) as detected by western blot. (D) Quantitative analysis of AKAP-9 expression shown in C by normalizing to α-Tubulin. **P<0.001 compared to normal tissue (Norm), n=9. (E) AKAP-9 expression in CRC tumor tissue was detected by immunohistochemistry staining.
3.6 AKAP-9 was essential for MALAT1-mediated CRC proliferation, migration and invasion
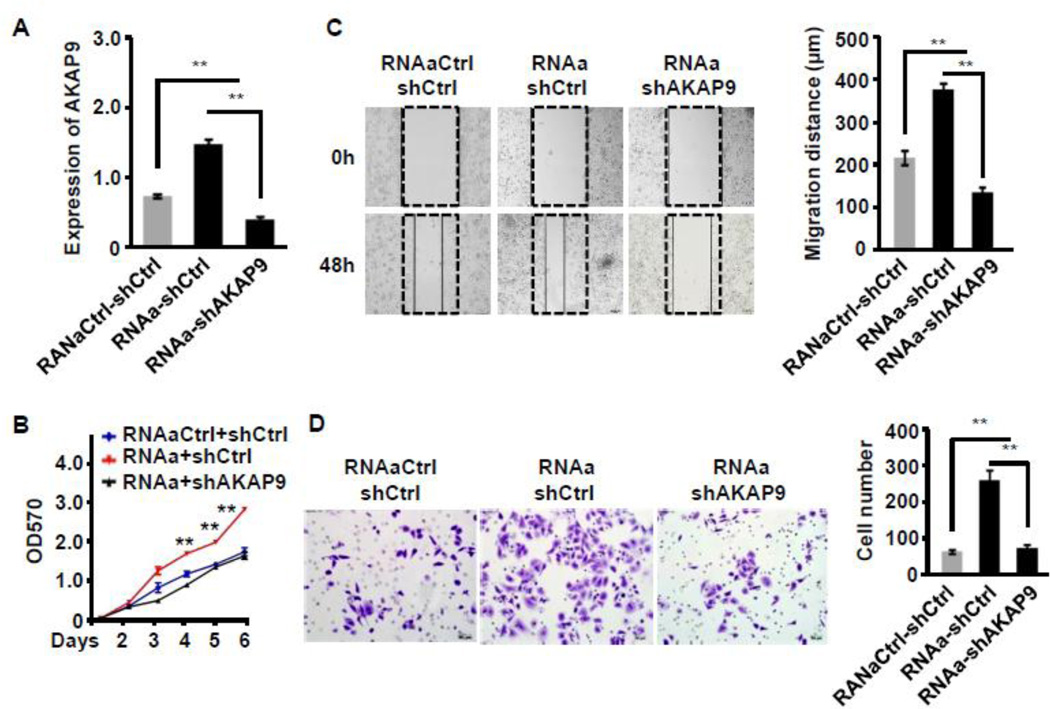
To test if MALAT1-mediated phenotype in CRC was indeed due to the induction of its target gene AKAP-9, we knocked down AKAP-9 by shRNA and tested if MALAT1-mediated CRC proliferation, migration, and invasion were altered in the absence of AKAP-9. As shown in Figure 6A, AKAP-9 shRNA effectively blocked AKAP-9 expression that was induced by MALAT1 activation in SW480 cells. Silencing of AKAP9 inhibited MALAT1-induced proliferation (Figure 6B), migration (Figure 6C) and invasion (Figure 6D) of SW480 CRC cells. These results demonstrate that MALAT1 promoted the proliferation, migration, and invasion of CRC through AKAP-9.
Figure 6. AKAP-9 was required for MALAT1-mediated CRC cell growth, migration and invasion.
SW480 cells were co-transfected control (RNAaCtrl) or MALAT1 saRNA (RNAa) with scramble (shCtrl) or AKAP-9 shRNA (shAKAP9) as indicated. Cell proliferation, migration and invasion were measured. (A) AKAP-9 expression was increased by RNAa activation but knocked down by shAKAP9. **P<0.01 (n=3). (B) Cell proliferation was detected by MTT assay. MALAT1 activation stimulated the SW480 cell proliferation, which was attenuated when AKAP-9 was knocked down. **P<0.01 compared to other two groups (n=3). (C) Cell migration was measured by wound-healing assay. Relative migration distance was 24 normalized to the original gap produced by inserts (0 h). **P<0.01 (n=3). (D) SW480 cell invasion was determined by matrigel invasion assay. MALAT1 induction enhanced SW480 cell invasion, which was attenuated by silencing of AKAP9. **P<0.01 (n=3).
