Abstract
The nuclear receptor NOR1 is an immediate-early response gene implicated in the transcriptional control of proliferation. Since the expression level of NOR1 is rapidly induced through cAMP response element binding (CREB) protein-dependent promoter activation, we investigated the contribution of histone acetylation to this transient induction. We demonstrate that NOR1 transcription is induced by histone deacetylase (HDAC) inhibition and by depletion of HDAC1 and HDAC3. HDAC inhibition activated the NOR1 promoter, increased histone acetylation and augmented the recruitment of phosphorylated CREB to the promoter. Furthermore, HDAC inhibition increased Ser133 phosphorylation of CREB and augmented NOR1 protein stability. These data outline previously unrecognized mechanisms of NOR1 regulation and illustrate a key role for histone acetylation in the rapid induction of NOR1.
Keywords: nuclear receptor, smooth muscle cell, histone deacetylase
1. Introduction
The neuron-derived orphan receptor-1 (NOR1) is an evolutionarily conserved member of the ligand-independent NR4A subfamily of the nuclear hormone receptor superfamily[1]. Members of this subfamily serve as immediate-early response genes to translate environmental cues into transcriptional programs of gene expression[2]. Being early response genes, the transcriptional activity of NR4A nuclear receptors is regulated by their rapid and transient expression as well as by posttranslational protein modification of the receptor[3, 4]. Although still in its infancy, emerging evidence has implicated NR4A receptors in the transcriptional control of proliferation, differentiation, survival, and inflammation[5]. Based on this evidence, characterizing the molecular mechanisms underlying inducible expression of NR4A receptors is critical for understanding their functional role in the transcriptional control of gene expression.
In vascular biology, we and others have previously characterized NOR1 as a mitogenic transcription factor[6, 7]. NOR1-deficiency limits aberrant vascular smooth muscle cell (SMC) proliferation, decreases pathological neointima formation in response to vascular injury, and reduces atherosclerosis formation in mice[8, 9]. In SMC, mitogen-induced NOR1 expression occurs rapidly, leading to a more than 600-fold increase in transcript levels[9]. This transient NOR1 expression is regulated by