Abstract
Objective
To assess the frequency of a novel prostate cancer-associated single nucleotide polymorphism (SNP), rs188140481, in a North American population and to evaluate the clinical significance of this variant including annotated prostatectomy pathology.
Subjects and Methods
We examined the frequency of the minor allele at rs188140481 in 4,299 North American men including 1,979 PC cases and 2,320 healthy volunteers.
We compared the clinico-pathologic features of PC between carriers and non-carriers of the SNP.
Results
The rs188140481[A] SNP was present in 1.6% of the cohort; it was significantly more likely to be carried by men with PC than healthy controls (OR 3.14; 95% CI 1.85-5.35).
After adjusting for age and PSA carriers were found 6.73-fold (95% CI 1.69-26.76) more likely to develop PC than non-carriers.
Age at diagnosis, frequency of a positive family history of PC, and biochemical recurrence rates were similar between SNP carriers and non-carriers.
Patients with the SNP had a proportionately higher frequency of stage ≥ T2c disease (29.5% vs. 20.1%; p = 0.13), Gleason ≥8 tumors (13.3% vs. 6.5%; p = 0.10), and extracapsular extension (28.9% vs. 18.8%; p = 0.12) compared to non-carriers.
Conclusions
rs188140481[A] is a rare SNP that confers greater risk of PC compared to SNPs identified by genome-wide association studies.
Because of its low frequency, larger studies are needed to validate the prognostic significance of this locus, and associations with adverse pathology.
Keywords: genetics, prostatic neoplasm, polymorphism, single nucleotide
Introduction
Prostate cancer (PC) is the most common non-skin cancer affecting men in the United States, with 238,590 new cases projected in 2013[1]. Genome-wide association (GWAS) and sequencing studies have recently generated rapid progress towards identifying genetic markers associated with increased PC risk[2]. Currently more than 77 different common single nucleotide polymorphisms (SNPs) have been identified on 11 different chromosomes that confer a modest risk of PC (OR: 1.12 - 2.04) in men of European ancestry[2-4]. The 8q24 region has emerged as a key region for PC susceptibility. In fact, it is often considered to be the genetic “hot-spot,” containing nine different PC-associated SNPs, within at least 5 distinct regions[5-9].
Recently, newer whole-genome sequencing (WGS) techniques have been used to identify rarer alleles associated with a far greater risk of developing PC and potentially more aggressive forms of the disease[10,11]. WGS has recently been employed to identify a novel SNP on 8q24 that is associated with PC in a predominantly European population[10]. This SNP was carried by only 1.1% of the men in that study. Although relatively rare, it was shown to confer a significantly greater risk of developing PC than previously identified loci (OR = 2.90). In the initial study, Gudmundsson and colleagues reported that this variant was associated with an earlier age of diagnosis (-1.26 years) and a trend toward more aggressive disease (p=0.08). Because this study focused on a cohort of European men undergoing various therapies for PC management, our objective was to further evaluate the clinical significance of this variant in a North American population, including annotated prostatectomy pathology and early follow-up for biochemical recurrence.
Methods
We performed a case-control study, including 1,979 men with PC who underwent radical prostatectomy at Northwestern Memorial Hospital between 2002 and 2012. These men were compared to a control group from a grass roots community screening study of 2,320 healthy volunteers with no known history of PC, serum PSA levels less than 2.5ng/ml and a normal digital rectal examination, as previously described[12]. The study was approved by the institutional review board at Northwestern University, and all participants provided informed consent. Serum PSA concentration, demographics, family history, prostate biopsy findings and prostatectomy pathology were recorded prospectively. Biochemical recurrence was defined as postoperative PSA greater than 0.2ng/ml. A Kaplan-Meier analysis was used to calculate the rate of progression-free survival after 90 months.