4. Discussion
Although the regulation of CRC growth are well understood, the mechanism governing CRC proliferation, migration, invasion or metastasis remain largely unknown. Our study demonstrates that MALAT1 is a novel pro-metastatic factor for CRC. Induction of MALAT1 in primary CRC strongly enhanced the tumor growth and metastasis in mice, underscoring the essential role of MALAT1 in CRC metastasis.
Because MALAT1 is a long (8 kb) gene, traditional expression clone vector is difficult to be efficiently transfected into cells. Thus, most of the previous functional studies are limited to the knockdown approach[22]. RNA activation-induced gene expression by targeting promoter sequence allowed us to physiologically and directly test the function of a given gene[23–26]. In fact, lentiviral-based delivery of dsRNAs has been reported to activate VEGF expression by targeting promoter sequences in an ischemic mouse model[27]. We found for the first time that RNA activation can up-regulate MALAT1 expression in CRC cells. MALAT1 overexpression via RNA activation significantly promoted CRC cell growth and colony formation, cell cycle G1/S transition in vitro, and the tumorigenesis of CRC xenograft in vivo. By using RNA activation, we were able to demonstrate that MALAT1 promotes colorectal carcinoma metastasis. MALAT1 expression level in CRC tissues of human patients with lymph node metastasis was higher than those without metastasis. Clinic pathological analysis also found that high-level of MALAT1 was associated with lymph node metastasis or distant metastasis. In animal models, increased expression of MALAT1 was sufficient to promote CRC metastasis to lung, liver and organs in the peritoneal cavity. This correlation indicates that MALAT1 may play a causal role in colorectal cancer metastasis, consistent with a recent study showing that MALAT1 is a potentially active player in the metastatic process of lung adenocarcinoma. MALAT1 promotes cell motility of lung cancer cells through transcriptional or post-transcriptional regulation of motility-related genes.
Although MALAT1 is involved in metastasis of different cancers, little is known about the underlying molecular mechanism. To begin to explore how MALAT1 regulates tumor invasion and metastasis in CRC, high-throughput gene array and proteomic techniques have identified AKAP-9 as the only gene that was induced in MALAT1-activated CRC cells. AKAP9 is linked to cancer development or metastasis including oral cancer[28], melanomas[29], thyroid carcinomas[30, 31], breast cancer[32], lung cancer[33]. However, the function of AKAP-9 in the onset of these tumors has not been experimentally established. There is also no data describing the role of AKAP-9 in the development of CRC cancer. Our results suggest that AKAP-9 is associated with MALAT1 in CRC tumor growth, consistent with a number of microarray dataset showing that AKAP-9 is correlated with MALAT1 in human CRC tissues, primarily cultured CRC, or CRC cell lines (http://www.ncbi.nlm.nih.gov/geoprofiles/advanced). In fact, AKAP-9 is critical for MALAT1-mediated CRC cell activities. Knockdown of AKAP-9 significantly attenuated MALAT1 function in CRC cell proliferation, migration and invasion, suggesting that MALAT1 promotes the proliferation and migration/invasion of CRC cells through targeting AKAP-9. In view of the important role of migration and invasion in metastasis, AKAP-9 is likely to be critical for MALAT1-mediated CRC metastasis, which would be an interesting and important subject for future investigation. Another question remaining to be explored is how AKAP-9 mediates MALAT1 function in CRC tumor growth. Despite of these limitations, this study has provided novel insight about CRC tumor development and metastasis.
Supplementary Material
Highlights.
MALAT1 is up-regulated in human primary CRC tissues with lymph node metastasis.
MALAT1 can be induced via RNA activation.
MALAT1 plays a critical role in CRC tumor growth and metastasis in mice.
AKAP-9 is correlated with MALAT1 in CRC tissues.
AKAP-9 mediates MALAT1 function in CRC proliferation/migration/invasion.
Acknowledgements
The authors are grateful to Professor Ding YQ and the researchers in the Key Laboratory of Molecular Tumor Pathology, Guangdong, China for providing technical assistance. We also thank the department of Bioinformatics at Southern Medical University of Guangzhou, China for the bioinformatics analysis.
Funding
National Natural Science Foundation of China (30770976. 81172054 to Z.G.L) and National Institutes of Health (HL123302, HL119053, HL107526 to S.Y.C)
Abbreviations
- MALAT1
metastasis associated lung adenocarcinoma transcript 1
- CRC
colorectal cancer
- AKAP-9
PRKA kinase anchor protein 9
- LncRNA
long Non-coding RNA
- NEAT2
nuclear-enriched transcript 2
- RNAa
RNA activation
- H&E
haematoxylin-eosin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: None declared.