Ser133 phosphorylation of CREB and its subsequent recruitment to cAMP response elements (CRE) in the NOR1 promoter[7]. The rapid kinetics underlying the induction of several immediate-early response genes are achieved by chromatin remodeling, including histone acetylation[10]. Because histone acetylation is also critical for the transcription of a variety of CREB target genes[11], we investigated in the present study whether inhibition of histone deacetylation activates NOR1 transcription. We provide direct evidence that inhibition of histone deacetylation augments CREB phosphorylation, increases histone acetylation and CREB recruitment to the NOR1 promoter, and supports posttranslational protein stability of NOR1.
2. Materials and Methods
Materials and Methods are available in the Supplementary Data.
3. Results
3.1. NOR1 expression is induced by the HDAC inhibitor Scriptaid in vascular SMC
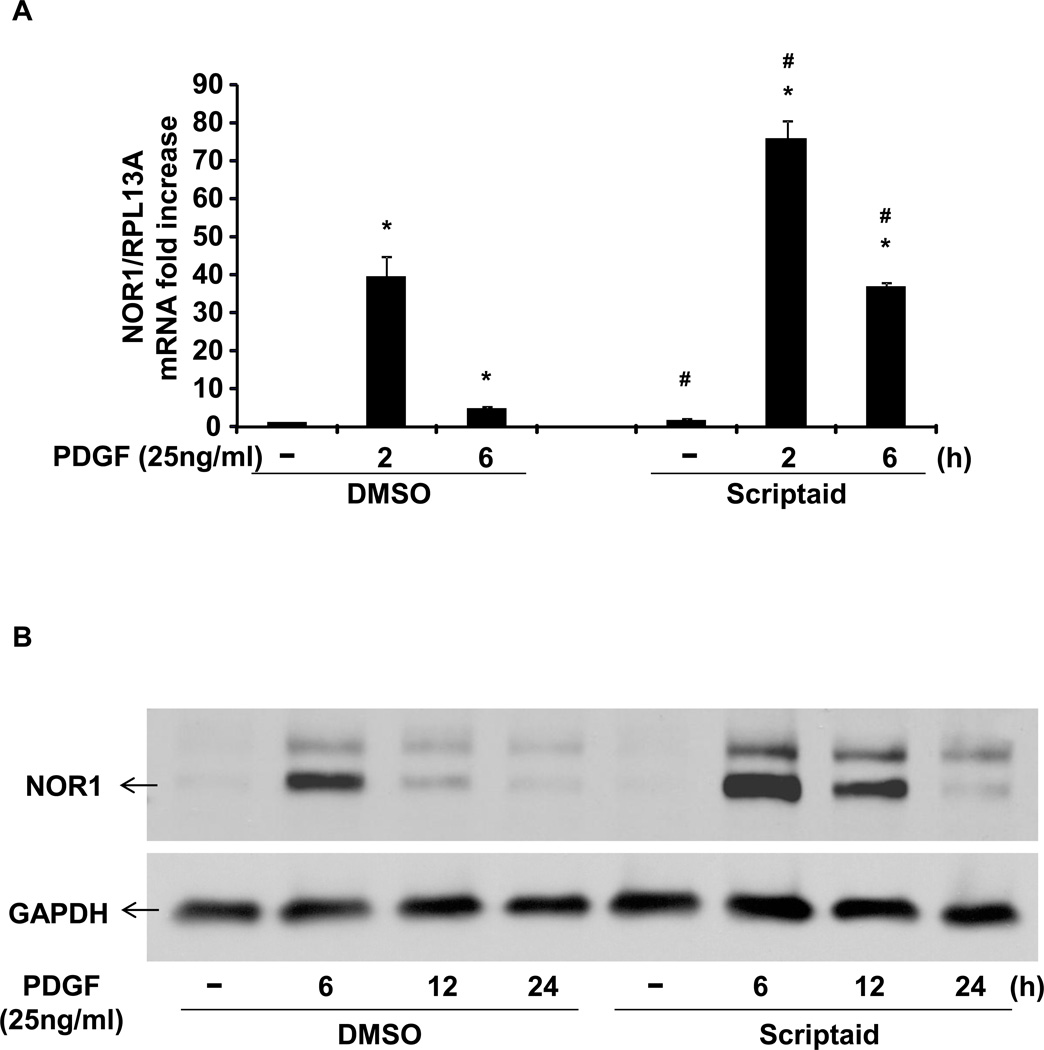
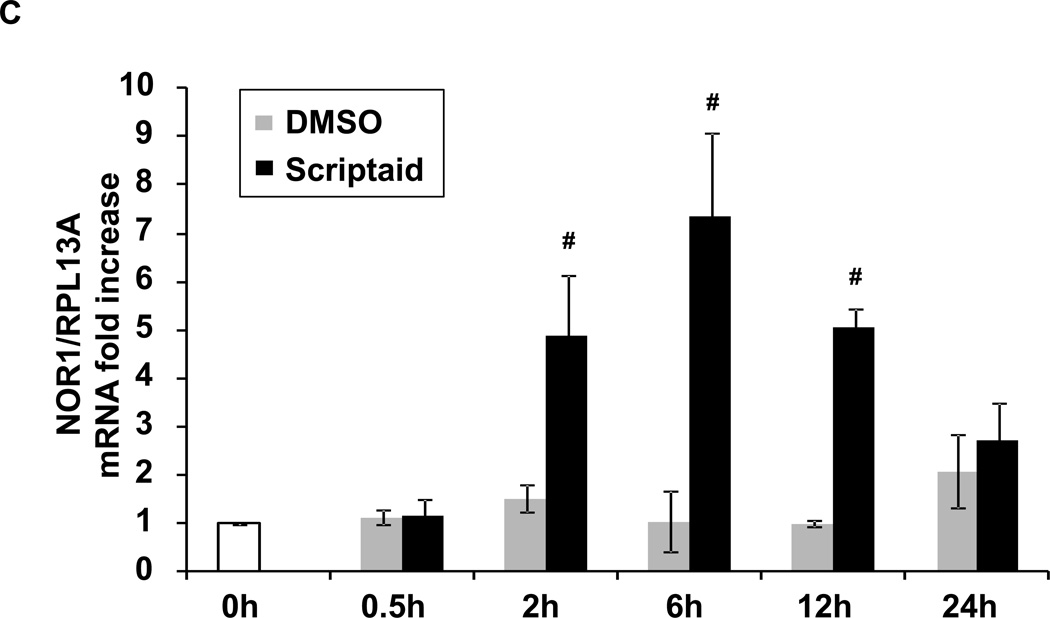
To investigate the epigenetic regulation of NOR1 expression by histone acetylation, we employed the HDAC inhibitor Scriptaid. Scriptaid is a hydroxamic acid-containing HDAC inhibitor and has been reported to increase histone acetylation with low cytotoxicity[12]. Pre-treatment of rat aortic SMC (RASMC) with Scriptaid (2 µg/ml) potentiated NOR1 mRNA and protein expression in response to PDGF (25 ng/ml). This effect was more apparent at the later time point after PDGF stimulation (Fig. 1A and B). Scriptaid alone also induced NOR1 mRNA expression in quiescent RASMC, although this induction was modest compared to co-stimulation with PDGF (Fig. 1A and C).
Fig. 1. The HDAC inhibitor Scriptaid induces NOR1 Expression in SMC.
(A–B) Quiescent RASMC were pretreated with Scriptaid (2 µg/ml) or DMSO for 30 min, and subsequently stimulated with PDGF (25 ng/ml). NOR1 mRNA (A) and protein expression (B) were analyzed at the indicated time points. (C) Quiescent RASMC were stimulated with Scriptaid (2 µg/ml) or DMSO for 0.5, 2, 6, 12 and 24 h for NOR1 mRNA analyses. mRNA expression was normalized to RPL13A and expressed as mean ± SEM fold increase over untreated (UT) cells (*P < 0.05 vs. unstimulated (−) in panel A; #P < 0.05 vs. DMSO in panel A and C). Immunoblotting for GAPDH was performed to assess equal loading. The autoradiograms shown are representative of three independently performed experiments.