DNA was extracted from whole blood at deCODE Genetics, Reykjavik, Iceland, using the Centarus (Nanogen) platform. All samples were genotyped for SNPs with a validated association to PC risk, including rs188140481 on 8q24, as previously described[10]. Logistic regression was performed to compare allele frequencies between cases and controls. Mann-Whitney U and chi-square tests were used to compare clinical variables between cases and controls, and between carriers and non-carriers. Multivariate analysis was also performed adjusting for age and PSA concentration. Statistical analyses were performed using SAS 9.3 software (Cary, NC).
Results
Our study population included 4,299 men, including 1,979 (46%) PC cases and 2,320 controls. The baseline clinical features of the cohort are shown in Table 1. Men treated for PC were significantly older (59.9 vs. 56.0 years, p < 0.001), had a higher serum PSA concentration than controls (4.9 vs. 0.8ng/ml, p<0.001), and were predominantly Caucasian (93% vs 59.2%). The frequency of a positive family history was similar between PC cases and controls (29.5% vs. 24.7%, p = 0.56). The minor allele (A) at rs188140481 was present in 1.6% of the study population; however, there was a significantly higher frequency of the SNP in cases compared to controls (2.5% vs. 0.8%; OR 3.14; 95% CI 1.85-5.35; p < 0.001). To account for the predominance of Caucasians among our patient population we compared the frequency of rs188140481 between cases and controls by race (Table 2). Among Caucasians the carriers of the SNP were again seen to be at a higher risk of being treated for PC (2.9% vs. 1.3%; OR 2.3; 95% CI 1.3-4.1). Only one non-Caucasian patient was a carrier of rs188140481 SNP. No individual was homozygous for the minor allele in our cohort. After adjusting for age and PSA concentration, rs188140481 increased the risk of PC overall by more than six fold (OR 6.73; 95% CI 1.69-26.76), however, when only white patients and controls were considered the risk of developing PC was almost twelve fold higher (OR 11.9; 95% CI 2.3-61.0).
Table 1. Demographic features of cases and controls in the study population.
| Controls | Prostate Cancer Cases | p value = | |
|---|---|---|---|
| n = | 2320 | 1979 | |
| Median Age (yrs) | 56.0 | 59.9 | <0.001 |
| Age (years) in Quartiles, n= (%) | |||
| ≤50 | 1090 (47.0) | 515 (26.1) | <0.001 |
| 51-57 | 487 (21.0) | 430 (21.8) | |
| 58-65 | 394 (17.0) | 613 (31.0) | |
| ≥66 | 349 (15.0) | 417 (21.1) | |
| Median PSA (ng/ml) | 0.8 | 4.9 | <0.001 |
| Race, n = (%) | |||
| Caucasian | 1270 (59.2) | 1613 (92.9) | <0.001 |
| African-American | 529 (24.7) | 55 (3.2) | |
| Latinos | 114 (5.3) | 25 (1.4) | |
| Others | 231 (10.8) | 43 (2.5) | |
| Unknown: | 176 (7.6) | 243 (12.3) |
Table 2.
Breakdown of carriers and non-carriers of rs18814048 by race. Only those patients that offered information about their race are included.
| Control Patients, n = (%) | PC Cases, n = (%) | p value: | |
|---|---|---|---|
| Caucasian | 16 (1.3) | 46 (2.9) | 0.004 |
| African-American | 0 | 0 | -- |
| Latinos | 0 | 0 | -- |
| Others | 1 (0.4) | 0 | 1.00 |
To determine whether the presence of the minor variant at rs188140481 predisposed patients to more aggressive forms of PC, we compared clinical and pathological features between carriers and non-carriers of the SNP (Tables 2). In our cohort, the age at diagnosis for cases was similar between carriers and non-carriers of rs188140481 SNP (59.9 yrs vs. 59.8, p = 0.93). Carriers of the rs188140481 SNP had a proportionately higher frequency of stage ≥T3 disease (29.5% vs. 20.1%; p = 0.13), Gleason ≥8 tumors (13.3% vs. 6.5%; p = 0.10), and extracapsular extension (28.9% vs. 18.8%; p = 0.12) compared to non-carriers, but none of these differences reached statistical significance (Table 3). After a median follow-up period of 55.7 months, no difference in the rates of biochemical recurrence between carriers and non-carriers was observed (14.6% vs. 12.7%; p = 0.64). Progression-free survival rates were also similar between patients carrying the minor and major variants of the SNP after 90 months (86% vs. 83%; log rank p = 0.83; Figure 1).