Reference
- 1.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 2.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 3.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci. 2013;14(8):16010–16039. doi: 10.3390/ijms140816010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schonrock N, Gotz J. Decoding the non-coding RNAs in Alzheimer's disease. Cell Mol Life Sci. 2012;69(21):3543–3559. doi: 10.1007/s00018-012-1125-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai Y, et al. Regulation of CARD8 expression by ANRIL and association of CARD8 single nucleotide polymorphism rs2043211 (p.C10X) with ischemic stroke. Stroke. 2014;45(2):383–388. doi: 10.1161/STROKEAHA.113.003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao XG, Touma M, Wang Y. Decoding the noncoding transcripts in human heart failure. Circulation. 2014;129(9):958–960. doi: 10.1161/CIRCULATIONAHA.114.007548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang T, et al. Noncoding RNAs in cancer and cancer stem cells. Chin J Cancer. 2013;32(11):582–593. doi: 10.5732/cjc.013.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt AM, Chang HY. Gene regulation: Long RNAs wire up cancer growth. Nature. 2013;500(7464):536–537. doi: 10.1038/nature12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu KH, et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiao HP, et al. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac J Cancer Prev. 2013;14(2):1077–1082. doi: 10.7314/apjcp.2013.14.2.1077. [DOI] [PubMed] [Google Scholar]
- 13.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilusz JE, Freier SM, Spector DL. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135(5):919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie L, et al. Expression of long noncoding RNA MALAT1 gene in human nasopharyngeal carcinoma cell lines and its biological significance. Nan Fang Yi Ke Da Xue Xue Bao. 2013;33(5):692–697. [PubMed] [Google Scholar]
- 16.Okugawa Y, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014 doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu XS, et al. MALAT1 promotes the proliferation and metastasis of gallbladder cancer cells by activating the ERK/MAPK pathway. Cancer Biol Ther. 2014;15(6):806–814. doi: 10.4161/cbt.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutschner T, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt LH, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6(12):1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 20.Rasool S, et al. A comparative overview of general risk factors associated with the incidence of colorectal cancer. Tumour Biol. 2013;34(5):2469–2476. doi: 10.1007/s13277-013-0876-y. [DOI] [PubMed] [Google Scholar]
- 21.Xu C, et al. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39(1):169–175. doi: 10.3892/ijo.2011.1007. [DOI] [PubMed] [Google Scholar]
- 22.Lai MC, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29(3):1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 23.Pushparaj PN, et al. RNAi and RNAa--the yin and yang of RNAome. Bioinformation. 2008;2(6):235–237. doi: 10.6026/97320630002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang V, et al. RNAa is conserved in mammalian cells. PLoS One. 2010;5(1):e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner MJ, Jiao AL, Slack FJ. Autoregulation of lin-4 microRNA transcription by RNA activation (RNAa) in C. elegans. Cell Cycle. 2014;13(5):772–781. doi: 10.4161/cc.27679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, et al. Prognostic value and function of KLF4 in prostate cancer: RNAa and vector-mediated overexpression identify KLF4 as an inhibitor of tumor cell growth and migration. Cancer Res. 2010;70(24):10182–10191. doi: 10.1158/0008-5472.CAN-10-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen R, et al. Up-regulation of VEGF by small activator RNA in human corpus cavernosum smooth muscle cells. J Sex Med. 2011;8(10):2773–2780. doi: 10.1111/j.1743-6109.2011.02412.x. [DOI] [PubMed] [Google Scholar]
- 28.Onken MD, et al. A surprising cross-species conservation in the genomic landscape of mouse and human oral cancer identifies a transcriptional signature predicting metastatic disease. Clin Cancer Res. 2014;20(11):2873–2884. doi: 10.1158/1078-0432.CCR-14-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kabbarah O, et al. Integrative genome comparison of primary and metastatic melanomas. PLoS One. 2010;5(5):e10770. doi: 10.1371/journal.pone.0010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caria P, Vanni R. Cytogenetic and molecular events in adenoma and well-differentiated thyroid follicular-cell neoplasia. Cancer Genet Cytogenet. 2010;203(1):21–29. doi: 10.1016/j.cancergencyto.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, et al. BRAF mutation and AKAP9 expression in sporadic papillary thyroid carcinomas. Pathology. 2006;38(3):201–204. doi: 10.1080/00313020600696264. [DOI] [PubMed] [Google Scholar]
- 32.Frank B, et al. Association of a common AKAP9 variant with breast cancer risk: a collaborative analysis. J Natl Cancer Inst. 2008;100(6):437–442. doi: 10.1093/jnci/djn037. [DOI] [PubMed] [Google Scholar]
- 33.Truong T, et al. International Lung Cancer Consortium: coordinated association study of 10 potential lung cancer susceptibility variants. Carcinogenesis. 2010;31(4):625–633. doi: 10.1093/carcin/bgq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.