3.2. HDAC Depletion increases PDGF-induced NOR1 mRNA expression
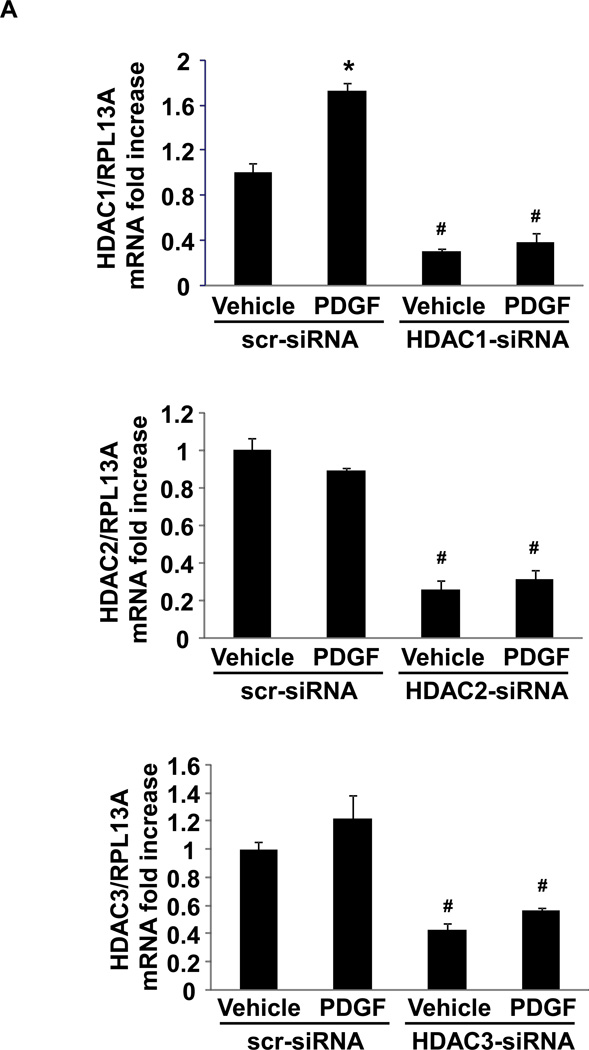
In order to identify selective HDACs that regulate NOR1 mRNA expression, RASMC were transiently transfected with siRNA against HDAC1, HDAC2 and HDAC3 followed by PDGF stimulation. As depicted in Fig. 2A, siRNA mediated depletion of HDAC1-3 was confirmed by quantitative RT-PCR. Primarily, depletion of HDAC3 enhanced both basal and inducible NOR1 mRNA expression following PDGF stimulation (Fig. 2B). In contrast, selective knock-down of HDAC1 or HDAC2 did not exhibit an overt effect on basal NOR1 transcription or NOR1 mRNA expression at 2 h. However, depletion of HDAC1 moderately increased NOR1 transcript at the analyzed 6 h time-point. These findings confirm that both non-selective pharmacological HDAC inhibition and selective depletion of predominantly HDAC3 enhance basal and mitogen-induced NOR1 expression.
Fig. 2. siRNA-mediated knockdown of HDAC3 expression increases PDGF-induced NOR1 mRNA expression.
(A–B) RASMC were transiently transfected with HDAC1, HDAC2, HDAC3 or scrambled (scr) siRNA (50 nM) for 6 h, and further cultured overnight. Transfected cells were starved in 0.01% FBS in DMEM for 48 hours and stimulated with PDGF (25 ng/ml) or vehicle (PBS) for 2 or 6 h as indicated for mRNA analyses. The expression of HDAC1, HDAC2, HDAC3 and NOR1 was normalized to RPL13A and expressed as mean ± SEM fold increase over scr-siRNA-transfected vehicle-treated cells (*P < 0.05 vs. vehicle, #P < 0.05 vs. scr-siRNA).
3.3. Scriptaid increases NOR1 promoter activity without affecting NOR1 transcript stability
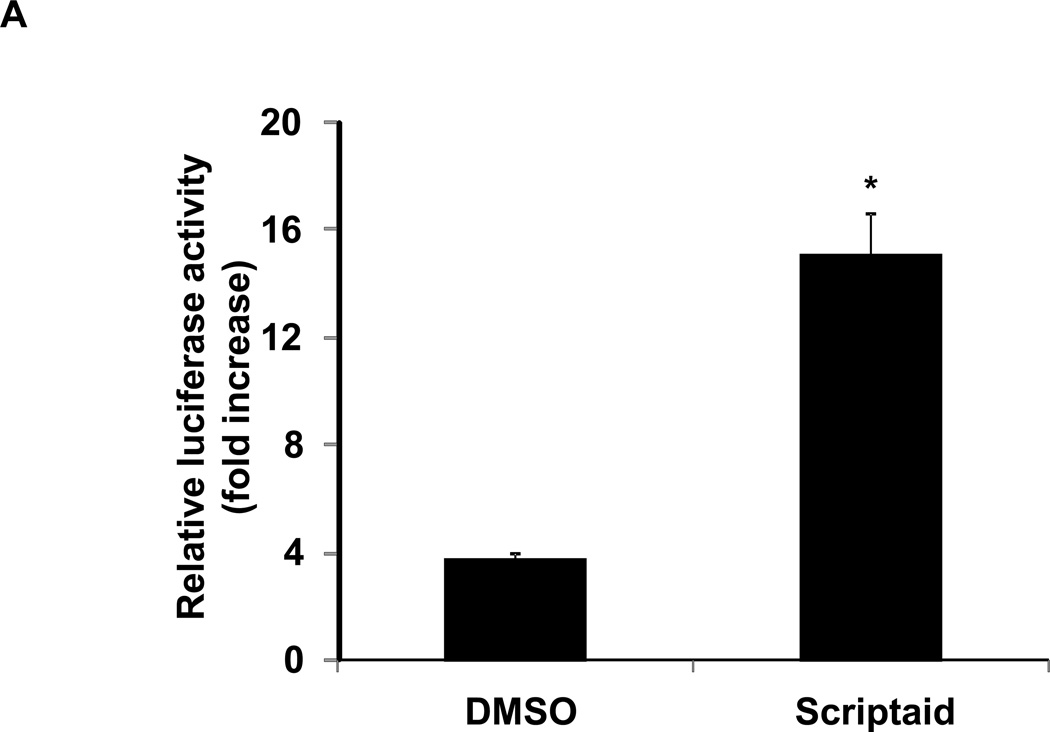
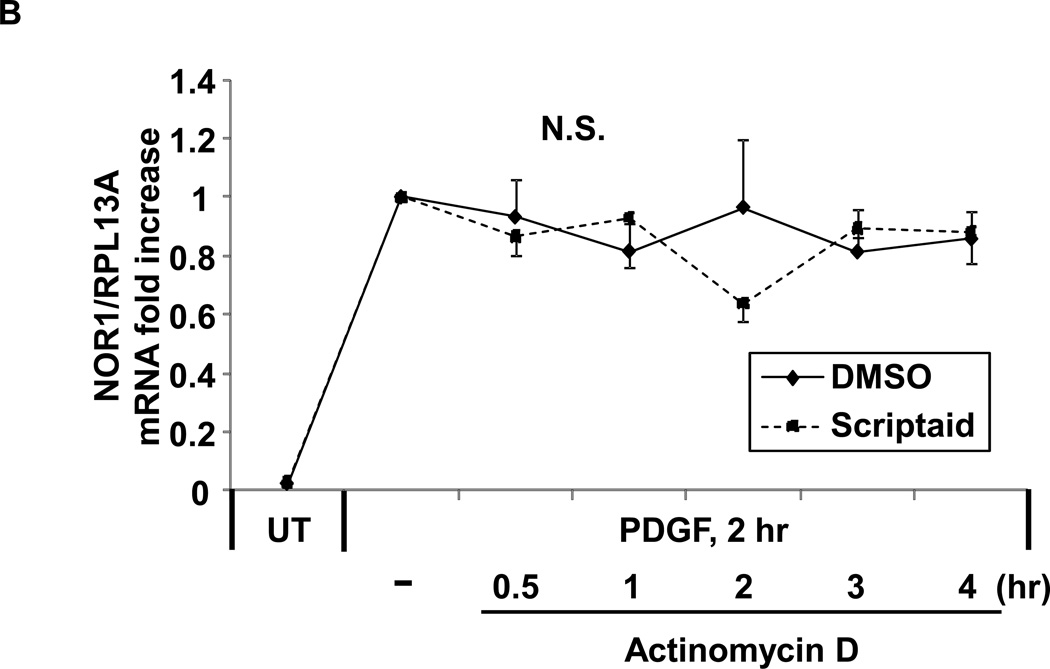
mRNA accumulation results from the net effect of de novo transcription as well as transcript stabilization. To further understand the mechanism by which NOR1 mRNA is induced in response to HDAC inhibition, we next analyzed NOR1 promoter activity in SMC. Using transient transfection of a luciferase reporter driven by the NOR1 promoter sequence, we first documented that Scriptaid induced NOR1 promoter activation in RASMC (Fig. 3A). This activation was potent and occurred even in the absence of mitogenic stimulation, indicating that HDAC inhibition alone is sufficient to increase