Table 3. Clinico-Pathologic Comparisons between Carriers and Non-Carriers of rs188140481[A].
| Carrier | Non-Carrier | P Value | |
|---|---|---|---|
| n= | 50 | 1925 | |
| Mean Age at Diagnosis | 59.9 | 59.8 | 0.93 |
| Family History of PC, n= (%) | 13 (29.5) | 400 (24.6) | 0.38 |
| Clinical Gleason Score, n= (%) | |||
| Gl≤ 6 | 27 (61.4) | 1099 (65.6) | 0.43 |
| Gl = 7 | 12 (27.3) | 468 (27.9) | |
| Gl≥ 8 | 5 (11.3) | 108 (6.5) | |
| Clinical Stage ≤ T1c, n= (%) | 35 (79.6) | 1203 (72.0) | 0.31 |
| Pathologic Gleason Score, n= (%) | |||
| Gl≤ 6 | 19 (42.2) | 826 (50.7) | 0.10 |
| Gl = 7 | 20 (44.5) | 706 (43.3) | |
| Gl≥ 8 | 6 (13.3) | 97 (6.0) | |
| Pathologic Stage ≤ T2c, n= (%) | 31 (70.5) | 1301 (79.9) | 0.13 |
| Extracapsular Extension, n= (%) | 13 (28.9) | 306 (18.8) | 0.12 |
| Seminal Vesicle Invasion, n= (%) | 2 (4.4) | 86 (5.3) | 1.00 |
| Lymph Node Metastases, n= (%) | 0 | 16 (1.0) | 1.00 |
| Follow-up, months, median = (range) | 69.9 (0.2-180) | 55.5 (0-324) | 0.25 |
| Biochemical Recurrence, n= (%) | 6 (14.6) | 201 (12.7) | 0.64 |
Figure 1.
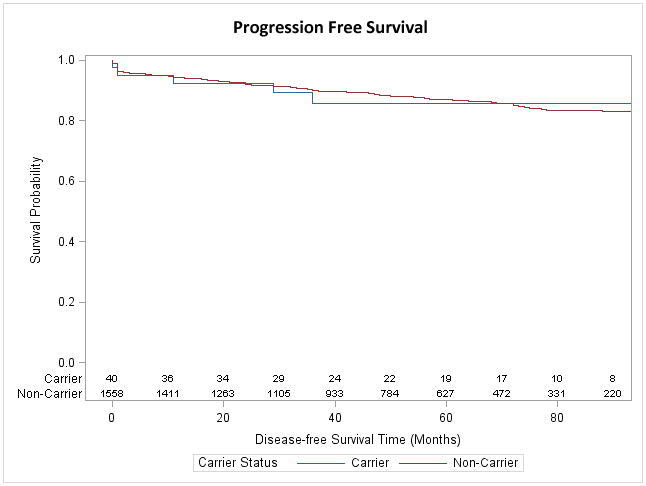
Kaplan-Meier plot of progression free survival of carriers of carriers and non-carriers of rs188140481[A]. Recurrence was defined as PSA ≥ 0.2ng/ml.
Discussion
We examined the frequency of a newly described PC-associated SNP in the 8q24 region (rs188140481) in a cohort of North American of PC cases and healthy volunteer controls. This population has been previously included in a previous study [10], however, here we present a more detailed analysis of the clinicopathological features of our North American population as well as multivariate analysis of the risk associated with the SNP. Previous studies have focused on European populations that did not include men of African, Asian, or Latino heritage[10]. In our population, the SNP was present in 2.5% of cases compared to only 0.8% of controls. Of note, this variant was not observed in African American or Latino men in our study, however, these groups were under represented in our patient population. Further studies that include a greater proportion of non-Caucasian men are needed to confirm this observation.