NOR1 transcription. We next evaluated the alternative induction of NOR1 transcript levels by HDAC inhibition through a potential stabilization of NOR1 mRNA. RASMC were pre-treated with Scriptaid and stimulated with PDGF for 2 h to induce NOR1 mRNA accumulation, followed by the addition of actinomycin D (10 µg/ml) to inhibit mRNA transcription. As depicted in Fig. 3B, chase experiments revealed that Scriptaid did not affect NOR1 transcript stability. In summary, these two experiments demonstrate that Scriptaid increases NOR1 mRNA expression by activating de novo NOR1 transcription.
Fig. 3. Scriptaid increases NOR1 promoter activity without affecting NOR1 transcript stability.
(A) RASMC were transiently transfected with a luciferase reporter construct (2 µg) driven by a 1.7kb NOR1 promoter fragment and stimulated with DMSO or Scriptaid (2 µg/ml) overnight. Protein lysate was collected and analyzed for luciferase activity. Data were normalized to Renilla luciferase activity and presented as mean ± SEM from three independently performed experiments (*P < 0.05 vs. 1 7 DMSO). (B) Quiescent RASMC (UT, untreated control) were pretreated with Scriptaid (2 µg/ml) or DMSO for 30 min and subsequently stimulated with PDGF (25 ng/ml) for 2 h. Actinomycin D (10 µg/ml) was added to inhibit transcription, and mRNA was collected at the indicated time points. NOR1 mRNA expression was normalized to RPL13A in three independent experiments. Data are expressed as mean ± SEM relative to samples stimulated with PDGF for 2 h without actinomycin. N.S. indicates that no statistical significance was detected between DMSO and Scriptaid treatments.
3.4. Scriptaid enhances CREB phosphorylation and its recruitment to the NOR1 promoter
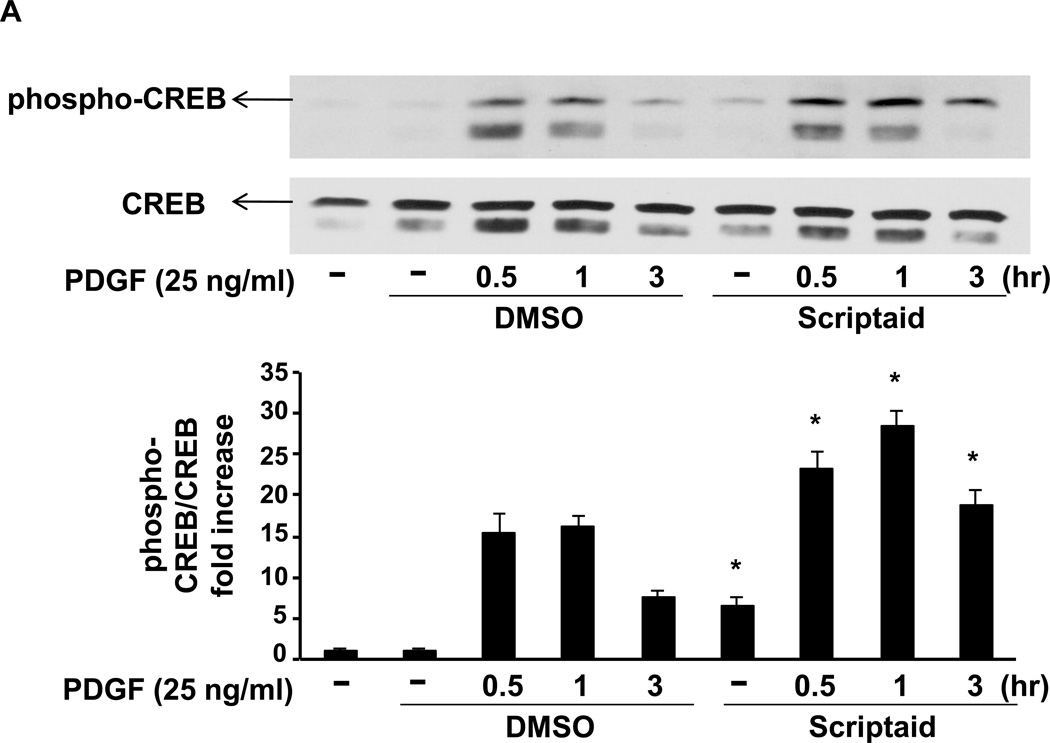
CREB Ser133 phosphorylation and its recruitment to CRE motifs within the NOR1 promoter mediate NOR1 transcriptional activation in response to PDGF[7)] To investigate the mechanism by which Scriptaid activates NOR1 transcription, CREB phosphorylation was next analyzed. Interestingly, Scriptaid alone rapidly induced CREB phosphorylation, although this effect was modest compared to PDGF stimulation (Fig. 4A). Similarly, PDGF-induced CREB phosphorylation was enhanced by Scriptaid at all time points analyzed (Fig. 4A).
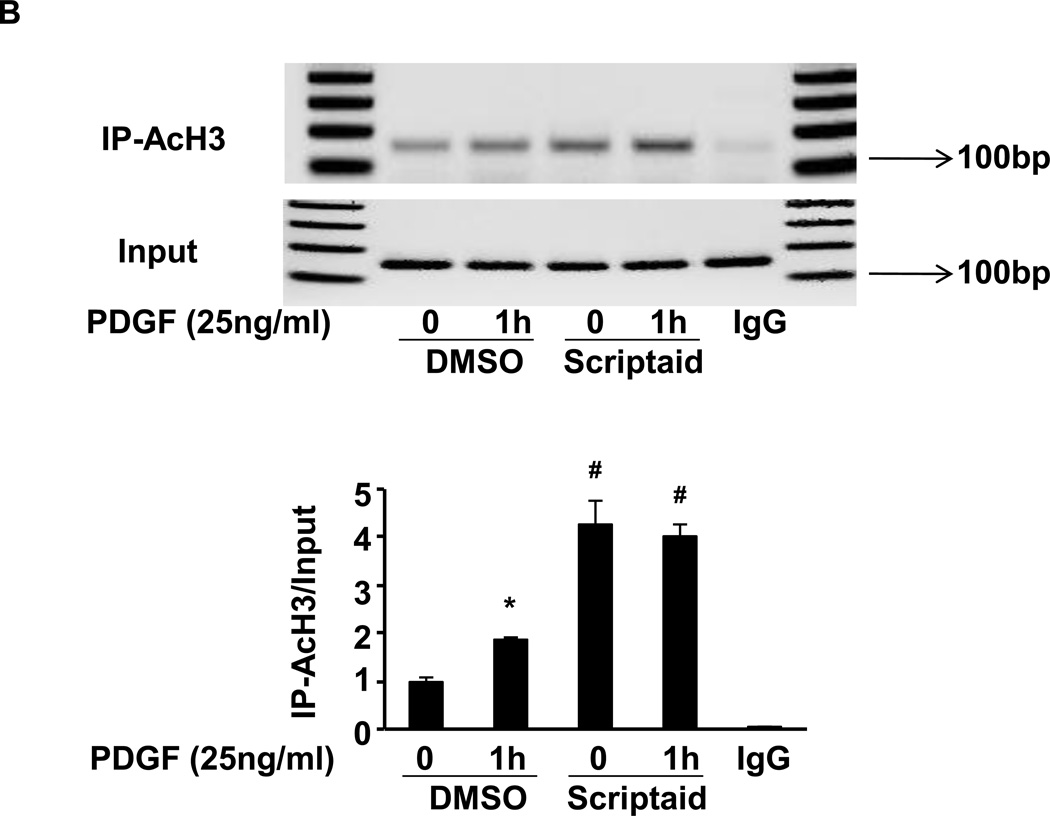
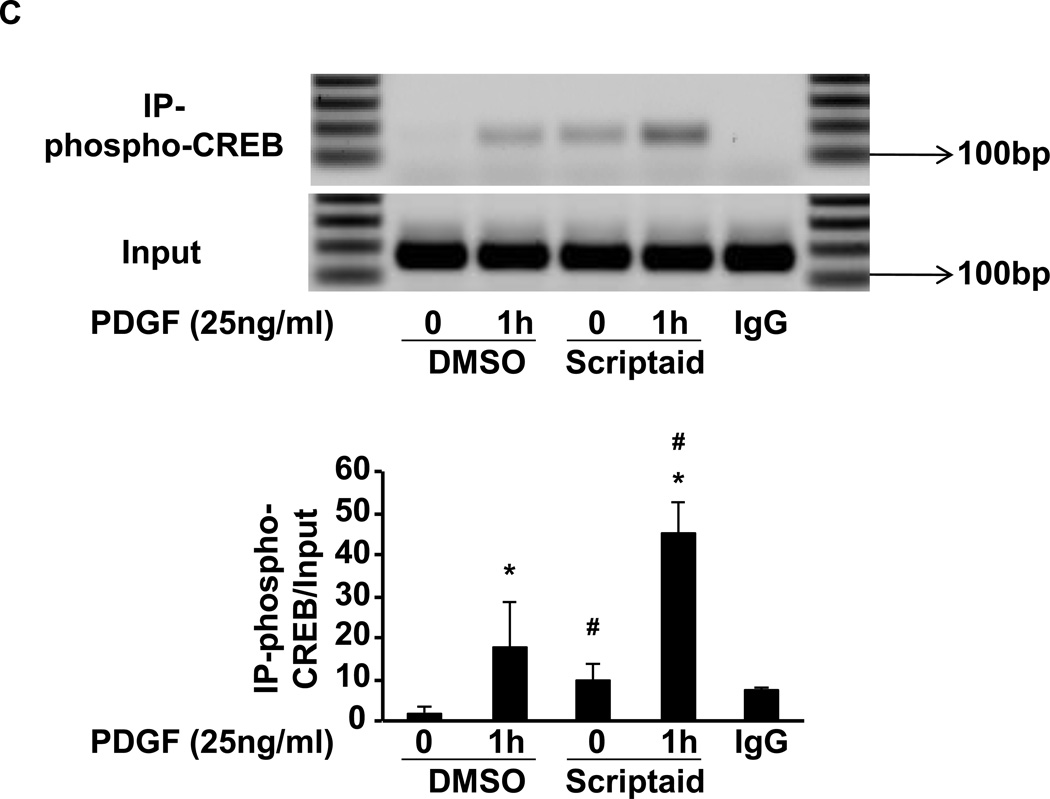
Fig. 4. Scriptaid enhances CREB phosphorylation and the recruitment of phospho-CREB to its binding sites in the NOR1 promoter.