Consistent with previous published investigations[10], we found a strong association between PC risk and carrier status of this SNP. However, we were unable to determine the risk contributed by individual alleles at rs188140481 since no individual in our cohort was homozygous at this locus. Overall, we did not find statistically significant differences in the age of diagnosis in carriers of the SNP compared to non-carriers, as was suggested by previous studies[10]. However, there were trends toward an association with higher tumor grade and disease stage, which could become more apparent with a larger sample size. Furthermore, the full impact of the rs188140481 SNP on disease aggressiveness may not be captured by our study, as it is limited to patients treated with surgical intervention with early follow-up. Older patients and those with more advanced clinical disease may have been treated by other forms of therapy, resulting in a selection bias. That notwithstanding, evaluation of a radical prostatectomy population with genetic data and annotated prostatectomy specimens also represents an important strength of the study. Another limitation of our study was its relatively small size in light of the low frequency of the allele studied.
Future studies employing WGS may uncover additional genetic loci with higher frequency. The presence of these SNPs may be used to target men for earlier and more aggressive screening and/or intervention and identify families at increased risk of PC, similar to the way that mutation in BRCA1 and BRCA2, are currently used to identify women at greater risk for breast cancer. In addition, as more genetic loci are found, it may be possible to combine them into screening panels that provide more accurate prognostic data than any individual SNP alone.
Acknowledgments
Research reported in this publication was supported by the National Institutes of Health, award numbers K07CA178258 to SL; P50CA090386 and 2R01CA089600 to WJC; a grant from the Urological Research Foundation to WJC, and the North Shore University Health System pilot award to BTH. The content is solely the responsibility if the authors and does not necessarily represent the official views of any funding institution.
Footnotes
Conflicts of Interest: Dr. Helfand reports grants from NorthShore University HealthSystem, during the conduct of this study
Dr. Loeb received an honorarium from Sanofi for speaking at the IGUCC meeting unrelated to the subject of this manuscript.
Dr. Catalona reports grants from Urological Research Foundation, non-financial support from deCODE genetics, Inc., grants from International Consortium for Prostate Cancer Genetics (ICPCG), during the conduct of the study; non-financial support from Nanosphere, Inc., personal fees and non-financial support from OHMX, Inc., personal fees from Beckman Coulter, Inc., outside the submitted work.
All other authors have no conflicts of interest to report.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013 Jan;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Kim ST, Cheng Y, Hsu FC, Jin T, Kader AK, Zheng SL, et al. Prostate cancer risk-associated variants reported from genome-wide association studies: meta-analysis and their contribution to genetic Variation. Prostate. 2010 Dec 1;70(16):1729–38. doi: 10.1002/pros.21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eeles RA, Olama AAA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013 Apr;45(4):385–91–391e1–2. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H, Wang B, Han C. Meta-analysis of genome-wide and replication association studies on prostate cancer. Prostate. 2011 Feb 1;71(2):209–24. doi: 10.1002/pros.21235. [DOI] [PubMed] [Google Scholar]
- 5.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007 May;39(5):631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 6.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008 Feb 28;358(9):910–9. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 7.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007 Apr 1;39(5):638–44. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishak MB, Giri VN. A systematic review of replication studies of prostate cancer susceptibility genetic variants in high-risk men originally identified from genome-wide association studies. Cancer Epidemiol Biomarkers Prev. 2011 Aug;20(8):1599–610. doi: 10.1158/1055-9965.EPI-11-0312. [DOI] [PubMed] [Google Scholar]
- 9.Hidorff LA, MacArthur J, Morales J, Junkins HA, Hall PN, Klemm AK, et al. Catalog of Published Genome-Wide Association Studies. [cited 2013 Jun 9]; [Internet]. genome.gov. Available from: http://www.genome.gov/gwastudies/
- 10.Gudmundsson J, Sulem P, Gudbjartsson DF, Masson G, Agnarsson BA, Benediktsdottir KR, et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet. 2012 Dec;44(12):1326–9. doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012 Jan 12;366(2):141–9. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007 Aug;39(8):977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]