(A) Quiescent RASMC were pretreated with Scriptaid (2 µg/ml) or DMSO for 30 min and subsequently stimulated with PDGF (25 ng/ml) for 0.5, 1 and 3 h for protein analyses. Immunoblotting for total CREB was performed to assess equal loading. The autoradiograms are representative of three independently performed experiments. Densitometric quantification is provided in the lower panel. (*P < 0.05 vs. DMSO). (B–C) Quiescent RASMC were pretreated with Scriptaid (2 µg/ml) or DMSO for 30 min and subsequently stimulated with PDGF (25 ng/ml) for 1 h for ChIP assays. Chromatin complexes were immunoprecipitated with antibodies against phospho-S133 CREB, AcH3K9, and species-matched IgG. PCR products were amplified using primers covering the CRE sites from −79 bp to −46 bp in the NOR1 promoter. The upper agarose gels shown are representative of three independently performed experiments. The lower graphs depict quantification of immunoprecipitated chromatin by real-time PCR. Cycle threshold (Ct) values were normalized to Ct values of input samples and presented as mean ± SEM fold increase over quiescent DMSO-treated samples, (*P < 0.05 vs. quiescent; #P < 0.05 vs. DMSO).
HDAC inhibition increases histone acetylation, and acetylated histone H3 lysine 9 (AcH3K9) has been implicated in gene activation[13]. Therefore, we next specifically analyzed histone acetylation at CRE sites within the endogenous NOR1 promoter using ChIP assays. As shown in Fig. 4B, PDGF stimulation induced histone H3 acetylation at the CRE motif in the NOR1 promoter. As expected, inhibition of HDAC activity by Scriptaid alone increased histone acetylation. This regulation of histone H3 acetylation was paralleled by the recruitment of Ser133-phosphorylated CREB to the same CRE motif (Fig. 4C). Collectively, these data establish that inhibition of HDAC activity induces histone acetylation, which is sufficient to facilitate binding of activated CREB to the CRE motif.
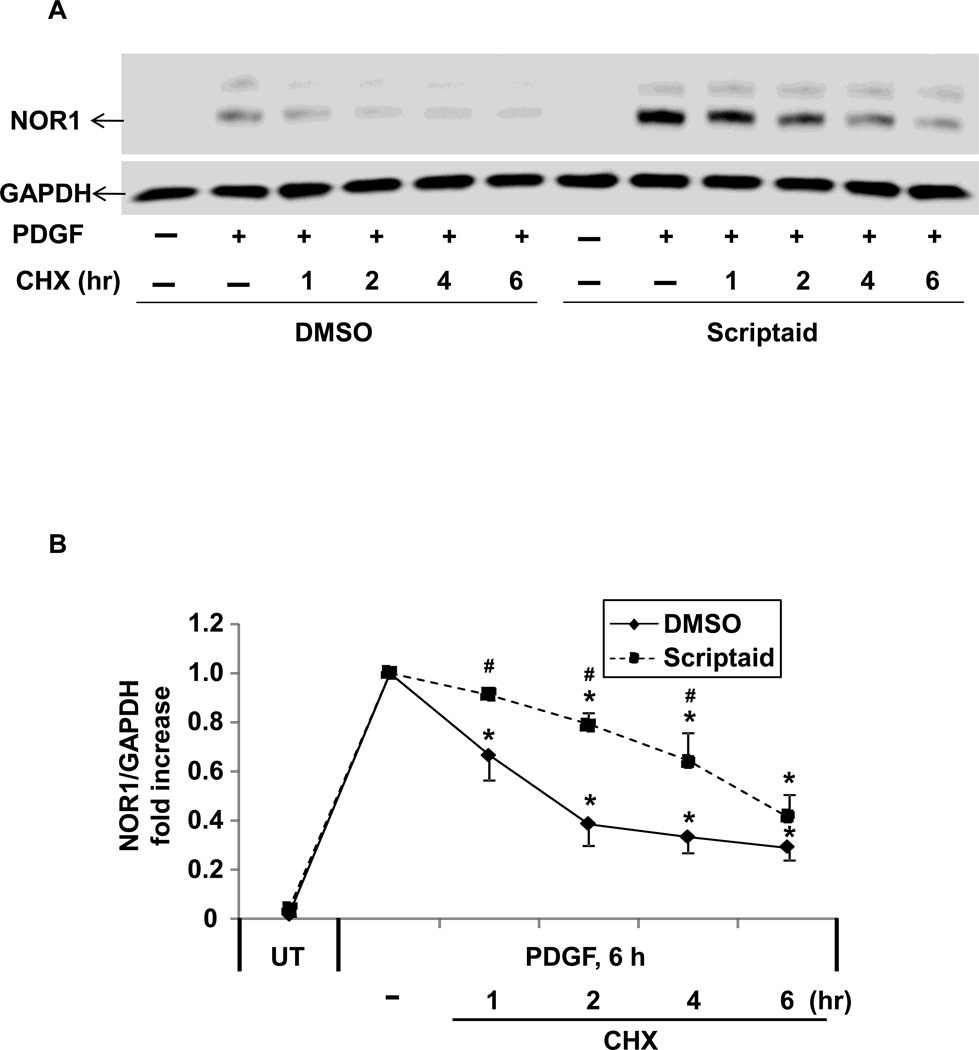
3.5. The HDAC inhibitor Scriptaid increases NOR1 protein stability
In addition to epigenetic regulation of transcription, HDAC inhibition induces posttranslational acetylation of nuclear receptors, which supports nuclear receptor protein stability and induces transcriptional activity[14]. To assess this possibility, RASMC were treated with vehicle or Scriptaid, and PDGF-stimulated protein levels were measured after inhibition of protein de novo synthesis with cycloheximide (Fig. 5A). As shown in Fig. 5B, quantification of protein levels revealed that Scriptaid significantly increased protein stability and half-life when compared to vehicle treatment. Therefore, HDAC inhibition not only induces NOR1 expression by generating permissive histone marks leading to increased transcription but also through posttranslational protein stabilization.
Fig. 5. The HDAC inhibitor Scriptaid increases NOR1 protein stability.
(A–B) Quiescent RASMC were pretreated with Scriptaid (2 µg/ml) or DMSO for 30 min and subsequently stimulated 1 8 with PDGF (25 ng/ml) for 6 h. Cycloheximide (CHX, 10 µg/ml) was added to inhibit protein synthesis and protein was collected at the indicated time points. (A) The autoradiograms are representative of three independently performed experiments. Immunoblotting for GAPDH was performed to assess equal loading. (B) Densitometric quantification of NOR1 expression was performed from three independent experiments and normalized to GAPDH expression. UT indicates quiescent cells. Results are expressed as mean ± SEM fold increase over samples stimulated with PDGF 6 h without cycloheximide; (*P < 0.05 vs. PDGF 6 h, # P < 0.05 vs. DMSO).
4. Discussion
Data presented here characterize NOR1 as an immediate-early response gene induced by histone acetylation in vascular cells. NOR1 has previously been characterized as a mitogenic transcription factor in SMC, and its deficiency attenuates SMC proliferation, atherosclerosis, and neointima formation[7, 9]. This activity of NOR1 as an effector of SMC proliferation is mediated by direct binding to its consensus elements in target promoters, including for example cyclin D and the S-phase-kinase-associated protein 2[9, 15]. Considering the rapid expression of NOR1, we sought to investigate whether chromatin modifications orchestrate its transient induction and the ensuing transcriptional activity to induce downstream target genes. In the present study, we demonstrate that inhibition of HDAC activity induces the transcriptional program underlying mitogen-induced NOR1 expression in SMC. In addition to this transcriptional mechanism of induction, HDAC inhibition stabilizes NOR1 protein through posttranslational modification of the receptor.
Inducible histone acetylation precedes the transcription of multiple immediate-early response genes[10]. To our knowledge, our data are the first to establish that pharmacological HDAC inhibition activates both basal and inducible expression of the immediate-early response gene NOR1 in SMC. Our observation of histone hyperacetylation-dependent NOR1 transcription in SMC is consistent with two prior studies, which identified increased NOR1 expression in response to treatment with HDAC inhibitors in hippocampal[16] and myeloid leukemia cells[17]. However, the regulation of NOR1 by HDAC inhibition remains controversial, since NOR1 expression has also been reported to be suppressed by the HDAC inhibitor trichostatin A in pheochromocytoma cells[18]. Therefore, these studies, along with our data, support the concept that NOR1 is regulated through distinct transcriptional mechanisms, which are likely both stimulus- and tissue-dependent.
Albeit these discrepant observations, the molecular mechanisms utlized by histone acetylation to regulate transcription factor-dependent NOR1 expression have not been investigated. Using siRNA technology, our experiments first indicated that primarily HDAC3 depletion activates NOR1 transcription in SMC. As mentioned previously, inducible NOR1 transcription in SMC is mediated by CREB binding to its responsive elements, which is activated following mitogen-induced Ser133 phorphorylation[7]. Interestingly, HDAC3 has been shown to repress CREB-dependent transcription in recent studies[19], and our ChIP experiments confirm that inhibition of HDAC activity results in increased histone acetylation and the subsequent recruitment of phosphorylated CREB to its consensus sites within the NOR1 promoter. However, in addition to this histone-dependent Ser133-phosphorylated CREB recruitment to the NOR1 promoter, HDAC inhibition also increased CREB phosphorylation in response to PDGF stimulation. These data point to a second mechanism of regulation and indicate that not only direct histone tail modification constitutes a target for HDAC-mediated repression of NOR1 but also the phosphorylation status of CREB itself. This notion is supported by previous data demonstrating that HDAC inhibitors augment CREB activity by altering the dephosphorylation of CREB[20]. Finally, many nuclear receptors are subject to acetylation, which regulates their stability, ligand sensitivity, and trans-activation potential[21]. Similarly, our experiments 1 0 revealed that HDAC inhibition enhances protein stability of NOR1. These studies provide evidence for a third and posttranslational mechanism regulating NOR1 expression in SMC, in which HDAC deacetylates NOR1 to maintain NOR1 hypoacetylated leading to its rapid degradation. Collectively, our data illustrate three novel mechanisms of transcriptional (i.e. histone deacetylation and dephosphorylation of CREB) and postranslational (i.e. protein degradation) regulation of NOR1, in which HDAC function coordinately in SMC to repress NOR1 transcription and facilitate its rapid degradation following mitogen stimulation.
Supplementary Material
Highlights.
-
*
NOR1 transcription is induced by histone deacetylase inhibition through chromatin modification
-
*
HDAC inhibition increases CREB phosphorylation and its recruitment to CRE consensus motifs within the NOR1 promoter
-
*
HDAC inhibition augments NOR1 protein stability
Acknowledgements
This work was supported by the NIH (R01HL084611, R01HL084611-04S1, and R01HL111040 to D.B.).
Abbreviations
- CREB
cAMP response element binding protein
- HDAC
histone deacetylase
- HAT
histone acetyl-transferase
- NOR1
neuron-derived orphan receptor 1
- PDGF
platelet-derived growth factor
- SMC
smooth muscle cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ohkura N, Hijikuro M, Yamamoto A, Miki K. Molecular cloning of a novel thyroid/steroid receptor superfamily gene from cultured rat neuronal cells. Biochem Biophys Res Commun. 1994;205:1959–1965. doi: 10.1006/bbrc.1994.2900. [DOI] [PubMed] [Google Scholar]
- 2.Pearen MA, Muscat GE. Minireview: Nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol. 2010;24:1891–1903. doi: 10.1210/me.2010-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis IJ, Hazel TG, Chen RH, Blenis J, Lau LF. Functional domains and phosphorylation of the orphan receptor Nur77. Mol Endocrinol. 1993;7:953–964. doi: 10.1210/mend.7.8.8232315. [DOI] [PubMed] [Google Scholar]
- 4.Zhang T, Jia N, Fei E, Wang P, Liao Z, Ding L, Yan M, Nukina N, Zhou J, Wang G. Nurr1 is phosphorylated by ERK2 in vitro and its phosphorylation upregulates tyrosine hydroxylase expression in SH-SY5Y cells. Neurosci Lett. 2007;423:118–122. doi: 10.1016/j.neulet.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, Bruemmer D. NR4A orphan nuclear receptors: transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler Thromb Vasc Biol. 2010;30:1535–1541. doi: 10.1161/ATVBAHA.109.191163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Gonzalez J, Rius J, Castello A, Cases-Langhoff C, Badimon L. Neuron-derived orphan receptor-1 (NOR-1) modulates vascular smooth muscle cell proliferation. Circ Res. 2003;92:96–103. doi: 10.1161/01.es.0000050921.53008.47. [DOI] [PubMed] [Google Scholar]
- 7.Nomiyama T, Nakamachi T, Gizard F, Heywood EB, Jones KL, Ohkura N, Kawamori R, Conneely OM, Bruemmer D. The NR4A orphan nuclear receptor NOR1 is induced by platelet-derived growth factor and mediates vascular smooth muscle cell proliferation. J Biol Chem. 2006;281:33467–33476. doi: 10.1074/jbc.M603436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y, Howatt DA, Gizard F, Nomiyama T, Findeisen HM, Heywood EB, Jones KL, Conneely OM, Daugherty A, Bruemmer D. Deficiency of the NR4A orphan nuclear receptor NOR1 decreases monocyte adhesion and atherosclerosis. Circ Res. 2010;107:501–511. doi: 10.1161/CIRCRESAHA.110.222083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nomiyama T, Zhao Y, Gizard F, Findeisen HM, Heywood EB, Jones KL, Conneely OM, Bruemmer D. Deficiency of the NR4A neuron-derived orphan receptor-1 attenuates neointima formation after vascular injury. Circulation. 2009;119:577–586. doi: 10.1161/CIRCULATIONAHA.108.822056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen TA, Allfrey VG. Rapid and reversible changes in nucleosome structure accompany the activation, repression, and superinduction of murine fibroblast protooncogenes c-fos and c-myc. Proc Natl Acad Sci U S A. 1987;84:5252–5256. doi: 10.1073/pnas.84.15.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 12.Su GH, Sohn TA, Ryu B, Kern SE. A novel histone deacetylase inhibitor identified by high-throughput transcriptional screening of a compound library. Cancer Res. 2000;60:3137–3142. [PubMed] [Google Scholar]
- 13.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 14.Kang SA, Na H, Kang HJ, Kim SH, Lee MH, Lee MO. Regulation of Nur77 protein turnover through acetylation and deacetylation induced by p300 and HDAC1. Biochem Pharmacol. 2010;80:867–873. doi: 10.1016/j.bcp.2010.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Gizard F, Zhao Y, Findeisen HM, Qing H, Cohn D, Heywood EB, Jones KL, Nomiyama T, Bruemmer D. Transcriptional regulation of S phase kinase-associated protein 2 by NR4A orphan nuclear receptor NOR1 in vascular smooth muscle cells. J Biol Chem. 2011;286:35485–35493. doi: 10.1074/jbc.M111.295840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hawk JD, Bookout AL, Poplawski SG, Bridi M, Rao AJ, Sulewski ME, Kroener BT, Manglesdorf DJ, Abel T. NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. J Clin Invest. 2012;122:3593–3602. doi: 10.1172/JCI64145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou L, Ruvolo VR, McQueen T, Chen W, Samudio IJ, Conneely O, Konopleva M, Andreeff M. HDAC inhibition by SNDX-275 (Entinostat) restores expression of silenced leukemia-associated transcription factors Nur77 and Nor1 and of key pro-apoptotic proteins in AML. Leukemia. 2013;27:1358–1368. doi: 10.1038/leu.2012.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fass DM, Butler JE, Goodman RH. Deacetylase activity is required for cAMP activation of a subset of CREB target genes. J Biol Chem. 2003;278:43014–43019. doi: 10.1074/jbc.M305905200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Henagan TM, Gao Z, Ye J. Inhibition of glyceroneogenesis by histone deacetylase 3 contributes to lipodystrophy in mice with adipose tissue inflammation. Endocrinology. 2011;152:1829–1838. doi: 10.1210/en.2010-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michael LF, Asahara H, Shulman AI, Kraus WL, Montminy M. The phosphorylation status of a cyclic AMP-responsive activator is modulated via a chromatin-dependent mechanism. Mol Cell Biol. 2000;20:1596–1603. doi: 10.1128/mcb.20.5.1596-1603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leader JE, Wang C, Fu M, Pestell RG. Epigenetic regulation of nuclear steroid receptors. Biochem Pharmacol. 2006;72:1589–1596. doi: 10.1016/j.bcp.2006.05.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.